TO THE EDITOR
BCR-ABL1, the product of the Philadelphia chromosome (Ph), is sufficient for inducing the chronic phase (CP) of chronic myeloid leukemia (CP-CML)1. Tyrosine kinase inhibitors (TKIs) suppress the Ph+ cell clone, restoring polyclonal hematopoiesis2. However, clonal cytogenetic abnormalities in Ph− cells become detectable in some patients achieving a cytogenetic response to TKIs, and slow evolution to myelodysplasia or acute myeloid leukemia (AML) has been observed2, 3. In rare cases, identical abnormalities were demonstrated both in Ph+ and Ph− cells, but most cases are consistent with independently acquired abnormalities, although an undetected common ancestral event cannot be excluded (reviewed in4). The clinical conundrum of coexisting leukemic disorders is that TKI suppression of CML may unmask a different, more aggressive disease. Here we report a patient who after starting imatinib rapidly converted from CML to fatal chronic myelomonocytic leukemia (CMML), demonstrating that this is not a theoretical consideration. Whole exome sequencing (WES) and genotyping of individual colonies revealed the clonal architecture during disease evolution and implicated TET2 and ASXL1 variants as early or germline events.
Case description
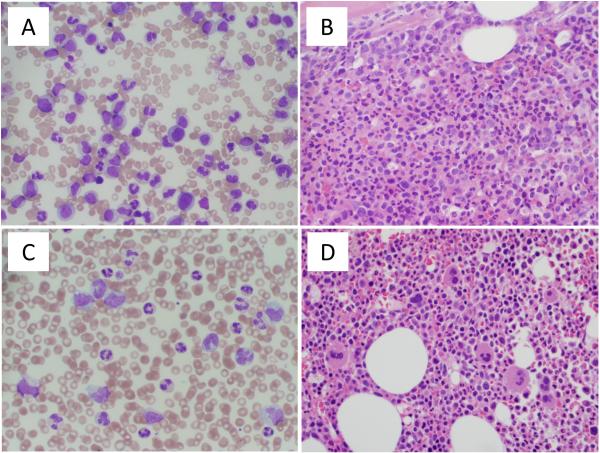
A 77-year-old man presented with fever and 16 kg weight loss. Clinical examination was unremarkable without splenomegaly. The white blood cell (WBC) count was 270,000/μL, with a myeloid left shift; hemoglobin was 9 g/dL and platelets were 55,000/μL (Supplementary Table 1). Bone marrow (BM) biopsy was 90% cellular with a left shift (Figure 1A and B). BM metaphase karyotyping was 46XY,t(9;22)(q34;q11.2)[20] and blood BCR-ABL1 mRNA (e13a2) was 10% on the international scale (IS). The patient was started on 400mg imatinib (considered day 1), and stayed on the same dose throughout the treatment. On day 67, partial hematological response was demonstrated, but a rise of monocytes was noted (Supplementary Table 1). At day 92, the WBC count rose to 73,000/μL, monocytes were 19%, hemoglobin was 9 g/dL and platelets were 80,000/μL. BM histology showed increased monocytes (Figure 1C and D), karyotyping was 46XY [30], and BCR-ABL1 was 0.12% IS. Sequencing was negative for BCR-ABL1 kinase domain mutations. A diagnosis of CMML was established. 5-azacytidine was added, with initial improvement of blood counts. The subsequent clinical course was complicated by sepsis; the patient declined further leukemia therapy and passed away.
Figure 1. Blood and bone marrow morphology.
(A) Peripheral blood smear at CML diagnosis demonstrating marked leukocytosis with granulocytic left shift and decreased platelets. (B) Bone marrow biopsy at CML diagnosis shows hypercellularity with granulocytic hyperplasia. (C) Peripheral blood smear on day 92 of imatinib therapy showing leukocytosis with monocytosis and (D) corresponding bone marrow biopsy showing hypercellular bone marrow with occasional hypolobated megakaryocytes.
Somatic mutations associated with phenotypic conversion to CMML
We performed WES (average read depth: 61x) on blood CD14+ cells from day 92, with CD3+ cells from the diagnostic sample as constitutional control. We identified four somatic single nucleotide variants (SNVs; EZH2I669M, KRASG12R, MSLNP462H and NTRK3V443I), all of which were confirmed by Sanger sequencing (Supplementary Table 2). Sequenom MassARRAY identified the same mutations in the day 67, 78, 92 and 124 samples, but not the diagnostic sample (Supplementary Table 3). In addition, we identified one nonsense variant in ASXL1 (c.24422delC→p.P808fs*10) and two nonsense variants in TET2 (TET2 c.1219delT→p.S407fs*20; c.4932delA→p.Y1645fs*50). Across all samples, including CD3+ and diagnostic CD14+ cells, ASXL1 c.24422delC and TET2 c.4932delA were detected at ~50%, while TET2 c.1219delT was detected at ~30% (Supplementary Table 3). ASXL1 c.24422delC and TET2 c.1219delT are listed in COSMIC and have been confirmed as somatic, while our findings are consistent with germline mutations or acquisition by a multipotent hematopoietic stem cell. While TET2 c4932delA has not been reported in COSMIC, a very similar variant (COSM4170135, c.4928delC, p.P1644fs*51) has. WES of the diagnostic sample at an average depth of 319x failed to identify additional mutations specific to the CML clone, but confirmed the presence of low level EZH2I669M, KRASG12R, MSLNP462H and NTRK3V443I.
Clonal architecture and evolution
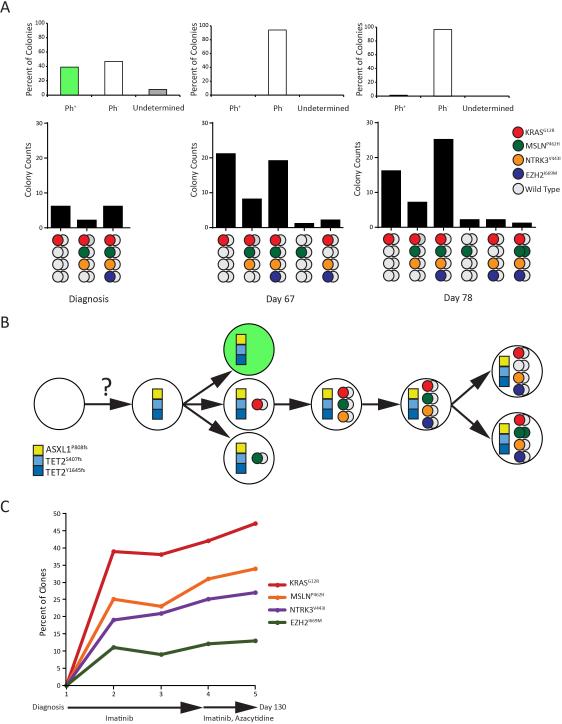
To unravel clonal relationships, we plated CD34+ cells from diagnosis, day 67 and 78 in colony assays (no viable cells available from days 92 and 124). Both DNA and RNA were extracted from ~100 single colonies and analyzed for EZH2I669M, KRASG12R, MSLNP462H and NTRK3V443I by MassARRAY and for BCR-ABL1 mRNA by RT-qPCR. In the diagnostic sample, 92/100 colonies were informative for BCR-ABL1 and all for DNA mutational analysis. Only 38% of informative colonies were BCR-ABL1-positive. This is unusual, as myeloid colonies from newly diagnosed CML patients are almost exclusively BCR-ABL1-positive5, but is consistent with the low BCR-ABL1 expression (10% IS). Altogether 14% of colonies were positive for at least one of the four somatic SNVs, all of which were BCR-ABL1-negative; 43% were wild type (Figure 2A). Genotypes included KRASG12R, KRASG12R/MSLNP462H/NTRK3V443I and KRASG12R/MSLNP462H/NTRK3V443I/EZH2I669M (Figure 2A). The failure of WES and MassARRAY to detect the SNVs in the diagnostic sample likely reflects their lower sensitivity; alternatively, in vitro culture with cytokines may favor CMML colonies due to their GM-CSF hypersensitivity6. No BCR-ABL1-positive colonies were detected on day 67, indicating effective CML therapy; however, 51/105 colonies (49%) were positive for at least one of the four SNVs. Most of the genotypes followed the patterns of the diagnostic sample (Figure 2A, lower panel); two colonies were KRASG12R/NTRK3V443I/EZH2I669M, and one was heterozygous MSLNP462H. Despite the increase of colonies with at least one SNV, the ratio between the various SNVs remained largely stable (Figure 2A). On day 78 colonies positive for all four SNVs were dominant; two colonies were KRASG12R/NTRK3V443I/EZH2I669M, two were heterozygous MSLNP462H only, and one was heterozygous for KRASG12R/NTRK3V443I/EZH2I669M and homozygous for MSLNP462H (Figure 2A). Altogether, these data are consistent with somatic acquisition of KRASG12R, followed by NTRK3V443I and MSLNP462H, and lastly EZH2I669M (Figure 2B). Whether MSLNP462H and NTRK3V443I were acquired successively or simultaneously cannot be distinguished. KRASG12R/NTRK3V443I/EZH2I669M colonies could reflect loss of the mutant MSLN allele in a side clone, or a sequencing error, which would also explain detection of MSLNP462H as the only SNV in two colonies. Since colony assays may skew clonal ratios present in vivo, we quantified KRASG12R, MSLNP462H, NTRK3V443I and EZH2I669M by pyrosequencing of MNCs from all five samples (Figure 2C). None of the SNVs was detected at diagnosis. KRASG12R occurred with at least 10% higher allelic frequency than the other SNVs in all subsequent samples, consistent with initial acquisition. EZH2I669M was 10-15% higher than MSLNP462H. This is at variance with the colony data suggesting EZH2I669M was acquired last, but could be explained by MSLNP462H loss from the clone harboring all 4 mutations (Figure 2B), via deletion or acquired uniparental disomy of the non-mutated allele. As shown in Figure 2A, we observed such a clone at low frequency in the colony assays (two colonies in the day 67 and 78 samples). Discrepancies in clonal representation between colony and sequencing data were also demonstrated in AML7.
Figure 2. Genotyping of successive blood samples.
(A) Colony genotyping at diagnosis, day 67 and day 78. (Upper panel) Proportion of BCR-ABL1-positive (green), BCR-ABL1-negative (white) and BCR-ABL1-undetermined colonies (gray). (Lower panel) Numbers of colonies with KRASG12R, MSLNP462H, NTRK3V443I, EZH2I669M alleles. Gray circles represent the non-mutant alleles. (B) Clonal architecture and evolution. The squares denote single nucleotide variants (SNVs) that were either present myeloid as well as T cells, suggesting they were germline variants or acquired by a progenitor cell with multilineage potential. Circles represent the four somatic SNVs, with color coding as in the previous panel. (C) Quantification of somatic SNVs by pyrosequencing.
While the presentation of our patient was consistent with CML, low hemoglobin and platelets were unusual in the absence of other high-risk CML features. On imatinib, the clinical phenotype rapidly morphed to CMML and four somatic point mutations were detected by WES of CD14+ cells on day 67. Although neither MassARRAY nor pyrosequencing had identified any of these variants at diagnosis, their rapid appearance suggested they predated imatinib therapy, which was confirmed by colony sequencing. BCR-ABL1 and the CMML-related point mutations may have arisen independently in different hematopoietic stem cells or may share a common abnormal ancestor. The latter is suggested by nonsense SNVs in ASXL1 and TET2, with identical allelic ratios in CD3+ and CD14+ cells and MNCs, and in all sequential samples, consistent with their presence in the germline or in a pluripotent hematopoietic stem cell. Somatic mutations in CMML T cells have been reported, although typically at a lower allelic ratio than in the myeloid lineage8. Verification would require an alternative source of germline DNA, which is not available. Irrespective of this limitation, it is likely that TET2S407fs*20 and ASXL1P808fs*10 contribute to CMML in this patient, as they have been reported as validated somatic variants in COSMIC. Moreover we have shown that TET2 (~50%) and ASXL1 (~40%) are amongst the most commonly mutated genes in CMML, while the frequency of NRAS or KRAS mutations is only 10-20%9. Another study identified TET2 mutations as founder mutations in CMML8, while KRASG12R was as a secondary event, and EZH2 mutations are acquired late9. This suggests that RASG12R and EZH2I669M were acquired by TET2/ASXL1 mutant cells. EZH2I669M is a recurrent CMML mutation and associated with poor outcome10. Neither MSLNP462H nor NTRK3V443I have been described in CMML or other cancers. MSLN encodes a precursor of two proteins, megakaryocyte potentiation factor (MPF), which enhances cytokine effects on megakaryocytes, and mesothelin, a cell adhesion molecule11, 12. p.P462T was previously reported in esophageal carcinoma13. Mutations of NTRK3 (also known as TrkC) have been described in medulloblastoma and other cancers14. In AML cell lines, NTRK3 enhances proliferation and inhibits apoptosis through activation of PI3K/AKT and AKT/mTOR15. Functional characterization will be required to determine whether MSLNP462H and NTRK3V443I contribute to disease progression or are bystanders.
Secondary Ph− leukemia after treatment for CML is rare. Prior to the introduction of imatinib, such cases were ascribed to cytotoxic chemotherapy. In the era of TKIs, effective suppression of the highly proliferative Ph+ clone may lead to rapid expansion of a previously unrecognized leukemic clone, as in our patient. It is conceivable that a non-specific agent, such as hydroxyurea, might have been a better option compared to the imatinib/5-azacytidine combination, illustrating the challenges of applying targeted therapy to clonally complex myeloproliferative neoplasms.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Derek Warner, director of Sequence and Genomics Core Facilities in the University of Utah and Jonathan Schumacher (ARUP) for their assistance with the pyrosequencing experiments. This work was supported by a Leukemia & Lymphoma Society (LLS) Translational Research Program Award (6086-12) (M.W.D.) and an LLS Specialized Center of Research Program Award (GCNCR0314A-UTAH) (M.W.D.). This work was also supported by the National Institutes of Health (NIH) National Cancer Institute (grants P01CA049639 [M.W.D.], R01CA178397 [M.W.D. and T.O.], and 5P30CA042014-24 [Huntsman Cancer Institute]), by the V Foundation for Cancer Research (M.W.D. and T.O.) and by U01HG006513 (G.T. Marth). J.S.K. was a Special Fellow of the LLS and was supported by a Translational Research Training in Hematology Award from the American Society of Hematology (ASH). A.M.E. was supported by the NIH National Cancer Institute (T32 CA093247) and a Career Development Award from LLS (5090-12), and is currently funded through a Scholar Award from ASH. A.M.E. also acknowledges support from the NIH Loan Repayment Program.
Footnotes
Conflict of Interest: Dr. Deininger is on the advisory board and is a consultant for Ariad, Incyte, Novartis, and Pfizer, and serves on the advisory board for CTI BioPharma Corp. His laboratory receives research funding from Bristol-Myers Squibb, Celgene, Gilead, and Novartis.
REFERENCES
- 1.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 2.Bumm T, Muller C, Al Ali HK, Krohn K, Shepherd P, Schmidt E, et al. Emergence of clonal cytogenetic abnormalities in Ph- cells in some CML patients in cytogenetic remission to imatinib but restoration of polyclonal hematopoiesis in the majority. Blood. 2003;101(5):1941–1949. doi: 10.1182/blood-2002-07-2053. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt M, Rinke J, Schafer V, Schnittger S, Kohlmann A, Obstfelder E, et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014 Dec;28(12):2292–2299. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- 4.Loriaux M, Deininger M. Clonal cytogenetic abnormalities in Philadelphia chromosome negative cells in chronic myeloid leukemia patients treated with imatinib. Leuk & Lymphoma. 2004;45(11):2197–2203. doi: 10.1080/10428190410001723278. [DOI] [PubMed] [Google Scholar]
- 5.Diamond J, Goldman JM, Melo JV. BCR-ABL, ABL-BCR, BCR, and ABL genes are all expressed in individual granulocyte-macrophage colony-forming unit colonies derived from blood of patients with chronic myeloid leukemia. Blood. 1995;85(8):2171–2175. [PubMed] [Google Scholar]
- 6.Padron E, Abdel-Wahab O. Importance of genetics in the clinical management of chronic myelomonocytic leukemia. J Clin Oncol. 2013 Jul 1;31(19):2374–2376. doi: 10.1200/JCO.2013.48.9120. [DOI] [PubMed] [Google Scholar]
- 7.Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014 Mar 17;25(3):379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013 Mar 21;121(12):2186–2198. doi: 10.1182/blood-2012-06-440347. [DOI] [PubMed] [Google Scholar]
- 9.Mason CC, Khorashad JS, Tantravahi SK, Kelley TW, Zabriskie MS, Yan D, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016 Apr;30(4):906–913. doi: 10.1038/leu.2015.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grossmann V, Kohlmann A, Eder C, Haferlach C, Kern W, Cross NC, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011 May;25(5):877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 11.Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onda M, Nagata S, Ho M, Bera TK, Hassan R, Alexander RH, et al. Megakaryocyte potentiation factor cleaved from mesothelin precursor is a useful tumor marker in the serum of patients with mesothelioma. Clinical cancer research. 2006 Jul 15;12(14 Pt 1):4225–4231. doi: 10.1158/1078-0432.CCR-06-0472. [DOI] [PubMed] [Google Scholar]
- 13.Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nature genetics. 2013;45(5):478–486. doi: 10.1038/ng.2591. 05//print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lannon CL, Sorensen PH. ETV6-NTRK3: a chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Seminars in cancer biology. 2005 Jun;15(3):215–223. doi: 10.1016/j.semcancer.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Kim MS, Kim GM, Choi YJ, Kim HJ, Kim YJ, Jin W. TrkC promotes survival and growth of leukemia cells through Akt-mTOR-dependent up-regulation of PLK-1 and Twist-1. Mol Cells. 2013 Aug;36(2):177–184. doi: 10.1007/s10059-013-0061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.