Abstract
Alterations in high-order chromatin, with concomitant modulation in gene expression, are one of the earliest events in the development of colorectal cancer (CRC). Cohesins are a family of proteins that modulate high-order chromatin, although the role in CRC remains incompletely understood. We, therefore, assessed the role of cohesin SA1 in CRC biology and as a biomarker focusing in particular on the increased incidence/mortality of CRC among African-Americans (AAs). Immunohistochemistry on tissue arrays revealed dramatically decreased SA1 expression in both adenomas (62%; p=0.001) and adenocarcinomas (75%; p=0.0001). RT-PCR performed in endoscopically normal rectal biopsies (n=78) revealed a profound decrease in SA1 expression in adenoma-harboring patients (field carcinogenesis) compared to those who were neoplasia-free (47%; p=0.03). From a racial perspective, CRC tissues from Caucasians had 56% higher SA1 expression than in AAs. This was mirrored in field carcinogenesis where healthy Caucasians expressed more SA1 at baseline compared to matched AA subjects (73%; p=0.003). However, as a biomarker for CRC risk, the diagnostic performance as assessed by area under ROC curve was greater in AAs (AUROC=0.724) than Caucasians (AUROC=0.585). From a biological perspective, SA1 modulation of high-order chromatin was demonstrated with both biophotonic (nanocytology) and chromatin accessibility (MNase) assays in SA1-knockdown-HT29 CRC cells. The functional consequences were underscored by increased proliferation (WST-1; p=0.0002, colony formation; p=0.001) in the SA1-knockdown-HT29 cells. These results provide the first evidence indicating a tumor suppressor role of SA1 in early colon carcinogenesis and as a risk-stratification biomarker giving potential insights into biological basis of racial disparities in CRC.
Keywords: Colorectal Cancer, Cohesin, SA1, Tumor Suppressor, Field Carcinogenesis, Partial Wave Spectroscopy (PWS), Higher-order Chromatin
Introduction
Colorectal cancer (CRC) remains the second leading cause of cancer deaths among Americans highlighting the need for more effective strategies [1]. The genomic era diagnosis has unprecedented promise of making significant impact on CRC mortality by employing powerful and increasingly affordable technologies such as next-generation sequencing to identify novel therapeutic targets. A complicating factor is that CRC is biologically heterogeneous at least four consensus molecular subtypes (CMS) and five proteomic pattern. [2] [3]. Moreover, the mutational load spectrum of CRC is generally quite high with MMR deficient CRCs (CMS1) having the most mutations of any major cancer [4]. Thus, discerning the driver mutations from the numerous passenger mutation background remains a challenging task. Further complicating the biology are the numerous epigenetic events that modify gene expression including methylation, copy number alterations etc. [5].
Recently, attention has focused on high order chromatin alterations as the fundamental event in carcinogenesis. Indeed, The Cancer Genome Atlas (TCGA) has revealed that some of the most commonly mutated genes related to higher-order chromatin structures are involved in nucleosome positioning and hence in modulation of gene expression (e.g., Arid 1a and other members of the SWI/SNF family) [6]. In the human genome, higher-order chromatin remodeling can be driven by self-associating topological domains that are enriched in cohesins which play an important role in governing long-range nuclear interactions and gene expression [7]. These cohesins comprise of two complexes that consist of Smc1, Smc3, Rad21/Scc1 and either of the stromal antigens (SA1 or SA2) which regulate chromatin looping, unmask promoters from the histone shield and control translation [8]. While cohesins have been found to be mutated in a small proportion of CRCs [9], a recent provocative report indicates that most CRCs possess higher frequency of mutations in specific DNA sites which are usually assigned to bind cohesin proteins [10]. These studies highlight the important role of alterations in the higher-order chromatin in colon carcinogenesis.
Our group has previously developed partial wave spectroscopic microscopy (PWS), a powerful modality to detect changes in the higher-order chromatin. By measuring fluctuations in the nanoscale parameters (disorder strength or Ld) with PWS, we have demonstrated that tumors have a large degree of chromatin heterogeneity [11]. An even more significant finding from a clinical perspective was that when endoscopically normal rectal mucosa from patients with colonic neoplasia was analyzed, they exhibited markedly altered higher-order chromatin [12]. These alterations in the higher-order chromatin represent a striking manifestation of field carcinogenesis which has previously also been noted through alterations in gene expression, methylation and proteomics along with ultrastructural changes as detected by transmission electron microscopy [13–15]. The potential clinical utility is for improved risk stratification for colon cancer screening through sampling the readily accessible mucosa (i.e. rectal examination). The clinical imperative is that while colonoscopy is recommended for the entire population at age fifty and older, the yield of advanced adenomas (adenomas ≥1 cm or high grade dysplasia or ≥25% villous features) is only ~7–8%, meaning that a vast majority of colonoscopies do not have any cancer preventive implications (through identification and removal of the biologically significant lesions) [16]. Therefore, developing a rectal risk stratification tool would help in identifying at-risk population where colonoscopy is more likely to be high-yield. This in particular is urgently needed in African Americans (AA) who are known to suffer disproportionately from CRCs with regards to both lifetime CRC incidence risk and mortality than Caucasians (by 23% and 53% in males; 22.5% and 45.5% in females respectively) [17, 18], with limited progress made in reducing such disparities over the last two decades [19]. Furthermore, data suggests that AAs develop CRC earlier than Caucasians [20]. Therefore, American College of Gastroenterology had recommended earlier screening for AAs [21] but other major societies did not endorse that view. Biologically, there are specific racial differences in CRCs with a next generation sequencing study with mutations in two genes (ephrin type A receptor and folliculin) were limited only AAs and not Caucasians [22]. However, to the best of our knowledge, there are no previous reports on race-specific alteration in CRC biomarkers, including during early field carcinogenesis. Thus, finding reliable biomarkers to better risk stratify this supposedly high-risk population (AAs) is important.
In this study, we explore the role of the higher-order chromatin modulator, cohesin SA1 in colon carcinogenesis. We demonstrate that downregulation of SA1 is an early alteration during neoplastic transformation and is a robust marker of field carcinogenesis. Importantly, the basal expression and diagnostic abilities differed by race providing the first demonstration of this concept. Finally we show that SA1 is biologically important altering phenotype of higher-order chromatin as demonstrated by the novel live-cell PWS and the classic MNase assay.
Materials and Methods
Human Subjects
All human studies were conducted with approval from the Institutional Review Board (IRB) guidelines of Boston University School of Medicine. Rectal biopsies were collected from patients undergoing screening or surveillance colonoscopy at Boston Medical Center. There were 40 males (51%) and 38 females (49%) with mean ages 54.5±4.3 and 52.3±1.3, respectively. The samples were distributed equally between AAs and Caucasians. In this cohort, 34% had adenomas and the remainder were considered controls (negative colonoscopies or left sided hyperplastic polyps). Given that this was an asymptomatic population no carcinomas were noted (estimated prevalence ~0.7% [23]). Exclusion criteria were: (a) incomplete colonoscopy (poor preparation, inability to intubate cecum or failure to recover polyps for pathological evaluation); (b) concurrent anticoagulant therapy; and (c) mucosal abnormalities (i.e. inflammatory bowel disease) (e) Isolated right sided proximal serrated polyps. Six biopsies were collected from endoscopically normal rectal mucosa. Biopsies were transported in PBS and 2 biopsies snap frozen in liquid nitrogen and stored at −80°C for RT PCR and 4 biopsies fixed in 10% buffered formalin for 24 h and stored in 70% alcohol for IHC.
Immunohistochemistry (IHC)
To determine the expression and localization of the cohesin SA-1, IHC was performed on (a) formalin fixed rectal biopsies (as above) that were embedded in paraffin blocks, sectioned (4µm thick) and mounted on Superfrost+ glass slides and (b) human CRC tissue arrays (US Biomax, Inc., Rockville, MD) assembled from 72 different grade tumors, 10 tumor-adjacent histologically normal tissues and 10 control samples (non-CRC tissue). The slides were deparaffinized by heating at 60°C (~1 hour) and two washes in xylene followed by rehydrating with graded alcohol washes. The tissue sections were subjected to antigen-epitope retrieval by pressure microwaving (Nordic-Ware) at high power setting (2 × 9 minutes) in antigen unmasking solution (Vector Laboratories, Burlingame, CA). After quenching the endogenous peroxidase activity in 3% hydrogen peroxide, the slides were incubated in 5% horse serum for 2–3 hours at room temperature to block non-specific binding. The sections were then incubated with anti-SA1 antibody (Abcam, Cambridge, MA; rabbit polyclonal; 1:200 dilutions) overnight at 4°C. After standard PBS washings, the sections were incubated with the universal biotinylated secondary antibody (1:2000) for 30min followed by avidin-biotin peroxidase using Vectastatin Elite ABC Reagent Kit (Vector Laboratories, CA). Finally the antigen-specific brown staining was developed by exposing the sections to 3, 3’-diaminobenzidine (DAB) as the chromagen substrate (for 1–3 minutes). For the negative controls, duplicate sections on the same slide were processed in the absence of the primary antibody. The color intensity was scored on a scale of 0–3 by an investigator blinded to colonoscopic findings for rectal biopsies.
RNA isolation and RT-PCR in rectal biopsies and isolated colonic tumors
In these studies, we compared the mRNA expression of SA1 in random rectal biopsies collected from AA or Caucasian patients with or without adenomas/polyps detected elsewhere in the colon. For these studies, we used frozen colonic tumors obtained from the Boston University Biospecimen Archive Research Core. RNA was extracted from the samples with TRI Reagent (Sigma Chemicals) following the manufacturer’s instructions (Molecular Research Center Inc., Cincinnati, OH). The samples were homogenized in a BeadBug homogenizer (Benchmark Scientific, Edison, NJ) and total RNA isolated using a RiboPure Kit (Ambion, Life Technologies; Woburn, MA) following the manufacturer’s instructions. After establishing the purity and concentration by spectrophotometry (OD 260/280), RNA was reverse transcribed with human SA1-specific TaqMan probes (Applied Biosystems, Carlsbad, CA) following the manufacturer’s protocol. Samples were processed using Step-One-Plus RT Thermo cycler (Life Technologies) using RT kit and Universal PCR Master Mix. All the samples were normalized to β-actin and the relative expression of SA1 was analyzed using the comparative (2−ΔΔCt) method as described previously [24]. The threshold of fold change significance was set as >1.5 (upregulation) and <0.67 (down-regulation).
SA1 knockdown assay
To further explore the functional consequences of SA1, we studied the effects of SA1 downregulation on cellular growth characteristics of human colon cancer cell line HT-29 cells (utilized because of relatively high level of basal SA1 expression). For these studies, we created an SA1 knockdown in HT-29 cells by stably transfecting them with SA1 specific short hairpin RNA (shRNA) vector (Santa Cruz Biotech) with a puromycin selection vector. We also performed transient transfections in cell lines with lower basal SA1 expression (e.g. SW480) using SiRNA (Santa Cruz Biotechnology) per manufacturer’s instructions.
Western blot analysis
Western blotting was done using standard techniques. Briefly, 30µg of sample protein was subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes (Amersham Pharmacia, Piscataway, NJ), blocked with 5% blotto and probed with specific antibody for PCNA (1:1000; Santa Cruz Biotech), β-catenin (1:500; Santa Cruz Biotech) and pβ-catenin (1:500; Cell Signaling) as described previously. Xerograms were developed with enhanced chemi-luminescence (Santa Cruz Biotechnology) and images acquired via UVP Bio-imaging Systems and the data processed using Labworks 4.6 software. The data was normalized to the expression of β-actin as loading control.
DNA methylation and acetylation assay
HT-29 cells were seeded in six-well plates at a density of 0.5×106. After 24h, media was replenished with 5-azadc (2.5mM) and sodium butyrate (1.0mM). After 48h of incubation at 37°C/5%CO2, the protein lysates were subjected to immunoblotting to measure SA1 expression.
Micrococcal Nuclease assay (MNase)
Since nucleosome positioning is critical for gene expression and most DNA-related processes, we performed the micrococcal nuclease digestion to examine SA1 nucleosomal positioning using the HT29-SA1ShRNA (Stable knockdown) and HT29-Scc (scramble vector) cells. The MNase assay was performed with minor modifications as previously reported by our group [15]. Fifty×106 cell suspensions were washed of media with ice-cold PBS (2000 RPM at 4°C for 10 minutes) and then resuspended in 5 ml of ice-cold NP-40 lysis buffer (10mM tris-HCl (pH 7.4), 10mM NaCl, 3mM MgCl2, 0.5% Nonidet P-40, 0.15mM spermine and 0.5mM spermidine) and incubated for 5 min on ice and the nuclear fraction isolated and washed with 2.5ml of MNase digestion buffer (10mM Tris-HCl, 15mM NaCl, 60mM KCl, 0.15mM spermine and 0.5mM spermidine) and re-suspended in 1 ml of MNase digestion buffer containing 2mM CaCl2. From the suspended solutions, 100µl aliquots were taken and incubated with increasing amounts of the MNase enzyme (0, 80 and 100 units) for 5 and 10 minutes. The reaction was stopped by adding 80µl of MNase digestion buffer, 20 µl of MNase stop buffer (100mM EDTA, 10mM EGTA), 3µl of Proteinase K (25mg/ml) and 10µl of 20% SDS and incubated overnight at 37°C. The samples were then extracted with 200 µl of phenol-chloroform solution, spun and the aqueous layer collected. Following that, 2 µl of RNAse-A (10 mg/ml) was added to the solution and incubated at 37°C for 2 hours and again phenol-chloroform extraction was performed. DNA was precipitated by adding 100% ethanol and after centrifugation and the pellet re-suspended in DNA hydration buffer or water and 5 µg of the isolated DNA was resolved in a 1.4% agarose gel and visualized by ethidium bromide staining.
Back-Scattering Interference Spectroscopic (BaSIS) microscopy and analysis
A full description of the BaSIS microscopy technique has been reported previously [25]. Briefly, the BaSIS instrument was built into a commercial inverted microscope (Leica DMIRB) equipped with a high NA oil immersion objective with broadband illumination provided by a Xenon lamp. Refractive index fluctuations are measured by sampling backscattered light at each wavelength 500–700nm using a combination of a liquid crystal tunable filter (LCTF) and a CMOS camera. Nanoscale differences in chromatin structure are measured as Σ, which quantifies the spatial fluctuations in the z direction [25]. HT29-SA1ShRNA and HT29-Scr cells were maintained at 37°C with CO2 and imaged in their normal media seeded onto petri dishes with coverslip bottoms (MatTek, Ashland, MA). To calculate the average Σ, nuclear ROIs were selected and averaged over all of the cells and biological repeats (n = ~80 nuclei, per group). Standardized student t-tests generated two-tailed P-value (assuming unequal variances) using Microsoft Excel.
Colony-forming assay
HT29-SA1ShRNA and HT29-Scr cells were seeded in triplicates at a density of 2.5 × 103 cells/10cm dishes in McCoy’s medium containing 1.0% FBS. After 2 week of growth, the cells were fixed and stained with crystal violet stain (0.1%, w/v) in 20nM 4-morpholinepropanesulfonic acid (Sigma Chemicals, St. Louis, MO). Colonies were counted and quantified as average of three independent experiments.
Proliferation/growth Assay
HT29-Scr and HT29-SA1 shRNA cells were seeded in multiples of eight in a 96-well plate (10,000 cells per well) in complete medium. After overnight incubation, the medium was replaced with 1% FBS and Penicillin/Streptomycin (100µl/ml) containing media. This was followed by the addition of 10µl of the Cell Proliferation Reagent, WST-1 (Roche, Indianapolis, IN), to each well and absorbance measured at 450nm after 4h of incubation [26]. From these measurements the values recorded at the reference wavelength (600nm) were subtracted, following the manufacturer’s guidelines. Data analysis was performed for three independent experiments, p**=0.0002.
Statistical analysis
Appropriate Excel 2010 statistical tools were used for determining statistical significance. AUROC curves were plotted using STATA 8 software. A two-tailed Student’s t-test was utilized as appropriate.
Results
Immunohistochemical loss of cohesin family member SA1 during colon carcinogenesis
IHC expression analysis for SA1 cohesin was performed on human tissue arrays with 92 samples as discussed in “Methods”. Ten 40× fields from each specimen were scored (0–3; with 0 being no intensity and 3 very strong). SA1 immunoreactivity was found to be expressed mostly in the nucleus and partially in the cytoplasm. As shown in Fig. 1A, out of the cohesins studied, SA1 was markedly reduced in adenomatous polyps (by 62%, p=0.001) with a progressive loss detected in adenocarcinomas (75%; p=0.0001; data not shown). To investigate SA1 as a marker of field carcinogenesis, IHC was performed in rectal biopsies obtained from patients with (n=33) or without (n=36) any concurrent neoplasia. As demonstrated in Fig. 1B), a marked downregulation was also observed in rectal mucosa from patients harboring colonic adenomatous polyps compared to no polyps (p=0.052). These studies demonstrate that SA-1 is progressively lost during colon carcinogenesis.
Figure 1.
Immunohistochemical Expression of SA-1. SA1 expression was measured in (A) Tissue Array sections with 20 normal and 20 adenomatous polyps and (B) rectal mucosal biopsies collected from patients with (9) or without (9) any detected adenomas (field effect) by immunohistochemical staining for SA1 as described in Methods Section. Ten optical fields from each specimen were scored (0–3; with 0 being no intensity and 3 very strong). Overall staining was mostly localized in nuclear and partly in cytoplasmic compartments as indicated by the arrows. As shown in panel (A) there was a significant decrease in SA1 intensity in adenomatous polyps compared to normal mucosa (p=0.001). Similar trend was observed in rectal mucosa from patients harboring adenomatous polyps elsewhere in their colon compared to patients who were adenoma-free (p=0.052).
Biomarker potential of SA1
To determine the performance of SA1 as a risk stratification biomarker, messenger RNA (mRNA) expression of SA1 was quantified by RT PCR in the rectal biopsies as described in “Methods”. As shown in Fig. 2A, baseline expression of rectal mucosal SA1 was observed to be significantly higher in Caucasians compared to AAs (73%; p=0.003). In the entire cohort, SA1 expression was significantly downregulated in subjects harboring colonic adenomas (by 47.3%; p=0.035) consonant with role as a biomarker for field carcinogenesis (Panel B). When stratified by race, AAs with adenoma manifested strikingly greater and statistically significant reduction in SA1 expression (by 60.6%; p=0.012) compared to AAs without adenomas. Caucasians on the other hand demonstrated lesser and statistically insignificant decrease in SA1 expression in subjects with adenomas compared those without (44.7%; p=0.113), suggesting a race-specific alteration. Likewise, as shown (panel C), SA1 mRNA expression was also found to be significantly lower in colon adenocarcinomas from AAs compared to Caucasians. Panel D depicts the ROC curve demonstrating diagnostic performance of rectal mucosal SA1 as a biomarker to risk-stratify subjects for presence of adenomas. Evidently, the biomarker sensitivity of SA1 was low for all subjects (AUROC = 0.606) or Caucasians (AUROC = 0.585), but the test performed significantly better when applied only in AAs (AUROC=0.724).
Figure 2.
Messenger RNA (mRNA) Expression of SA-1 in Field Carcinogenesis. SA1 mRNA was measured in rectal mucosal biopsies collected from patients with (n=33) or without (n=36) any adenomas (field effect) by RT PCR as described in Methods Section. Panel A shows that baseline expression of rectal mucosal SA1 was significantly higher in Caucasians compared to American Americans (73%; p=0.003). Panel B demonstrates that SA1 expression was found to be significantly decreased in patients bearing colonic adenomas versus those who are adenoma free (by 47.3%; p=0.035). When analyzed by race, the suppression of SA1 in adenoma harboring patients was markedly greater in AA rather than Caucasians (60.6%; p=0.012 versus 44.7%; p=0.113, respectively). Panel C SA1 mRNA demonstrates that SA1 mRNA expression in frank CRCs (frozen specimens) also markedly decreased in AAs compared to Caucasians. Panel D depicts the ROC analysis of SA 1 suppression in endoscopically-normal rectal mucosa showing superior performance in AAs than Caucasians.
Epigenetic modulations of SA1
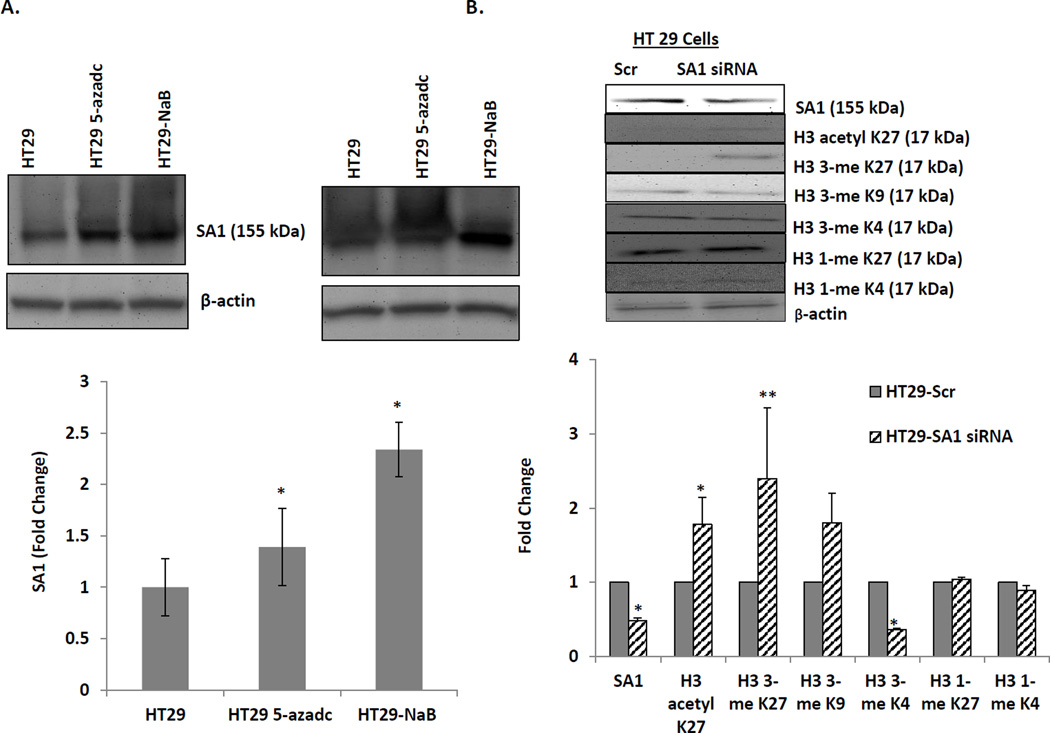
Furthermore, to elucidate the mechanism of SA1 gene silencing during colon carcinogenesis, we investigated potential epigenetic modifications such as DNA methylation and/or histone deacetylation as catalyzed by DNA methyl transferases (DNMTs) and histone deacetylases (HDACs) respectively (Fig 3a). For these studies, we exposed HT-29 cells to either a demethylating agent (5-Aza-dC) or an HDAC inhibitor (sodium butyrate; NaB). After 48h, SA1 was found to be overexpressed in both 5-Aza-dC or NaB treated HT29, suggesting important relevance of epigenetic modulations in SA1 repression. Furthermore, since methylation of histones can modify chromatin structure by increasing or decreasing transcription depending on which amino acids are methylated and how many methyl groups are attached, we analyzed the effect of SA1 ablation on such methylation patterns. We observed that while SA1 knockdown in HT-29 cells specifically reduced the tri-methylation at histone mark H3 lysine 4 (H3K4), it increased tri-methylation at H3 lysine 27 (H3K27) residues without altering mono-methylation marks at either site. (Fig 3b) Histone mark H3K27 when trimethylated are known to shut down transcription by tightly associating with inactive gene promoters while trimethylation of H3K4 produces an opposite effect. Also, tri-methylations of another mark H3 Lysine 9 (H3K9) showed no modifications by SA1 knockdown.
Figure 3.
Role of epigenetic modulations in the loss of SA1 in CRC cells. (A) HT29 cells were treated with 2.5mM 5-Aza-dC (DNMT inhibitor) and 1.0mM NaB (HDACs inhibitor) for 24h before subjecting to immunoblot analysis. As shown in the histograms, there was a significant up-regulation of SA1 by 5-Aza (*p=0.014) as well as NaB (**p=0.01) (B) siRNA-mediated down-regulation of SA1 in HT29 cells caused alterations in both acetylation (*p=0.005) and tri-methylation (**p=0.001) at Histone marks H3K27 and demethylation at H3K4 (*p=0.004) as measured by immunoblotting. Tri-methylation at H3K9 and mono-methylation at H3K27 and H3K4 showed no differences. β-actin was used as an internal control and the quantification was performed using VisionWorks LS, represented as the fold change in protein expression compared to HT29-Scr. The error bars represent standard error calculated from three independent experiments.
Effect on nucleosome positioning/occupancy
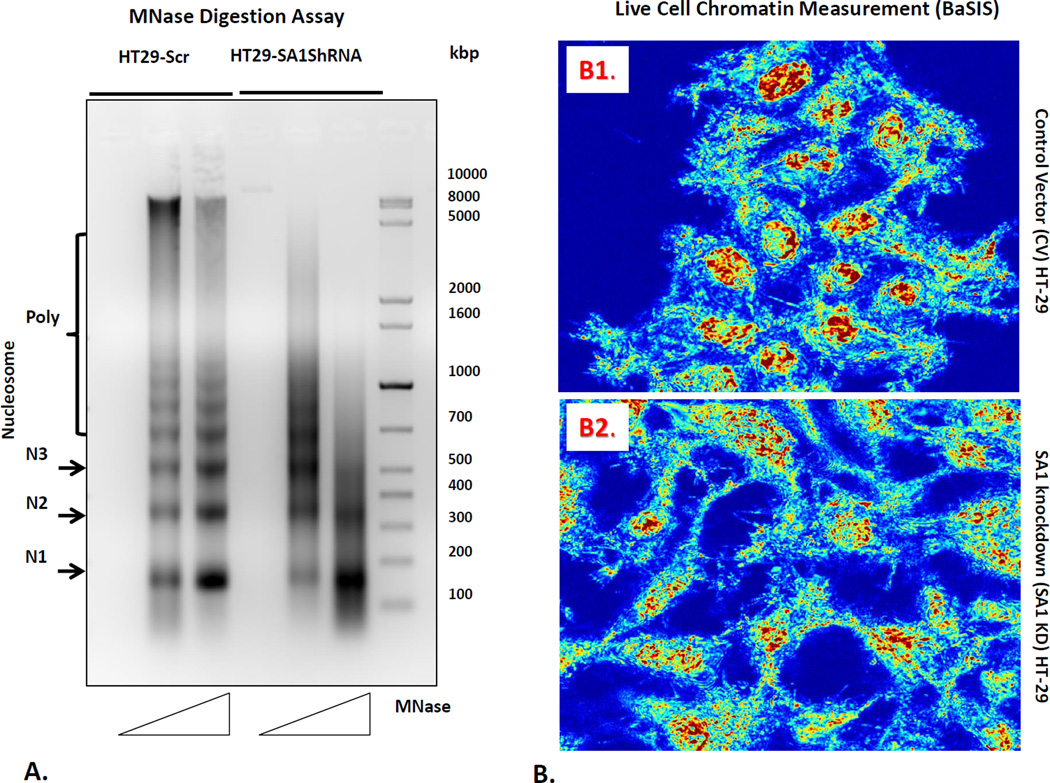
Nucleosomes, which encapsulate 147bp segment of DNA wrapped around the histone octamer 1.65 times [27] are not randomly distributed but carefully positioned in certain genomic regions to impact transcriptionally activity. Nucleosome positioning is determined by the combination of DNA sequence, nucleosome remodeling enzymes and transcription factors. To verify that SA1 has a direct bearing on the higher-order chromatin structure as indicated by chromatin accessibility to nuclease digestion, we examined the susceptibility of HT29 cells (control) and SA1 shRNA transfected HT29 cells to MNase digestion (Fig. 4A). Comparison of the control and SA1 shRNA transfected HT29 cells showed different MNase digestion patterns. The variation in the pattern of the digested DNA ladder is reflective of changes in the nucleosomal occupancy. The DNA ladder bands correspond to either the mononucleosome (147bp) or polynucleosomes, in successively increasing size. The ladder shows an increase in intensity with increasing concentration of the MNase enzyme, indicating a higher degree of digestion as the ratio of enzyme to DNA increases. The densitometric analysis shows the difference in intensity of the low molecular weight bands corresponding to mono-, di- and tri-nucleosomes, marked as N1, N2 and N3 respectively, between control and SA1 shRNA transfected HT29 cells. The results indicate that the inter-nucleosomal DNA linker may be less accessible for MNase digestion in theSA1 shRNA transfected HT29 cells compared to controls.
Figure 4.
Chromatin modifications related to SA1 downregulation. For these studies, we first performed (panel A) micrococcal nuclease digestion assay to determine the nucleosome positioning pattern of the nuclei isolated from HT 29 cells and SA1 shRNA transfected HT29 cells. Nuclei were digested with increasing concentrations of the MNase enzyme (50 and 100 units) for 5min followed by DNA extraction using phenol-chloroform. 5µg of DNA was then separated on a 1.5% agarose gel. Mono (N1), -di (N2), -tri (N3) and poly-nucleosomes are indicated on the left. Furthermore, we measured nanoscale chromatin alterations (panel B) in live cells using the novel technique, Back-Scattering Interference Spectroscopic (BaSIS) microscopy. BaSIS quantifies the spatial fluctuations in the z direction, measured as Σ, with sensitivity to structures between 20–200nm that can quantify the dynamics of the nano-molecular organization in live cells without using exogenous labels. Control (Panel B1) and SA-1 shRNA knockdown (Panel B2) cells were imaged in glass bottom petri dishes. The nanoscale spatial fluctuations (Σ) was calculated for each nucleus and averaged over 3 experiments (.n=~80 cells/group). SA1 loss decreased nuclear Σ (p<0.01), demonstrating that SA1 is key in maintaining nanoscale higher-order chromatin leading to more homogeneity of nuclear structures.
Effect of SA1 depletion on chromatin structure
Nanocytology (PWS) can be measured on live cells using the next generation iteration, Back-Scattering Interference Spectroscopic (BaSIS) microscopy [13]. BaSIS quantifies the spatial fluctuations in the z direction, measured as Σ, with sensitivity to structures between 20–200nm that can quantify the dynamics of the nano-molecular organization in live cells without using exogenous labels. Control and SA1 shRNA transfected HT29 cells were imaged in glass bottom petri dishes (Fig. 4B1 & B2). The Σ was calculated for each nucleus and averaged over 3 experiments. We found that the nuclear Σ decreases in SA1 shRNA transfected HT29 cells SA1-KD cells compared to control cells (−11%, n=~80 cells per group, p<0.01). These data suggest that depletion of SA1 alters the nanoscale higher-order chromatin structure, leading to more homogeneity of nuclear structure.
Effect of SA1 depletion on cellular proliferation
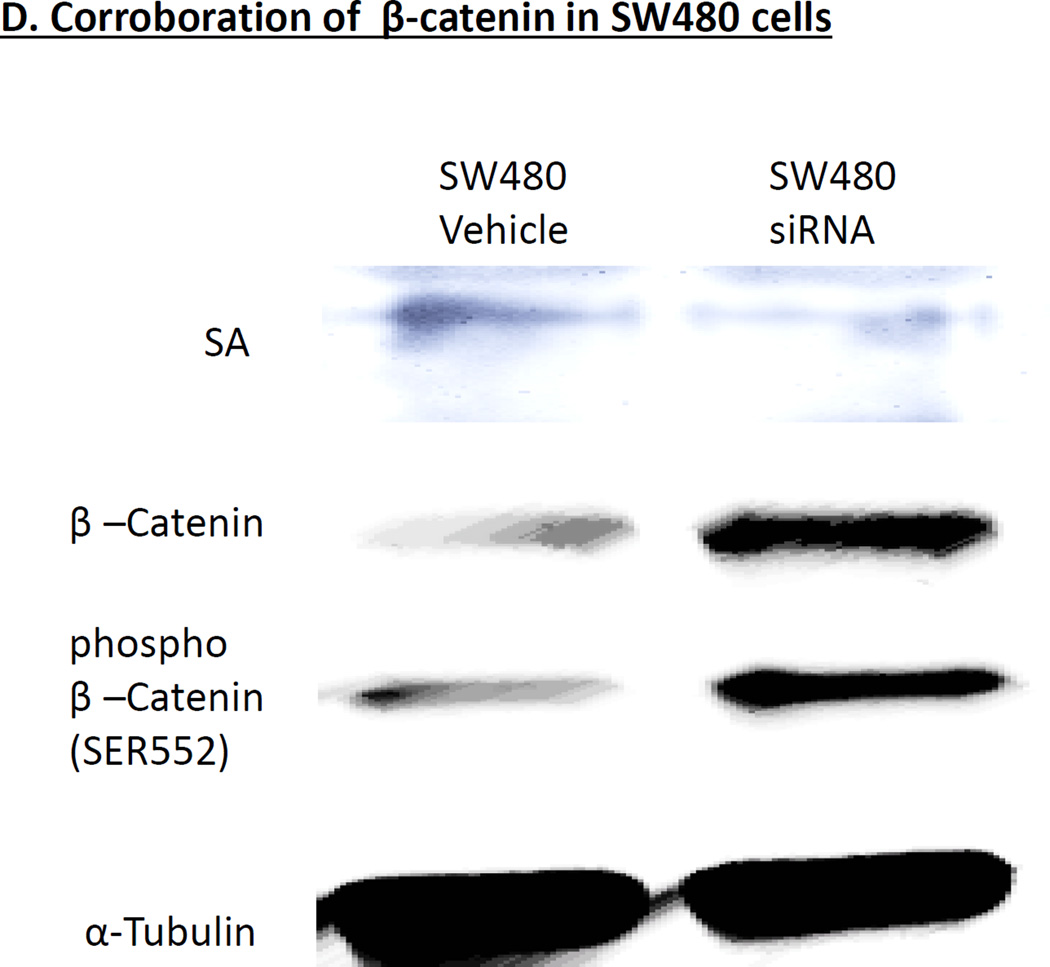
For these studies, we first performed a colony formation assay by separately seeding 2,500 HT29 cells and SA1 shRNA transfected HT29 cells in 10-cm dishes that were cultured for 2 weeks. Cells were fixed and stained with 0.1% crystal violet and representative photographs were taken for HT29 and SA1 shRNA transfected HT29 cells. As shown in Fig. 5a, SA1 shRNA transfected HT29 cells showed significant increase in colony formation as compared to control (HT29 cells), *p = 0.001. Next we performed WST-1 proliferation assay-HT29 and SA1 shRNA transfected HT29 cells were seeded in multiples of eight in 96-well plates and their growth was monitored by measuring absorbance after incubation with WST-1 reagent. Data is represented as the mean absorbance (O.D450 nm − O.D(Ref=600 nm)) of replicates for three independent experiments. The SA1 shRNA transfected HT29 cells showed significant increase in proliferation as compared to control HT29 cells, *p = 0.0002 (Fig. 5b). Lastly, we performed Western blot analysis to measure the expression of proliferation marker PCNA and β-Catenin. Immunoblotting revealed significant up-regulations in protein levels of PCNA and both phosphorylated (pS552) β-Catenin and total-β-Catenin with siRNA mediated SA1 down-regulations in HT29 cells (Fig 5c). We replicated the studies in another human CRC cell line SW480 and found similar results (Fig 5d).
Figure 5.
SA1 Modulates Proliferation in CRC cells= (A) Colony formation assay was performed by seeding 2500 cells in 10-cm dishes and grown for 2 weeks. Cells were fixed and stained with 0.1% crystal violet and representative photographs were taken for HT29 and SA1 shRNA transfected HT29 cells. The SA1 shRNA transfected HT29 cells showed significant increase in colony formation as compared to control HT29 cells, *p = 0.001. (B) HT29 and SA1 shRNA transfected HT29 cells were seeded in multiples of eight in 96-well plates and their growth was monitored by measuring absorbance after incubation with WST-1 reagent. Data is represented as the mean absorbance (O.D 450 nm − O.D Ref=600 nm) of replicates for three independent experiments. The SA1 shRNA transfected HT29 cells showed significant increase in proliferation as compared to control HT29 cells, *p = 0.0002. (C) Immunoblotting revealed significant up-regulations in protein levels of PCNA and both phosphorylated (pS552) β-Catenin and total-β-Catenin with siRNA mediated SA1 down-regulations in HT29 cells. β-actin was used as an internal control. Quantification was performed using VisionWorkLS, represented as the fold change in protein expression as compared to that of the control (1.0). The error bars represent standard error calculated from three independent experiments. * represents significant change (p<0.05) in the protein levels. (D) Corroboration of findings with different cell line. SW480 cells Immunoblotting revealed significant up-regulations in protein levels of PCNA and both phosphorylated (pS552) β-Catenin and total-β-Catenin with siRNA mediated SA1 down-regulations in HT29 cells. β-actin was used as an internal control. Quantification was performed using VisionWorkLS, represented as the fold change in protein expression as compared to that of the control (1.0). The error bars represent standard error calculated from three independent experiments. * represents significant change (p<0.05) in the protein levels.
Discussion
We demonstrate herein, for the first time, that cohesin SA1 is lost early during colon carcinogenesis. Our data indicates that SA1 expression is suppressed in field carcinogenesis potentially through epigenetic events. Intriguingly, the SA1 expression appeared to have a racial predilection with lower expression in endoscopically-normal rectal mucosa and CRCs in AAs compared to Caucasians. For risk stratification, rectal SA1 performed somewhat better in AAs than Caucasians underscoring the need to evaluate biomarkers in context of race. From a biological perspective, the cell culture studies suggested that SA1 loss may modulate key pathways in colon carcinogenesis including β-catenin signaling. Finally using the novel BaSIS (live cell PWS) technology, we show that SA1 altered higher-order chromatin and the transcriptional implications are further supported by histone mark analysis.
Higher-order chromatin is increasingly realized to be a major determinant of transcriptional activation and abnormalities in its organization are linked to cancer [28]. In a mammalian cell, for the chromatin to fold into a relatively smaller nuclear confines (~10µM), DNA (~2 meters long) undergoes several levels of compactions. Nucleosomes form the fundamental units of compaction in which DNA is tightly wound around (1.7 turns) the histone core (11nm) resembling beads on a string that densely fold in the nucleus. However, despite the enormous level of compaction and organization, chromatin selectively makes DNA accessible to interact with specific protein regulators to regulate transcription. The structure is complex with several groups suggesting that chromatin architecture may represent a compact polymer state under many levels of organization defined as a fractal globule [29, 30]. For instance, SWI-SNF family of proteins (such as Arid1a which is well established in colon carcinogenesis) are thought to expose promoter regions through moving nucleosomes [31]. The cohesins, on the other hand, serve as modulators of chromatin looping which allows the placement of enhancer elements upstream of promoters to enhance gene transcription. To initiate appropriate gene functions, cohesin proteins, such as SA1, complexed with CTCF (a zinc-finger DNA binding protein) at the promoter region [32]. Indeed, in many cancers including CRCs higher mutation frequency has been attributed to the CTCF/cohesin-DNA binding sites rather than in cohesin proteins alone [10]. While there have been relatively few studies demonstrating changes in the protein expression, some reports suggest cohesin levels may have prognostic value [33]. To our knowledge, this study is the first to link cohesin levels to initiation of carcinogenesis.
It is striking that these changes in SA1 occur in the histologically normal mucosa consonant with a role in field carcinogenesis. Field carcinogenesis leads to a permissive milieu for tumorigenesis that reflects the molecular interplay between both genetic susceptibility as well as impact of exogenous risk factors (such as diet; smoking that introduces toxins in the fecal stream). Indeed, despite being histologically normal, there are profound molecular alterations (genomic, methylation, microRNA, proteomic etc) in patients harboring neoplasia. A parsimonious hypothesis assumes a central role for alterations in the higher-order chromatin as evidenced by PWS, TEM, karyometric measurements etc.[34–36]. The biological determinants of these changes are varied but proteins that modulate higher-order chromatin including cohesins may have an important role. Indeed, in keeping with this, our group has previously noted that PWS alterations from the endoscopically normal rectal mucosa had excellent diagnostic potential [37], supporting our results from the diagnostic studies with SA1. While others have shown that cohesin loss occurs at the adenoma stage [38], ours is the first study to show modulation at the earliest stages of carcinogenesis. A complexity in unraveling the biological determinants is the pleotropic effects of cohesins including sister chromatid segregation. The relative importance of cohesin induced alterations in gene expression versus aneuploidy in carcinogenesis is unclear. For instance, some reports in bladder cancer linked SA1 to aneuploidy [39] whereas others found that this tumor suppressor gene promoted bladder malignancies regardless of ploidy [40]. SA1 may have other functions such as being downstream of important drivers in colon carcinogenesis such as Wnt signaling etc. [41]. The functional importance is underscored by the multiple developmental abnormalities in germline mutations such as noted in Cornelia de Lange Syndrome [42].
Detection and identification of markers of field carcinogenesis may have strong clinical ramifications. For instance, field carcinogenesis is the biological underpinning for the clinical guidelines on post-polypectomy surveillance [43]. Several groups, including ours, have focused on exploiting the field carcinogenesis for risk stratification in order to assist personalized screening given its remarkable inefficiency. Utilizing novel array of optical modalities, our group has provided ample evidence that minimally invasive interrogation of the rectal epithelium could risk-stratify patients for CRC [44, 45]. Furthermore, field carcinogenesis detected in the uninvolved rectal mucosa was reported to be indicative of nanoscale changes in higher-order chromatin and early tumorigenesis [12, 13].
The mechanism of SA1 loss in colon carcinogenesis is uncertain. Previous reports suggest that mutational inactivation in cohesin proteins as such may be infrequent, alternatively making epigenetic modulations of this tumor suppressor gene a cogent focus a logical candidate. Our data indicates that SA1 may be impacted upon by both histone acetylation as well as methylation, two common themes in cancer biology. Methylation is particularly attractive given its well established role in field carcinogenesis and several reports suggest race based modulation although it needs to be emphasized that our data only relates to cell culture [46, 47]. However, the precise modalities of gene regulation are involved in SA1 loss in early colonic neoplastic transformation remains to be elucidated and other transcriptional or post-translational processes may be involved.
To understand the functional biology of reduced SA1 levels, we found that SA1 knockdown resulted in increased cellular proliferation. The knockdown model was in a way utilized to recapitulate the observed lower basal levels of SA1 in AAs. The increase in cell proliferation in SA1 knockdown HT29 cells (increased WST and PCNA) was corroborated by activation of key signaling pathways including β-Catenin. Future studies will focus on further understanding the biological implications of SA1 loss.
One of the important facets of this work stems from the fact that there has been limited previous appreciation of the demographic considerations in biomarker discovery. On these lines we have previously demonstrated that specific microRNAs in the field carcinogenesis have gender-specific implications [48]. Thus, while the performance is modest, to our knowledge this is the first demonstration of racial differences in cancer biomarkers for screening. Furthermore, since there is dramatically less basal expression of SA1 in AAs, it may suggest a potential mechanism for the disproportionate toll of CRC in AAs.
With regards to potential long term clinical implications, these center on the paramount nature of risk stratification is paramount for CRC screening especially in disadvantaged populations. Finding less invasive techniques and tissue acquisition potentially at the point of care (rectal swab) to help identify relatively high-risk individuals may improve compliance of the minority patients with more invasive testing such as colonoscopy. This report may herald translation of recent knowledge regarding biological differences in the colon carcinogenesis between races to clinical applications such as risk analysis.
This study has many strengths including the novelty of our findings with SA1 in specific and cohesins in general. The observation that SA1 was lost at the earliest stages (field carcinogenesis) was striking and its profound nature strongly implicates not only high order chromatin regulators as drivers of colon carcinogenesis. The combination of protein and message in a prospectively collected cohort undergoing colonoscopy is a powerful resource and the diverse cohort enables insights into racial disparities. Moreover, the demonstration of the impact of SA1 on high order chromatin is compelling since we utilized both conventional (MNase) and novel (BaSIS) techniques.
Our study has several limitations that should be acknowledged. First, the number of patients is not large and thus we were unable to perform subgroup analysis on different adenoma types (serrated, advanced etc.). Second, the expression data was not coupled with mutational data although this is mitigated by the relative rarity of cohesin mutations in CRC (~2% [49]). Third, SA1 was taken as a candidate approach although as noted here and in a previously reported pilot study, and although the expression of other cohesins including SA2, SMC3 and CTCF were also measured, it was not an exhaustive evaluation in patient populations. Fourth, racial data was self-reported and recent studies have impugned reliability [50]. However, while imperfect this is currently the state of the art and should not bias our results. Finally, from a clinical perspective, SA1 performance as a biomarker was modest but the vast majority of the neoplastic lesions were small adenomas. Our previous work has indicated that field carcinogenesis is more pronounced with more significant neoplasia (e.g. advanced adenomas) with diminutive adenomas engendering limited alteration [34]. Furthermore, since the major novelty of the work is the demonstration of race-specific biomarkers, the actual performance of SA1 is secondary.
In conclusion, we demonstrate for the first time that cohesin SA1 is an important tumor suppressor gene in early colon carcinogenesis. SA1 modulates higher-order chromatin alterations that are the hallmark of field carcinogenesis. Furthermore, lower expression of cohesin SA1 in AAs versus Caucasians may provide a potential biological explanation for the increased incidence and mortality of CRC in AAs. Finally, this provides the proof-of-concept of the need to factor race into biomarker development. Future studies are needed to understand the ramifications of cohesin dysregulation for cancer screening, prevention and therapeutics.
Acknowledgments
The authors thank Ms. Beth Parker for excellent support in manuscript preparation.
Drs. Roy and Backman are co-founders and shareholders in Nanocytomics LLC which has licensed the PWS technology.
Funding: H. K. Roy & V. Backman were awarded R01 CA165309, R01 CA156186, 1 R01 CA200064, R01 CA183101. H. K. Roy and R.K. Wali were awarded R03 CA195143.
Footnotes
Competing Interests: All other authors do not have any competing interests.
Author Contributions: HKR developed the concept and the study design and provided overall supervision; RKW, CRW, AC, performed immunohistochemical analyses; YS-C, LA and VB performed the PWS analyses; NM performed the cell culture with the knock-down, MNase assay and analyzed other cell culture parameters; MDC, AKT and BL performed the RT-PCR in race specific samples; AHC, AC and HKR contributed to patient recruitment. AR performed statistical analysis. Initial drafts of the manuscript were written by HKR and RKW and reviewed by all co-authors.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. PubMed PMID: 26742998. [DOI] [PubMed] [Google Scholar]
- 2.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. PubMed PMID: 26457759; PubMed Central PMCID: PMCPMC4636487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. doi: 10.1038/nature13438. PubMed PMID: 25043054; PubMed Central PMCID: PMC4249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. PubMed PMID: 23539594; PubMed Central PMCID: PMC3749880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. PubMed PMID: 22810696; PubMed Central PMCID: PMC3401966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. PubMed PMID: 24132290; PubMed Central PMCID: PMC3927368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuguchi T, Fudenberg G, Mehta S, Belton JM, Taneja N, Folco HD, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature. 2014;516(7531):432–435. doi: 10.1038/nature13833. Epub 2014/10/14. doi: nature13833 [pii] 10.1038/nature13833. PubMed PMID: 25307058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D. The translational landscape of the mammalian cell cycle. Mol Cell. 2013;52(4):574–582. doi: 10.1016/j.molcel.2013.09.018. Epub 2013/10/15. doi: S1097-2765(13)00710-7 [pii] 10.1016/j.molcel.2013.09.018. PubMed PMID: 24120665; PubMed Central PMCID: PMC3959127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mannini L, Menga S, Musio A. The expanding universe of cohesin functions: a new genome stability caretaker involved in human disease and cancer. Hum Mutat. 2010;31(6):623–630. doi: 10.1002/humu.21252. Epub 2010/06/01. PubMed PMID: 20513141. [DOI] [PubMed] [Google Scholar]
- 10.Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47(7):818–821. doi: 10.1038/ng.3335. Epub 2015/06/09. doi: ng.3335 [pii] 10.1038/ng.3335. PubMed PMID: 26053496. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian H, Roy HK, Pradhan P, Goldberg MJ, Muldoon J, Brand RE, et al. Nanoscale cellular changes in field carcinogenesis detected by partial wave spectroscopy. Cancer Res. 2009;69(13):5357–5363. doi: 10.1158/0008-5472.CAN-08-3895. PubMed PMID: 19549915; PubMed Central PMCID: PMC2802178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damania D, Roy HK, Subramanian H, Weinberg DS, Rex DK, Goldberg MJ, et al. Nanocytology of rectal colonocytes to assess risk of colon cancer based on field cancerization. Cancer Res. 72(11):2720–2727. doi: 10.1158/0008-5472.CAN-11-3807. Epub 2012/04/12. doi: 0008-5472.CAN-11-3807 [pii] 10.1158/0008-5472.CAN-11-3807. PubMed PMID: 22491589; PubMed Central PMCID: PMC3557939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherkezyan L, Stypula-Cyrus Y, Subramanian H, White C, Dela Cruz M, Wali RK, et al. Nanoscale changes in chromatin organization represent the initial steps of tumorigenesis: a transmission electron microscopy study. BMC Cancer. 14:189. doi: 10.1186/1471-2407-14-189. Epub 2014/03/19. doi: 1471-2407-14-189 [pii] 10.1186/1471-2407-14-189. PubMed PMID: 24629088; PubMed Central PMCID: PMC3995586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pradhan P, Damania D, Joshi HM, Turzhitsky V, Subramanian H, Roy HK, et al. Quantification of nanoscale density fluctuations by electron microscopy: probing cellular alterations in early carcinogenesis. Physical biology. 2011;8(2):026012. doi: 10.1088/1478-3975/8/2/026012. PubMed PMID: 21441647; PubMed Central PMCID: PMC3332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stypula-Cyrus Y, Damania D, Kunte DP, Cruz MD, Subramanian H, Roy HK, et al. HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure. PLOS One. 2013;8(5):e64600. doi: 10.1371/journal.pone.0064600. Epub 2013/06/01. PONE-D-13-03055 [pii] PubMed PMID: 23724067; PubMed Central PMCID: PMC3665824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rex DK, Bond JH, Winawer S, Levin TR, Burt RW, Johnson DA, et al. Quality in the technical performance of colonoscopy and the continuous quality improvement process for colonoscopy: recommendations of the U.S. Multi-Society Task Force on Colorectal Cancer. The American journal of gastroenterology. 2002;97(6):1296–1308. doi: 10.1111/j.1572-0241.2002.05812.x. PubMed PMID: 12094842. [DOI] [PubMed] [Google Scholar]
- 17.DeSantis C, Naishadham D, Jemal A. Cancer statistics for African Americans, 2013. CA Cancer J Clin. 63(3):151–166. doi: 10.3322/caac.21173. Epub 2013/02/07. PubMed PMID: 23386565. [DOI] [PubMed] [Google Scholar]
- 18.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. PubMed PMID: 23335087. [DOI] [PubMed] [Google Scholar]
- 19.Aizer AA, Wilhite TJ, Chen MH, Graham PL, Choueiri TK, Hoffman KE, et al. Lack of reduction in racial disparities in cancer-specific mortality over a 20-year period. Cancer. 2014;120(10):1532–1539. doi: 10.1002/cncr.28617. Epub 2014/05/28. PubMed PMID: 24863392. [DOI] [PubMed] [Google Scholar]
- 20.Zisman AL, Nickolov A, Brand RE, Gorchow A, Roy HK. Associations between the age at diagnosis and location of colorectal cancer and the use of alcohol and tobacco: implications for screening. Arch Intern Med. 2006;166(6):629–634. doi: 10.1001/archinte.166.6.629. Epub 2006/03/29. doi: 166/6/629 [pii] 10.1001/archinte.166.6.629. PubMed PMID: 16567601. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal S, Bhupinderjit A, Bhutani MS, Boardman L, Nguyen C, Romero Y, et al. Colorectal cancer in African Americans. The American journal of gastroenterology. 2005;100(3):515–523. doi: 10.1111/j.1572-0241.2005.41829.x. discussion 4. PubMed PMID: 15743345. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman DA, Williams JL, Holub JL, Morris CD, Logan JR, Eisen GM, et al. Race, ethnicity, and sex affect risk for polyps >9 mm in average-risk individuals. Gastroenterology. 2014;147(2):351–358. doi: 10.1053/j.gastro.2014.04.037. quiz e14–5. Epub 2014/05/03. doi: S0016-5085(14)00593-9 [pii] 10.1053/j.gastro.2014.04.037. PubMed PMID: 24786894; PubMed Central PMCID: PMC4121117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. PubMed PMID: 24645800. [DOI] [PubMed] [Google Scholar]
- 24.Cruz MD, Wali RK, Bianchi LK, Radosevich AJ, Crawford SE, Jepeal L, et al. Colonic mucosal fatty acid synthase as an early biomarker for colorectal neoplasia: modulation by obesity and gender. Cancer Epidemiol Biomarkers Prev. 23(11):2413–2421. doi: 10.1158/1055-9965.EPI-14-0026. Epub 2014/08/27. doi: 1055-9965.EPI-14-0026 [pii] 10.1158/1055-9965.EPI-14-0026. PubMed PMID: 25155760; PubMed Central PMCID: PMC4470400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherkezyan L, Capoglu I, Subramanian H, Rogers JD, Damania D, Taflove A, et al. Interferometric spectroscopy of scattered light can quantify the statistics of subdiffractional refractive-index fluctuations. Phys Rev Lett. 2013;111(3):033903. doi: 10.1103/PhysRevLett.111.033903. Epub 2013/08/06. PubMed PMID: 23909326; PubMed Central PMCID: PMC4123763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wali RK, Kunte DP, Koetsier JL, Bissonnette M, Roy HK. Polyethylene glycol-mediated colorectal cancer chemoprevention: roles of epidermal growth factor receptor and Snail. Mol Cancer Ther. 2008;7(9):3103–3111. doi: 10.1158/1535-7163.MCT-08-0434. PubMed PMID: 18790788; PubMed Central PMCID: PMC2547487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. PubMed PMID: 9305837. [DOI] [PubMed] [Google Scholar]
- 28.Misteli T. Higher-order genome organization in human disease. Cold Spring Harbor perspectives in biology. 2010;2(8):a000794. doi: 10.1101/cshperspect.a000794. PubMed PMID: 20591991; PubMed Central PMCID: PMC2908770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. PubMed PMID: 19815776; PubMed Central PMCID: PMC2858594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2011;19(1):37–51. doi: 10.1007/s10577-010-9177-0. PubMed PMID: 21274616; PubMed Central PMCID: PMC3040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33(2):265–280. doi: 10.1128/MCB.01008-12. PubMed PMID: 23129809; PubMed Central PMCID: PMC3554127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105(24):8309–8314. doi: 10.1073/pnas.0801273105. Epub 2008/06/14. doi: 0801273105 [pii] 10.1073/pnas.0801273105. PubMed PMID: 18550811; PubMed Central PMCID: PMC2448833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Repo H, Loyttyniemi E, Nykanen M, Lintunen M, Karra H, Pitkanen R, et al. The Expression of Cohesin Subunit SA2 Predicts Breast Cancer Survival. Applied immunohistochemistry & molecular morphology : AIMM / official publication of the Society for Applied Immunohistochemistry. 2015 doi: 10.1097/PAI.0000000000000240. PubMed PMID: 26447899. [DOI] [PubMed] [Google Scholar]
- 34.Damania D, Roy HK, Subramanian H, Weinberg DS, Rex DK, Goldberg MJ, et al. Nanocytology of rectal colonocytes to assess risk of colon cancer based on field cancerization. Cancer Res. 2012;72(11):2720–2727. doi: 10.1158/0008-5472.CAN-11-3807. PubMed PMID: 22491589; PubMed Central PMCID: PMC3557939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherkezyan L, Stypula-Cyrus Y, Subramanian H, White C, Dela Cruz M, Wali R, et al. Nanoscale changes in chromatin organization represent the initial steps of tumorigenesis: a transmission electron microscopy study. BMC Cancer. 2014;14(189) doi: 10.1186/1471-2407-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alberts DS, Einspahr JG, Krouse RS, Prasad A, Ranger-Moore J, Hamilton P, et al. Karyometry of the colonic mucosa. Cancer Epidemiol Biomarkers Prev. 2007;16(12):2704–2716. doi: 10.1158/1055-9965.EPI-07-0595. PubMed PMID: 18086777. [DOI] [PubMed] [Google Scholar]
- 37.Roy HK, Damania DP, DelaCruz M, Kunte DP, Subramanian H, Crawford SE, et al. Nano-architectural alterations in mucus layer fecal colonocytes in field carcinogenesis: potential for screening. Cancer Prev Res (Phila) 6(10):1111–1119. doi: 10.1158/1940-6207.CAPR-13-0138. Epub 2013/08/29. doi: 1940-6207.CAPR-13-0138 [pii] 10.1158/1940-6207.CAPR-13-0138. PubMed PMID: 23983085; PubMed Central PMCID: PMC3873747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Yan Y, Deb S, Rangasamy D, Germann M, Malaterre J, et al. Cohesin Rad21 Mediates Loss of Heterozygosity and Is Upregulated via Wnt Promoting Transcriptional Dysregulation in Gastrointestinal Tumors. Cell Rep. 2014;9(5):1781–1797. doi: 10.1016/j.celrep.2014.10.059. Epub 2014/12/04. doi: S2211-1247(14)00928-0 [pii] 10.1016/j.celrep.2014.10.059. PubMed PMID: 25464844. [DOI] [PubMed] [Google Scholar]
- 39.Remeseiro S, Cuadrado A, Carretero M, Martinez P, Drosopoulos WC, Canamero M, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31(9):2076–2089. doi: 10.1038/emboj.2012.11. Epub 2012/03/15. doi: emboj201211 [pii] 10.1038/emboj.2012.11. PubMed PMID: 22415365; PubMed Central PMCID: PMC3343459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nature genetics. 2013;45(12):1464–1469. doi: 10.1038/ng.2799. PubMed PMID: 24121791; PubMed Central PMCID: PMC3840052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghiselli G, Coffee N, Munnery CE, Koratkar R, Siracusa LD. The cohesin SMC3 is a target the for beta-catenin/TCF4 transactivation pathway. J Biol Chem. 2003;278(22):20259–20267. doi: 10.1074/jbc.M209511200. Epub 2003/03/26. M209511200 [pii] PubMed PMID: 12651860. [DOI] [PubMed] [Google Scholar]
- 42.Remeseiro S, Cuadrado A, Gomez-Lopez G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31(9):2090–2102. doi: 10.1038/emboj.2012.60. Epub 2012/03/15. doi: emboj201260 [pii] 10.1038/emboj.2012.60. PubMed PMID: 22415368; PubMed Central PMCID: PMC3343463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capoglu IR, Rogers JD, Taflove A, Backman V. The Microscope in a Computer: Image Synthesis from Three-Dimensional Full-Vector Solutions of Maxwell's Equations at the Nanometer Scale. In: Wolf E, editor. Progress in Optics, Vol 57. Progress in Optics. 572012. pp. 1–91. [Google Scholar]
- 44.Damania D, Bleher R, Wu J, Rogers J, Subramanian H, Roy HK, et al. Visualizing Native Cell Nano-architecture during Early Carcinogenesis using Scanning Transmission Electron Microscopy. Microscopy & Microanalysis Conference. 2012 [Google Scholar]
- 45.Roy HK, Hensing T, Backman V. Nanocytology for field carcinogenesis detection: novel paradigm for lung cancer risk stratification. Future oncology. 2011;7(1):1–3. doi: 10.2217/fon.10.176. PubMed PMID: 21174531; PubMed Central PMCID: PMC4123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso S, Dai Y, Yamashita K, Horiuchi S, Dai T, Matsunaga A, et al. Methylation of MGMT and ADAMTS14 in normal colon MUCOSA: biomarkers of a field defect for cancerization preferentially targeting elder African-Americans. Oncotarget. 2015;6(5):3420–3431. doi: 10.18632/oncotarget.2852. PubMed PMID: 25638164; PubMed Central PMCID: PMCPMC4413663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashktorab H, Daremipouran M, Goel A, Varma S, Leavitt R, Sun X, et al. DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics. 2014;9(4):503–512. doi: 10.4161/epi.27644. PubMed PMID: 24441198; PubMed Central PMCID: PMCPMC4121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wali RK, Hensing TA, Ray DW, Dela Cruz M, Tiwari AK, Radosevich A, et al. Buccal microRNA dysregulation in lung field carcinogenesis: gender-specific implications. International journal of oncology. 2014;45(3):1209–1215. doi: 10.3892/ijo.2014.2495. PubMed PMID: 24919547; PubMed Central PMCID: PMC4144027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.075. PubMed PMID: 27149842; PubMed Central PMCID: PMCPMC4850357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mersha TB, Abebe T. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum Genomics. 2015;9:1. doi: 10.1186/s40246-014-0023-x. PubMed PMID: 25563503; PubMed Central PMCID: PMCPMC4307746. [DOI] [PMC free article] [PubMed] [Google Scholar]