Summary
Src kinase activity is controlled by various mechanisms, involving a coordinated movement of kinase and regulatory domains. Notwithstanding the extensive knowledge related to the backbone dynamics, little is known about the more subtle side-chain dynamics within the regulatory domains and their role in the activation process. Here, we show through experimental methyl dynamic results and predicted changes in side-chain conformational couplings that Fyn’s SH2 structure contains a dynamic network capable of propagating binding information. We reveal that binding the phosphorylated tail of Fyn perturbs a residue cluster near the linker connecting Fyn’s SH2 and SH3 domains, which is know to be relevant regulating Fyn’s activity. Biochemical perturbation experiments validate that those residues are essential for Fyn’s inhibition, leading to a gain of function upon mutation. These findings reveal how side chain dynamics may facilitate the allosteric regulation of the different members of the Src kinase family.
Keywords: Fyn, SH2, methyl dynamics, prediction, long-range communication, regulation, Src-like kinases
Graphical Abstract
eTOC Blurb
Huculeci et al. combine NMR sidechain relaxation experiments with predictions to reveal how binding to the phosphorylated tail of the Src Family Kinase Fyn transduces a signal through its SH2 domain, regulating activity. The results support a lock hypothesis that explains the rigidification of the SH2-SH3 linker.
Introduction
As protein tyrosine kinases (PTK) (Manning et al., 2002) play often an essential role in different cellular functions, significant attention has been given to the elucidation of their structure and dynamics. Among them, the Src family kinases (SFK) are maybe the most substantially analyzed PTK as they have been implicated in a wide variety of diseases (Boggon and Eck, 2004; Parsons and Parsons, 2004). The up-regulation of the SFK Fyn, for instance, has been associated with prostate (Posadas et al., 2009), ovarian (Huang et al., 2008), breast (Charpin et al., 2009; Kostic et al., 2009) and pancreatic (Chen et al., 2011) cancers, while other studies reported its involvement in chronic myeloid leukemia (Singh et al., 2012) and in Alzheimer’s disease (Lee et al., 2004; Nygaard et al., 2014; Yang et al., 2011). Like all other family members, Fyn is composed of an N-terminal membrane localization motif followed by a so called unique domain, which is linked to the SH3 and SH2 regulatory domains, the actual kinase domain (SH1) and a C-terminal tail (Boggon and Eck, 2004). The catalytic activity of Fyn is down-regulated through the coordinated binding of the SH3-SH2 domain pair with specific intramolecular ligands (Engen et al., 2008). The coordinated action of both domains displaces an α-helix in the kinase domain, blocking the catalytic site and rendering Fyn inactive. Releasing the interaction between the SH2 domain and the pY-tail disrupts the contacts between the SH3 domain and the SH2-kinase linker, opening the active site (Engen et al., 2008). As SH3 and SH2 domains may have diverse roles in the regulation of SFK activity (Arold et al., 2001; Moarefi et al., 1997) and both are required to deactivate the members of that family (Cowan-Jacob et al., 2005; Nagar et al., 2006), it is crucial to understand which dynamic mechanisms are in place to facilitate the switching.
Different studies have aimed to address this question (Arold et al., 2001; Engen et al., 1999; Engen et al., 2008; Hochrein et al., 2006; Tessari et al., 1997; Ulmer et al., 2002; Young et al., 2001). Overall, those studies suggest that at the level of large (backbone) changes, binding a peptide to one domain does not influence the other domain and vice versa. Nevertheless, it was shown that when the SH2 domain is bound to the pY-tail, the motions of the SH2 and SH3 domains are dynamically coupled, forming a “snap-lock” configuration (Young et al., 2001). Specifically, when the SH2 domain is docked to the pY-tail of the kinase, the SH2-SH3 linker is rigidified, coupling the movement of the two domains. This dynamical change facilitates the binding of the SH3 domain to the SH2-kinase linker, thus blocking the kinase domain in its inactive conformation (Young et al., 2001).
As the inactive state of the enzyme requires the binding of Fyn’s pY-tail to the SH2 module, one can postulate that the SH2 domain is actively propagating the binding information to the SH3-SH2 linker in order to lock the SH3 domain in place. To verify such a hypothesis one needs to identify which mechanisms are at work in the SH2 domain that can inform the linker to “open” (activate Fyn) or “close” (inactivate Fyn) the lock. Yet, as binding a phosphorylated peptide to the Fyn SH2 domain has been shown to initiate no significant backbone changes in the domain structure (Ulmer et al., 2002), other more subtle mechanisms may be at work. As was argued before (Tsai et al., 2008), the lack of change in the overall shape of the domain upon binding does not necessarily mean that allostery is not at work. So far, none of the studies involving the SH2 domain from SFK investigated changes in side-chain dynamics, which can provide substantial information about intra-molecular communication even when there is no change in backbone conformation (Daily et al., 2008; Whitley and Lee, 2009).
Here we propose a mechanism on how Fyn’s SH2 domain could propagate information resulting in the rigidification of the SH3-SH2 linker, by employing a hybrid approach that combines experimental results with computational predictions. NMR deuterium-based methyl side-chain relaxation experiments (Houben et al., 2007; Kay et al., 1996; Kay et al., 1998; Millet et al., 2002; Skrynnikov et al., 2002) on the Fyn SH2 form in isolation and when it is bound to the pY-tail of Fyn, allow us to identify the dynamic properties of the different methyl-group containing residues within the protein. Concretely, they show which methyl-group bearing residues experience a change in their dynamics upon binding and could hence be involved in transducing the binding information to distal locations in the structure (Fuentes et al., 2004; Whitley and Lee, 2009). Yet, as less than one third of the Fyn SH2 residues have methyl groups, these results provide only a partial image of the transduction mechanism. In combination with our Monte Carlo sampling – Information Theoretical method (MCIT) we extend the side-chain relaxation results to include also the non-methyl-group bearing residues, while minimizing the violations with the experimental data. The MCIT takes as input NMR solution structures and provides predictions that have been shown to accurately reflect the results of side-chain relaxation experiments (Cilia et al., 2012) as well as trans-hydrogen bond scalar couplings (Zafra Ruano et al., 2016). For this analysis, we used two solution structures of Fyn SH2 in its free form and bound to the pY-tail of Fyn (see Supplemental Experimental Procedures 1, Figure S1, Table S1 and Table S2). The integration with the experimental results produces a network of dynamically affected residues, linking the Fyn SH2 binding site to residues located throughout the structure with significant changes directed to the SH3-SH2 linker. This network reveals a detailed and experimentally supported picture of Fyn SH2’s intra-domain communication, allowing us to postulate the hypothesis that the locking mechanism is mediated through dynamic changes in a small cluster of residues close to the SH2 N-terminal region in direct contact with the SH3-SH2 linker. We validate this hypothesis using a coupled kinase assay that studies how mutants in identified residues close to the SH3-SH2 linker affect kinase inhibition through Csk, showing that the residues are essential for the down-regulation of Fyn and therefore potentially also for other SFK.
Results
Methyl dynamics changes spread through the Fyn SH2 structure to distant locations
To reveal the potential of the side-chain dynamics in regulating the activity of Fyn, we determined the side-chain dynamics in the free and bound states at the picosecond to nanosecond time scale, focussing particularly on the motions of the methyl groups (Experimental procedures and Supplemental Experimental Procedures 3). Five deuterium spin relaxation rates were measured for both free (Table S4) and pY-tail bound states (Table S5) (Houben et al., 2007; Millet et al., 2002; Muhandiram et al., 1995), while the model-free formalism was used to facilitate the interpretation of the dynamical data (Lipari and Szabo, 1982a, b; Skrynnikov et al., 2002). The latter approach determines the dynamics via an order parameter (S2axis) and its corresponding internal correlation time (τe). The values of S2axis (see Table S6) lie in the range [0,1], where 0 refers to an isotropic motion with no restrictions and 1 indicates a fixed orientation. As the S2axis values of each methyl type are influenced by the distance from the backbone (Mittermaier et al., 1999), the values need to be normalized in order to determine their significance. To perform this normalization the average S2axis values (<S2axis>) per methyl type were determined using the S2axis values from both free and bound states (see Supplemental Experimental Procedures 3, Figure S2, Table S6) and then subtracted from each S2axis values in either state. Positive normalized S2axis values indicate a decrease in flexibility in relation to the mean or a more restricted motion at the side-chain level, while negative values account for the opposite.
Figure 1 shows the normalized S2axis values annotated on the conformer with the lowest energy from the ensembles of free (Figure 1a) and bound (Figure 1b) Fyn SH2 (see Figure S1 for the ensembles). As a general trend for both states, residues located in the α-helix regions show either more restricted motions (e.g. see A159-β, L163-δ1 from αA-helix and T223-γ2, L227-δ1/δ2 from αB-helix in Figure 1a–b, right panels) or motions similar to the mean for their methyl type. Equivalent behaviour is observed for the V200-γ2 methyl group located on the βD-sheet and oriented towards the core of the structure.
Figure 1.
Normalized methyl axis order parameters for the free (a) and pY-tail bound Fyn SH2 domain (b) mapped onto the corresponding lowest energy structures from the NMR ensembles (see Figure S1). Methyls are shown as solid spheres and coloured in function of their flexibility with respect to the average <S2axis> per methyl type: red represents enhanced flexibility and blue highlights increased rigidity. The methyl groups are colour-coded according to the legend below the panels, while the methyls lacking experimental data are represented as smaller spheres, in white. The pY-tail peptide is shown in orange.
In contrast, some methyl groups located in the specificity binding site region and in its vicinity (L208-δ1/δ2, T216-γ2 and T217-γ2) are characterized by enhanced flexibility, indicated by negative deviations from the average, in both free and bound forms. These data are in line with our earlier observation that the specificity-binding site is not engaged in strong connections with the pY-tail peptide (Bradshaw et al., 1998).
To identify the residues involved in transducing the binding information, the differences between both the S2axis of the pY-tail complex and the free form were calculated (Figure 2a) and the ΔS2axis were mapped onto the structure (Figure 2b). In Figure 2a a change is considered to be significant when the absolute value of ΔS2axis (|ΔS2axis |) is higher than the uncertainty (error bars) determined for those differences. |ΔS2axis| values for pY-tail binding to Fyn SH2 range from 0.03 to 0.49 and comparable values were observed upon peptide binding to CaM (Lee et al., 2000), while much lower values |ΔS2axis| in the range of 0.02 to 0.17 were found after phosphotyrosine peptide binding to the PLCC SH2 domain (Kay et al., 1996). The internal correlation time τe for the Fyn SH2 domain, free and upon pY-tail binding, are discussed in Supplemental Experimental Procedures 4. On average, a rigidification of the methyl bearing side-chains is observed with an average increased ΔS2axis value of 0.03 per affected residue. For comparison, an 0.07 average increase per residue was observed for peptide binding to CaM (Lee et al., 2000), corresponding to a total reduction of conformational entropy by 35 kcal/mol.
Figure 2.
The dynamical effects induced by interaction of the pY-tail peptide on the Fyn SH2 domain. (a) Differences in methyl axis order parameter, S2axis, upon pY-tail binding on the Fyn SH2 domain. The values are represented as a function of methyl position, with significant changes shown as filled bars. The corresponding secondary structure elements are depicted at the top of the figure. The two methyl groups marked with star (T172-γ2, T181-γ2) are overlapping in the free form, where they carry the same S2axis values. (b) The differences in the S2axis between the bound and the free states are mapped onto the lowest energy SH2:pY-tail complex structure. Methyls are shown as solid spheres and coloured in function of their change in flexibility upon binding: red represents enhanced flexibility and blue highlights decrease in flexibility. Only significant changes are coloured in red or blue, while the other methyl groups are coloured in grey. The methyls lacking experimental data are represented as white smaller spheres. The pY-tail peptide is shown in orange.
As can be seen in Figure 2a, Fyn SH2 methyl-containing residues experience mostly an increase in rigidity (blue) with only few isolated side-chains characterized by enhanced flexibility (red) upon binding. The loss of flexibility in several areas can be attributed to direct interaction with the peptide. The pY residue is packed for instance against the γ2 methyl groups of T180 and T181, which exhibit positive ΔS2axis values. Increase in S2axis values upon binding are also observed for T216-γ2, T217-γ2 and A236-β, which are located in the specificity binding area.
Another interesting result is represented by the enhanced flexibility of L238-δ2, located in the specificity pocket area. When Fyn SH2 binds to a natural target or a high affinity peptide, L238 interacts with the binder and stabilizes the complex (Bradshaw et al., 1998; Kaneko et al., 2010). Even with weak interactions between the pY-tail and the specificity pocket observed here, a general increased rigidity of the side-chains in this area is observed (Figure 2a and 2b). However, the side-chain of L238 is pointing towards the interior of the structure, where it is buried in a hydrophobic core and the δ2 methyl of L238 experiences an increase in flexibility. Interestingly, in the same hydrophobic core L187-δ1, located on the βC-sheet, also displays enhanced dynamics in the presence of the pY-tail. An increase of dynamics displayed by side-chains distal from the binding-site has been observed for other proteins upon peptide binding and has been proposed to provide an entropic compensation to the free energy of binding (Igumenova et al., 2006). Only one methyl-bearing residue in the same hydrophobic core is largely rigidified (L242-δ2), while others only experience a slight rigidification (I189-γ/δ, I205-γ, L224-γ2) or do not display a signification change in dynamics (I175-γ, I215-γ/δ, L224-δ1). V228-γ1, which is located at the edge of this hydrophobic core far away from the binding pocket, is partially solvent exposed and experiences an enhanced flexibility.
In summary, the data show that changes in dynamics are not only experienced by methyl groups located in close proximity to the peptide, but also by methyls positioned at large distances from the peptide binding pockets (more than 18 Å), providing evidence for long-range communication through the SH2 domain structure.
Hybridization of experimental and computational results paints the full picture
As only 30% of the residues bear methyls in Fyn SH2, insufficient information is available on how the system transduces the binding information throughout the domain structure. To overcome this issue the experimental observations are here extended with computational predictions provided through our MCIT method (Cilia et al., 2012; Lenaerts et al., 2008; Zafra Ruano et al., 2016), a method we developed to predict from structure the residues experiencing dynamic changes induced by binding. For details on the MCIT method we refer to the Experimental Procedures (which explains the scheme in Figure 3a) as well as the previously published papers. MCIT uses here two NMR ensembles, one for the free form and another for the form bound to the pY-tail (see Supplementary Experimental Procedures 1 and Figure S1 for information on these structures). Here we assume that if one can deploy the MCIT predictor so that it predicts most of the experimentally identified methyl-bearing residues correctly, then the associated non-methyl-bearing residues identified at the same time, are also highly relevant and provide hence a more complete view of the residues involved in the transduction process. In other words, they are predicted-by-association. Yet, one needs to remember that the experimental and predictive approach do not identify the same kind of data: whereas the side-chain relaxation experiments provide insight into the changes in the methyl dynamics located on individual side-chains, MCIT predicts changes in the conformational coupling between side-chains. Notwithstanding this difference, prior work has shown that the MCIT accurately predicts the residues that are identified by the former (Cilia et al., 2012).
Figure 3.
Flow charts describing the MCIT approach and the iterative procedure for the integration of experimental and predictive data (see Experimental procedures for a detailed explanation). a) visualizes the MCIT approach producing the matrix of changes in dynamic couplings on which CAST operates. b) CAST uses a threshold CT to include more and more residues in the results, which generates a ROC curve that provides insight in the quality of the predictions. c) shows the iterative procedure to screen different AL values in order to find the best one (AL*). Branching conditions are represented in diamond shapes, the top one represents the termination condition and the two in the lower righ express the two criteria used to select the best AL* on the experimental data. The procedure takes as inputs the experimental data (bar plot at |ΔS2axis| at top) and computational results (represented by the matrix of absolute changes in mutual information reported in part a). d) a plot showing for different AL values (which correspond to different |ΔS2axis|) what AUC can be achieved.
In order to retrieve the network that follows the experimental results in Figure 2 as closely as possible, a parameter screening is required to determine for which clustering threshold CT (see Experimental Procedures) most of the methyl-bearing residues are part of the clusters identified by the cluster analysis technique CAST (Ben-Dor et al., 1999) while at the same time excluding the insignificant dynamic changes (|ΔS2axis|). As argued earlier, |ΔS2axis| values are considered insignificant when they are smaller than the derived error. Yet too small |ΔS2axis| values may in themselves be irrelevant, requiring us to identify another suitable threshold that will provide a classification of the experimentally identified residues into those that we accept and those exclude as relevant for the internal dynamics (see also the discussion on the ΔS2axis values in the previous section). This second threshold will separate the experimental results in positive instances when they are above this cut-off and negative instances when they are below. We refer here to this threshold as the acceptance level (AL) This categorization allows one to quantify whether the methyl-bearing residues are predicted correctly (true positives (TP) and true negatives (TN)) or incorrectly for a given value of CT, where false-positives (FP) are those residues predicted as being relevant without sufficient experimental support (i.e. high enough |ΔS2axis|) and false-negatives (FN) are the residues that were determined to be relevant experimentally but not predicted as such. Thus decreasing CT systematically will allow us to draw a ROC curve (see Figure 3b), providing an objective approach to determine which CT to use for a given separation of experimental results in positive and negative instances. In practice this CT is given by the point the closest to the upper-left corner of the ROC curve (i.e. closest to ideal performance maximum true positive rate or TPR (=1) and minimum false positive rate or FPR (=0)) (see Figure 3b). The Area Under the ROC Curve (AUC), which measures the quality of the prediction with respect to the experimental data, is a general measure to evaluate the quality of the prediction (see Figure 3d). In summary, the parameter screening will calculate for a range of AL values the predictive quality (see Figure 3c), allowing us to select the one that provides the most meaningful results, i.e. AL*. The procedure is summarized in Figure 3c and explained in detail in Experimental Procedures. By iterating over the AL values, we determined that an AL* of 0.08 (orange lines in Figure 4a and orange circle in Figure S5) provides a good trade-off between accuracy of the predictions measured by the AUC (condition 1 in Figure 3c) and balance between the number positive and negative instances (condition 2 in Figure 3c), which is particularly important when extending the prediction from the methyl-bearing residues to all the residues of the Fyn SH2 domain. Given the so determined AL*, three residues (M195, T217 and L224) are considered as negative instances (too small |ΔS2axis|) out of the eighteen methyl-bearing residues, for which experimental relaxation data were considered significant (see Figure 2). It is worth noting that I189 and L208 are considered relevant residues even if one of the two methyl groups (I189-γ2 and L208-δ1) is classified not relevant by the acceptance level.
Figure 4.
(a) All the methyl groups with significant ΔS2axis values and the selected acceptance level (orange lines). The lines represent the boundaries between residues most affected by the binding event (relevant residues) and those that are barely affected (irrelevant residues). (b) ROC curve of the Fyn SH2 predictions at the selected acceptance level (blue line) and a random prediction (grey line). The point indicated by the orange circle corresponds to the best predictive threshold CT* (the point closest to the top-left corner).
Once AL* is determined (Figure 4a) the procedure also determines from the corresponding ROC curve the best threshold CT* (see Figure 4b), which jointly maximises the correct classifications of relevant residues (TPR = 0.67) while minimizing incorrect predictions of non-relevant residues (FPR = 0.0). We hence avoid false positives with respect to the experimental data by selecting CT*=0.044 (see orange circle in Figure 4b). CT* determines now which residues the CAST algorithm will incorporate in the cluster identified from the data in the matrix of Figure S4. The resulting cluster is guaranteed to cover most of the relevant changes visualized in Figure 2a, while adding non-methyl bearing residues that are detected at the same time as the relevant methyl-bearing residues.
A cluster of residues near SH3-SH2 linker may play a role in regulating Fyn activity
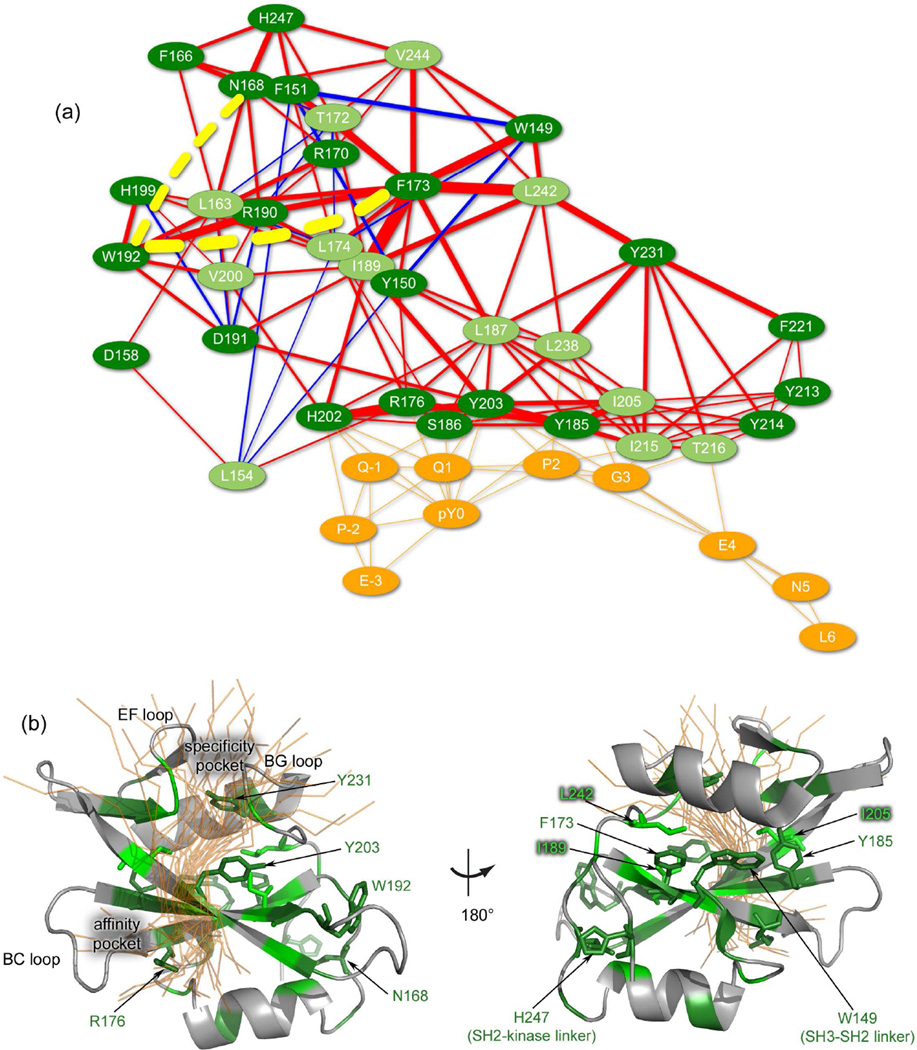
Using this CT*=0.044, the network of most affected residues within Fyn SH2 can be determined, as is shown in Figure 5a (see also Figure S6). The network consists of 35 residues, where 13 of them have methyl groups with a ΔS2axis value above AL*, while the links represent changes in coupling, ΔMI, between residues within 5Å distance (see Experimental procedures). Long distance connections exist, as for instance the two dashed lines in the figure show, but are not visualized for clarity. Mapping the predicted residues onto the structure of the Fyn SH2:pY-tail complex (see Figure 5b) shows that the network extends from the two binding regions over the central β-sheet towards isolated positions at both termini, i.e. the SH3-SH2 linker and the SH2-kinase linker.
Figure 5.
(a) Dynamical changes network in Fyn SH2 domain induced by pY-tail binding. Only the residues predicted as most affected by the binding event are represented (informative group). Light and dark nuances of green differentiate between the methyl bearing and non-methyl bearing residues, respectively. Red edges represent an increase in coupling between two residues from the informative group upon peptide binding, whereas blue ones show a reduction in coupling. The magnitude of the coupling is indicated by the thickness of the edges. Note that only the short-range couplings are represented, while the yellow-dotted lines illustrate examples of weighted long-range dynamical effects: W192 - F173 and W192 -N168. The peptide residues are depicted in orange, while the orange edges represent the contact network (not weighted) between the pY-tail peptide and the SH2 domain. (b) Fyn SH2:pY-tail complex and the residues predicted as the most dynamically affected by the binding. The informative group is coloured in green: light nuances depict the methyl bearing residues (predicted as being relevant and observed also experimentally) and the dark nuances show the non-methyl bearing residues (predicted as being relevant). The rest of the residues are shown in grey. Only the subgroup of residues predicted as most affected by the binding event is shown in sticks representation and labelled. The residues are mapped on the lowest energy Fyn SH2:pY-tail complex structure. The pY-tail peptide is illustrated in orange and all the peptide conformations from the ensemble are shown in transparent ribbon representation. For clarity, the loops involved in the two binding sites and the termini regions are indicated.
Figure 6a visualizes twelve residues identified in our data using the CAST algorithm that experience the most significant changes in the sidechain conformation. This set includes residues from the binding site (R176, Y185, Y203 and I205), a couple close to the SH2-kinase linker (N168 and H247) and a cluster of residues in proximity of the SH3-SH2 linker (W149, F173, I189 and L242). Given the hypothesis discussed in the introduction our main interest in this article is in the four latter residues. Figure 6b visualizes these residues within the context of the structure, combining both the sidechain conformations of W149, F173 and I189 in the free and bound NMR ensembles with the relaxation changes induced by binding at the level of the sidechains in some neighbouring residues (red and blue balls). In this figure we see some interesting effects in F173 and W149, which are separated by approximately 6 Å between their Cα atoms.
Figure 6.
The top 12 highly coupled residues upon pY-tail binding at Fyn SH2. (a) The dynamical changes network from Figure 6a, where only the top 12 highly affected residues by the pY-tail binding event are highlighted using the same colouring scheme as in 6a. (b) The dynamic effects propagating through the structure towards the SH3-SH2 linker. All the conformations for the residues composing this cluster are shown in narrow lines representation: blue for the Fyn SH2:pY-tail complex and red for the free Fyn SH2 domain. The bidirectional arrows indicate the two populations observed for the I189 and F173 in the complex state. The spheres represent methyl groups dynamically affected by the pY-tail binding event: red indicates increased flexibility, while intense blue increased rigidity upon binding. For clarity, the stick representations depict the conformations from the lowest energy structure from the corresponding ensembles. The Fyn SH2 secondary structure elements are depicted in transparent cartoon representation.
In the Fyn SH2:pY-tail complex, which we call the locked state (corresponding to the inactive kinase), the two aromatic rings of F173 and W149 have a parallel orientation, with the aromatic ring of W149 pointing towards the linker. In the free form, the aromatic rings are organized in a perpendicular manner, which will be referred to as the “open” state (corresponding to the active kinase). We hypothesize that the switching between both states depends on the conformation of the residues within this cluster and more specifically the W149 side-chain. This hypothesis gains initial support from the observation that the aromatic rings of F173 and W149 are part of a π-stacking interaction (Burley and Petsko, 1985), which are non-covalent attractive interactions that lead to structure stabilization, especially when the aromatic rings are within a distance of 4.5 to 7 Å. Disturbing the π-stacking interaction results in the opening of the lock, hence rendering the linker more flexible, allowing the protein to become active.
A second support for the locking hypothesis is provided by the sidechain relaxation data discussed earlier. As can be observed, F173 has two conformation populations in the peptide bound state (blue lines), while in the free form (red lines) the same residue is represented by only one population. These 2 populations are in agreement with the cross-peak signal originating in the CζHζ atoms, which is visible in the free state and it disappears in the bound conformation, likely due to conformational exchange. In addition, this structural information is confirmed by the dynamical behaviour of the nearby methyl groups, I189-δ1 and L242-δ2. Both methyl groups experience a decreased flexibility on the ps-ns time scale (intense blue spheres) upon the binding event as a result of the steric effects induced by the conformational variability of F173. The reorientation of the aromatic side-chain of F173 restricts the motion of the two methyl side-chains located in proximity. The dynamical behaviour of F173 may represent an important element that facilitates the transmission of the side-chain perturbation to in the direction of the SH3-SH2 linker. In addition, the detailed in Figure 6b view also shows the higher variability of the W149 aromatic side-chain in the free form (red lines), when compared with the peptide bound state (blue lines). The different ring orientation between the two states provides additional freedom to the close side-chain V228-γ1 (red sphere). The experimental results confirmed this enhanced flexibility of the methyl side-chain upon peptide binding. As in the case of F173, W149 side-chain dynamics is predicted to be altered upon peptide binding, which points to an entropic mechanism of transferring the information. To a lesser extent, side-chain perturbations were predicted also for Y203, which is characterized by a slightly different orientation and increased variability in the bound state, in respect to the free form. This behaviour reduces the spatial restrictions for L238-δ2 methyl group, which shows an enhanced flexibility (red sphere).
An activity assay reveals that the residue cluster near the SH3-SH2 linker is essential for the down-regulation of Fyn
All previous result led us to hypothesize that the residues near the SH3-SH2 linker may be involved in regulating the activity of Fyn Kinase. To validate this hypothesis, we need to determine whether mutating those residues influences Fyn’s activity. The idea is that, if these residues are crucial for stabilizing the linker and ensuring the inhibition of Fyn then mutating them should activate the kinase or make it more difficult to maintain the inhibited form. To answer this hypothesis, we produced mutants of the residues closest to the linker, i.e. W149A, I189A, F173A, and L242A, and determined the functional effect of these mutations using in vitro biochemical assays (see Experimental Procedures). These assays quantify the amount of C-terminal Src kinase (Csk), which is known to phosphorylate the C-terminal tail of Src-family kinases (SFK), necessary to deactivate Fyn kinase. The inhibition of SFK activity is dependent on the SH3-SH2 domains and the conserved tyrosine in the tail of SFK (Figure S8). Csk titrations are performed to determine the concentration at which 50% of the initial Fyn kinase activity is inhibited (IC50) (see Figure S9 and Table S8). This validation shows that all but one of the Fyn mutants (I189A) are resistant to Csk inhibition, resulting in a 30- to 50- fold increase in IC50 (Figure 7).
Figure 7.
Mutations in SH2 domain make Fyn resistant to inhibition by Csk. The enzymatic kinase activity of Fyn SH3-SH2-Kinase domain constructs was determined in a continuous spectrophotometric assays and the concentration of recombinant Csk (IC50) that is required to inhibit 50% of Fyn kinase activity is reported. Addition of recombinant Csk leads to Fyn tail phosphorylation and inhibition of Fyn kinase activity. Error bars represent the standard deviation and *** indicate a p-value < 0.001.
This validation study shows that three of the identified residues within the SH2 regulatory domain of Fyn kinase and close to the SH3-SH2 linker (W149, F173, and L242) are involved in regulating Fyn’s activity and potentially have a role transmitting binding information through the SH2 regulatory domain of SFK Fyn. This is a first step towards revealing the subtle intra-molecular dynamical changes that are fundamental to Fyn regulation.
Discussion
As mentioned in the introduction, different articles (Arold et al., 2001; Boggon and Eck, 2004; Cowan-Jacob et al., 2005; Superti-Furga et al., 1993; Young et al., 2001) have argued that Src kinases are maintained in their inactive form through a cooperative mechanism involving both SH3 and SH2 domains (Young et al., 2001). An analysis using molecular dynamics highlighted the role of the linker in maintaining this inactive conformation (Young et al., 2001), leading to the suggestion that a snap-lock mechanism exists, which is triggered by the binding of the pY-tail to the SH2 domain. As no dynamic effects have been observed at the level of the backbone that could explain the rigidification of the linker (Arold et al., 2001; Tessari et al., 1997; Ulmer et al., 2002), the question of which mechanisms could potentially induce the opening and closing of this lock remained unclear. The results provided here suggest that the sidechain dynamics of specific residues (see Figure 6) located close the SH3-SH2 linker (W149, F173, I189 and L242) may play a role in this process, providing an entropy-based locking mechanism.
We disrupted those four residues by mutation to alanine and tested the ability of Csk to inhibit these mutant proteins. We found that all but one tested mutation in the core of the channel showed a significantly increased resistance towards inhibition by Csk. Only mutation I189A showed similar sensitivity to Csk as Fyn WT. This could be because I189A is the most conservative mutation tested here. Additionally, I189 experiences a smaller ΔS2Axis value compared to L242 (see Figure 2). The effect of mutation of regulation by Csk, indicates that these residues are in fact necessary to communicate the inhibitory effect of tail phosphorylation to the kinase domain. We therefore predict that such deregulated mutants in the SH2 domain could perturb cellular SFK signalling pathways.
Next to the validation experiments we performed here, the relevance of the residues in the Fyn SH2 network is further supported by several known mutations that have been shown to strongly affect SFK kinase functioning. For instance, a mutation of the highly conserved residue W460C (corresponding to W149 in Fyn) located in the N-terminal part of the SH2 domain of the Fer kinase has been identified in lung cancer (Richardson et al., 2009). Similarly, multiple mutations in the Btk’s SH2 domain are responsible for X-linked agammaglobulinemia (Lappalainen et al., 2008). Some of these mutations were observed at residues proposed to be part of the top 12 highly coupled residues (see Figure 7a): R307G/K/T (corresponding to R176 in Fyn), Y334S (corresponding to Y203 in Fyn) or H362Q/R (corresponding to Y231 in Fyn) (Lappalainen et al., 2008).
In conclusion, we argued here that fast sidechain dynamics may play an important regulatory role in Fyn, which may most likely be extended to closely related SFK. Future evidence collected within the context of the interacting SH3 domain or the full protein will either confirm or refute the hypothesis we suggest here. Nonetheless the evidence of the activity assays clearly shows that the identified residue cluster is essential for regulating Fyn’s activity.
Experimental procedures
2H Relaxation
The five side-chain deuterium relaxation rates [RQ(Dz), RQ(D+), RQ(3Dz2-2), RQ(D+Dz+DzD+) and RQ(D+2)] were measured for the 13CH2D methyl groups on a Varian NMR Direct-Drive System 800 MHz spectrometer. The rates were recorded and analyzed according to the published procedures (Houben et al., 2007; Millet et al., 2002). “On the fly” correction of the CzHz remote magnetization was carried out in all the cases. Additionally, R(CzHz) rates were recorded to correct the double-quantum relaxation rate for the contributions from the dipolar interactions at the methyl group level between deuterium and protons as well as between deuterium and carbon atom. The delays, the scans and the number of complex points used to record the 2H relaxation rates and R(CzHz) rates are listed in Table S3. The relaxation rates and their experimental errors were extracted by fitting the peak intensity to a mono-exponential function. The peak intensities for some of the methyl groups were weak and could not be measured reliably. All the methyl groups for which at least one of the five relaxation rates had an experimental error higher than 1.5 times actual value of the rate were removed from the analysis. Among the five rates, double quantum and quadrupolar order relaxation rates are less sensitive and generate noisy signal decays. Often, these two rates contribute to methyl group exclusion. This is the case of L174-δ1, L174-δ2, V200-γ1, L238-δ1 and L244-δ1 in free SH2 domain and of L174-δ1, L174-δ2, L187-δ2, V200-γ1, L227-δ1, L238-δ1 and L242-δ1 in the bound form. The relaxation data for V243-γ1 and V244-γ1 in both free and bound forms could not be measured due to resonance overlap with methyl groups originating from the His-tag region. Only in the case of the free form, the T172-γ2 completely overlaps with the T181-γ2. The relaxation decay is characterized by a proper fit and the same relaxation rate was assigned to both methyl groups. Additionally, the five different deuterium relaxation rates must fulfil the following ordering: RQ(Dz) ≤ 5/3RQ(D+2) ≤ 5/3RQ(3Dz2-2) < RQ(D+) ≤ 5/3RQ(D+Dz+DzD+), if J(0) ≥ J(ωD) ≥ J(2ωD) (Jacobsen et al., 1976). As shown in Supplemental materials (Figure S7), most of the relaxation decays fulfil the trend. The larger errors for some rates are originating in peaks with low intensity, broad signal in the 1H-13C correlation spectrum. The rates not respecting the inequity within their errors were removed from the analysis. The following methyls were in this situation: L164-δ1 and L227-δ2 in free SH2 domain and L163-δ2, L164-δ1, L164-δ2, I175-δ1 and I205-γ2 in the bound form. Overall, reliable deuterium relaxation rates were measured for 37 out of the 48 methyl groups in the free form and 34 of the 48 methyl groups in case of the SH2:pY-tail complex. Between these two sets of relaxation parameters, 32 methyls are in common.
The Lipari-Szabo model-free formalism was used to fit the deuterium relaxation rates and to extract the motional parameters: S2axis and τe (Lipari and Szabo, 1982a, b). S2axis represents the order parameter of the vector between the methyl carbon and the adjacent carbon atom, while τe is the corresponding internal correlation time. To discard the contribution from the free methyl rotation, S2axis was obtained by scaling S2 with α=[(3cos2θ-1)/2]2, where θ is the angle between the 13C-2H bound and the methyl symmetry axis. Considering a tetrahedral geometry for the methyl group: S2 = 0.111S2axis (Mittermaier and Kay, 1999). Additionally, as the five relaxation rates were recorded, an extended model-free analysis allowed us to investigate possible contributions from nanosecond time scales, by extracting two additional parameters: Ss2 and τs (Skrynnikov et al., 2002). Ss2 represents the order parameter for slow internal motions and τs the corresponding correlation time. The overall correlation time, τc, used for analysis was fixed to 12.13ns and 13.13ns, for the free and complex form respectively. These values were extracted independently from backbone amide 15N relaxation measurements, assuming an isotropic motion. 15N R1 and R2 relaxation data were measured on the same sample used for recording the five deuterium relaxation rates. τc was calculated with TENSOR2 software from R1 and R2 rates after removing all the data for residues with high internal flexibility, typically the loop regions (Dosset et al., 2000). Since SH2 domains are prone to aggregate (Farrow et al., 1994; Finerty et al., 2002; Pintar et al., 1996; Siegal et al., 1998; Zhang et al., 1998) the sample stability was checked with R2 measurements, recorded at different time intervals. At 1 mM the free Fyn SH2 sample without the His-tag showed a significant self-association after 48 hours at 25°C (data not shown). In contrast, in similar conditions the His-tagged version of the Fyn SH2 was stable for more than 20 days. Consequently, all the NMR measurements of both the free and the pY-tail complex states were performed on the His-tag version of the construct. The use of long His-tag construct explains the higher τc values, when compared with the typical overall correlation time for Fyn SH2 domains (Farrow et al., 1994; Finerty et al., 2002). All the NMR data were processed using NMRPipe (Delaglio et al., 1995). Fitting the 2H relaxation rates to extract the model-free parameters, as well as model selection using an F-test were performed in GNU Octave version 3.4.0. Error estimates were determined from 500 Monte Carlo simulations.
Prediction of dynamical changes induced by the binding event
Our predictive method takes as input the NMR ensembles of the free and bound state of the Fyn SH2 domain (see Figure S1) and samples the conformational freedom of the side-chains, separately in each state, using a Monte-Carlo sampling process (Cilia et al., 2012) (see Figure 3a). Given these data, the method then determines in each state whether there exist conformational dependencies between the side-chain conformations of all residue pairs, quantifying this dependency through mutual information (MI). Whereas a high MI value indicates that the movement of two side-chains happens in a coordinated manner, a low MI value demonstrates that their movement is independent. Calculating the absolute change in mutual information, which is the absolute value of the difference in MI between bound and unbound states (|ΔMI|=|MI_bound-MI_free|), provides insight into the conformational dependencies that have changed due to binding. This step produces a matrix of changes, visualized in detail in Figure S4. The majority of the values in this matrix are close to zero (blue), indicating that the main dynamic changes are limited to a number of specific residues, which is expected. Prior predictions for the PDZ domain of PTPN11 (Cilia et al., 2012) and different SH3 domains (Zafra Ruano et al., 2016) have shown that such sparse effects are to be expected as only small effects at the level of the backbone are observed in these small domains.
Once this matrix is obtained, the Cluster Affinity Search Technique (CAST) (Ben-Dor et al., 1999) is used to identify residue groups whose members experience strong dynamic changes with those belonging to the same group (see Figure 3a). CAST identifies these clusters of residues using a threshold parameter (CT), which expresses a minimal coupling level within each cluster. For decreasing CT values, the size of the cluster(s) increases as more and more residues are accepted to be part of them until finally, for a low enough threshold, all elements are merged into one big cluster.
Network representation of the dynamical changes
The clusters retrieved for a particular value of CT can be mapped on a network of structurally interacting residues, providing information on the dynamic changes localized in the protein structure. In Figures 5a and 6a we represent this network for Fyn SH2 by discarding the long-range effects for clarity of the representation. We only highlight changes in coupling between residues in contact, where a contact is present when two residues have at least one pair of side-chain atoms at a distance less or equal to 5 Å in more than 50% of the structures of the SH2:pY-tail NMR ensemble.
Combing experimental and predictive data
In this paper we merge the above-described predictive approach with the data obtained by NMR side-chain relaxation experiments, in order to complement the experimental results on methyl-bearing residues with predictions for the non-methyl bearing residues. To do so, we devised an iterative approach that seeks for a subset of relevant methyl-bearing residues for which, starting from the predicted network, a corresponding subset of relevant non-methyl bearing residues can be derived with maximal accuracy on the experimental data. The procedure is illustrated in Figure 4c.
The reasoning proposed in this study assumes that not all the experimental ΔS2axis values, defined as significant based on the experimental conditions (Figure 3a), are also relevant for understanding the Fyn SH2 domain internal dynamics induced by the peptide binding. For instance, M195-ε is a very flexible methyl group that respects the significance criteria, but due to its highly solvent surface accessibility and isolated location in respect to the Fyn SH2 functional regions, it is less probable to have relevant dynamical changes upon peptide binding.
We first defined an acceptance level (AL) on the experimental |ΔS2axis| values, which determines the number of positive (relevant for the internal dynamics of the domain) and negative residue instances (non-relevant for the domain internal dynamics). As the predictive approach is not able to differentiate between the two methyl groups on the same residue, such as is the case for L238 and I189, we considered them as relevant residues when at least one of the |ΔS2axis| values is above AL.
We then repeated the following steps (see Figure 4c) until a termination criterion is met (namely AL is above the maximum |ΔS2axis|): i) to increase AL and consequently produce a labeling of the methyl-bearing residues into positive (P) and negative instances (N); ii) to perform a Receiver Operating Characteristics (ROC) analysis of the predictions by varying the CAST threshold (CT) and to compute the Area Under the ROC Curve (AUC), which then measures the quality of the predictions with respect to the labeling of the experimental data given in (i) in a way that is independent of the chosen CT; iii) to keep the computed AUC and AL values when they meet two combined criteria that we fixed: first, AUC must be high enough (at least 0.75 considering that 0.5 would be the performance of a random predictor); second, AL gives a better balanced ratio between positive and negative residue instances with respect to the best-so-far AL* value. In Figure 4 a cascade of two yellow diamond shapes (condition 1 and 2) represent these two criteria. For the latter criterion, we compare the current AL dependent ratio between number of positive (P) and negative examples (N) with the best-so-far one in order to privilege the higher one (i.e. the closest to 1 assuming the lowest between N or P is at the numerator). Figure S5 shows the trace of the iterative algorithm in terms of AUC (y axis) for each AL affecting the labeling of the experimental data (x axis).
Upon termination, the ROC curve corresponding to the chosen AL* is analyzed to determine the best threshold CT* on the predictions, which corresponds to the closest point to the top-left corner of the ROC plot. This threshold is then used to identify the informative group of methyl- and non-methyl bearing residues, which are predicted as most affected by changes in coupling induced by the binding event.
Fyn kinase assays
Human Fyn kinase (corresponding to residues 1–537) was cloned into vector 2BT (Addgene plasmid # 29666) and expressed in E.coli BL21DE3 with an aminoterminal TEV cleavable hexa-histidine tag. Fyn was purified as described before for Src kinase (Seeliger et al., 2005). Csk was expressed and purified from E.coli as described before (Levinson et al., 2008). Kinase activity was tested against a Src kinase optimal substrate peptide (EAIYAAPFAKKK) in a continuous spectrophotometric assay at 30 °C (Barker et al., 1995). Since Csk has low activity against substrate peptides, even high concentrations of Csk only yielded a small background kinase activity that could be subtracted from the activity of Fyn kinase. We had shown before that Src kinase does not trans-autophosphorylate under the conditions of the kinase assay and we therefore exclude any contribution of autophosphorylation on the assay (Seeliger et al., 2007).
Highlights.
Binding to Fyn’s pY-tail rigidifies the SH3-SH2 linker, inhibiting activity.
We identify a sidechain dynamics network that explains the process.
A residue cluster near the linker appears to act via a locking mechanism.
Kinase assays validate the cluster’s importance.
Acknowledgments
RH, NvN, LB and TL acknowledge the support of the F.W.O. for this research (grants G011609N and G025915N) . EC and TL also acknowledge the support of the F.R.S.-F.N.R.S (grant FRFC 2460611F), of which EC is a postdoctoral researcher. KH, NvN and TL also acknowledge the FP7 BioNMR project. The VIB and the Hercules Foundation support the research in NvN group. We thank Evgeny Tischenko (Agilent) and Adrien Favier (IBS, Grenoble) for implementing the relaxation experiments, the latter supported by the FWO-MULTIMAR. AL and MAS are supported by NIGMS 2T32GM008444 and NIH 1R35GM119437. AL and MAS would like to thank W. Todd Miller for helpful discussion and Weibing Zhang for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceived and designed this study: RH, EC NVN and TL. Performed and analyzed the NMR-related experiments: RH, LB, KH, NvN and TL. Performed and analyzed the computational research EC, RH, NvN and TL. Performed and analyzed the kinase assay: AL and MAS. Wrote the paper: RH, EC, AL, LB, KH, MAS, NvN and TL.
References
- Arold ST, Ulmer TS, Mulhern TD, Werner JM, Ladbury JE, Campbell ID, Noble MEM. The Role of the Src Homology 3-Src Homology 2 Interface in the Regulation of Src Kinases. Journal of Biological Chemistry. 2001;276:17199–17205. doi: 10.1074/jbc.M011185200. [DOI] [PubMed] [Google Scholar]
- Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c–src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. Journal of computational biology : a journal of computational molecular cell biology. 1999;6:281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Eck MJ. Structure and regulation of Src family kinases. Oncogene. 2004;23:7918–7927. doi: 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- Bradshaw JM, Grucza RA, Ladbury JE, Waksman G. Probing the “two-pronged plug two-holed socket” model for the mechanism of binding of the Src SH2 domain to phosphotyrosyl peptides: a thermodynamic study. Biochemistry. 1998;37:9083–9090. doi: 10.1021/bi973147k. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Charpin C, Secq V, Giusiano S, Carpentier S, Andrac L, Lavaut MN, Allasia C, Bonnier P, Garcia S. A signature predictive of disease outcome in breast carcinomas, identified by quantitative immunocytochemical assays. Int J Cancer. 2009;124:2124–2134. doi: 10.1002/ijc.24177. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li ZH, Liu XD, Wang SG, Bie P, Jiang P, et al. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis. 2011;32:1419–1426. doi: 10.1093/carcin/bgr088. [DOI] [PubMed] [Google Scholar]
- Cilia E, Vuister GW, Lenaerts T. Accurate Prediction of the Dynamical Changes within the Second PDZ Domain of PTP1e. PLoS Computational Biology. 2012;8:e1002794. doi: 10.1371/journal.pcbi.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan-Jacob SW, Fendrich G, Manley PW, Jahnke W, Fabbro D, Liebetanz J, Meyer T. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–871. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Daily MD, Upadhyaya TJ, Gray JJ. Contact rearrangements form coupled networks from local motions in allosteric proteins. Proteins: Structure, Function, and Bioinformatics. 2008;71:455–466. doi: 10.1002/prot.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J Biomol NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Comparison of SH3 and SH2 domain dynamics when expressed alone or in an SH(3+2) construct: the role of protein dynamics in functional regulation. Journal of Molecular Biology. 1999;287:645–656. doi: 10.1006/jmbi.1999.2619. [DOI] [PubMed] [Google Scholar]
- Engen JR, Wales TE, Hochrein JM, Meyn MA, Banu Ozkan S, Bahar I, Smithgall TE. Structure and dynamic regulation of Src-family kinases. Cellular and molecular life sciences : CMLS. 2008;65:3058–3073. doi: 10.1007/s00018-008-8122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JDLEK. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Finerty PJ, Jr, Muhandiram R, Forman-Kay JD. Side-chain dynamics of the SAP SH2 domain correlate with a binding hot spot and a region with conformational plasticity. Journal of Molecular Biology. 2002;322:605–620. doi: 10.1016/s0022-2836(02)00803-3. [DOI] [PubMed] [Google Scholar]
- Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. Journal of molecular biology. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Hochrein JM, Hochrein JM, Lerner EC, Schiavone AP, Smithgall TE, Engen JR. An examination of dynamics crosstalk between SH2 and SH3 domains by hydrogen/deuterium exchange and mass spectrometry. Protein science : a publication of the Protein Society. 2006;15:65–73. doi: 10.1110/ps.051782206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben K, Blanchard L, Blackledge M, Marion D. Intrinsic dynamics of the partly unstructured PX domain from the Sendai virus RNA polymerase cofactor P. Biophys J. 2007;93:2830–2844. doi: 10.1529/biophysj.107.108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RY, Wang SM, Hsieh CY, Wu JC. Lysophosphatidic acid induces ovarian cancer cell dispersal by activating Fyn kinase associated with p120-catenin. Int J Cancer. 2008;123:801–809. doi: 10.1002/ijc.23579. [DOI] [PubMed] [Google Scholar]
- Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chem Rev. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen JP, Bildsøe HK, Schaumburg K. Application of density matrix formalism in NMR spectroscopy. II. The one-spin-1 case in anisotropic phase. J Magn Reson. 1976;23:153–164. [Google Scholar]
- Kaneko T, Huang H, Zhao B, Li L, Liu H, Voss CK, Wu C, Schiller MR, Li SS. Loops govern SH2 domain specificity by controlling access to binding pockets. Sci Signal. 2010;3:ra34. doi: 10.1126/scisignal.2000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L, Muhandiram DR, Farrow NA, Aubin Y, Forman-Kay JD. Correlation between Dynamics and High Affinity Binding in an SH2 Domain Interaction. Biochemistry. 1996;35:361–368. doi: 10.1021/bi9522312. [DOI] [PubMed] [Google Scholar]
- Kay LE, Muhandiram DR, Wolf G, Shoelson SE, Forman-Kay JD. Correlation between binding and dynamics at SH2 domain interfaces. Nature Structural Biology. 1998;5:156–163. doi: 10.1038/nsb0298-156. [DOI] [PubMed] [Google Scholar]
- Kostic A, Lynch CD, Sheetz MP. Differential matrix rigidity response in breast cancer cell lines correlates with the tissue tropism. PLoS One. 2009;4:e6361. doi: 10.1371/journal.pone.0006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen I, Thusberg J, Shen B, Vihinen M. Genome wide analysis of pathogenic SH2 domain mutations. Proteins. 2008;72:779–792. doi: 10.1002/prot.21970. [DOI] [PubMed] [Google Scholar]
- Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat Struct Biol. 2000;7:72–77. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- Lee G, Thangavel R, Sharma VM, Litersky JM, Bhaskar K, Fang SM, Do LH, Andreadis A, Van Hoesen G, Ksiezak-Reding H. Phosphorylation of tau by fyn: implications for Alzheimer’s disease. J Neurosci. 2004;24:2304–2312. doi: 10.1523/JNEUROSCI.4162-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaerts T, Ferkinghoff-Borg J, Stricher F, Serrano L, Schymkowitz JWH, Rousseau F. Quantifying information transfer by protein domains: analysis of the Fyn SH2 domain structure. BMC structural biology. 2008;8:43. doi: 10.1186/1472-6807-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson NM, Seeliger MA, Cole PA, Kuriyan J. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134:124–134. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. 1982a;104:4546–4559. [Google Scholar]
- Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. 1982b;104:4559–4570. [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science (New York, NY) 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Millet O, Muhandiram DR, Skrynnikov NR, Kay LE. Deuterium spin probes of side-chain dynamics in proteins. 1. Measurement of five relaxation rates per deuteron in (13)C-labeled and fractionally (2)H-enriched proteins in solution. J Am Chem Soc. 2002;124:6439–6448. doi: 10.1021/ja012497y. [DOI] [PubMed] [Google Scholar]
- Mittermaier A, Kay LE. Measurement of Methyl 2H Quadrupolar Couplings in Oriented Proteins. How Uniform Is the Quadrupolar Coupling Constant? J Am Chem Soc. 1999;121:10608–10613. [Google Scholar]
- Mittermaier A, Kay LE, Forman-Kay JD. Analysis of deuterium relaxation-derived methyl axis order parameters and correlation with local structure. J Biomol NMR. 1999;13:181–185. doi: 10.1023/A:1008387715167. [DOI] [PubMed] [Google Scholar]
- Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Yamazaki T, Sykes BD, Kay LE. Measurement of H-2 T-1 and T-1p Relaxation-Times in Uniformly C-13-Labeled and Fractionally H-2-Labeled Proteins in Solution. J Am Chem Soc. 1995;117:11536–11544. [Google Scholar]
- Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, Kuriyan J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Molecular Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Nygaard HB, van Dyck CH, Strittmatter SM. Fyn kinase inhibition as a novel therapy for Alzheimer’s disease. Alzheimers Res Ther. 2014;6:8. doi: 10.1186/alzrt238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- Pintar A, Hensmann M, Jumel K, Pitkeathly M, Harding SE, Campbell ID. Solution studies of the SH2 domain from the fyn tyrosine kinase: secondary structure, backbone dynamics and protein association. Eur Biophys J. 1996;24:371–380. doi: 10.1007/BF00576709. [DOI] [PubMed] [Google Scholar]
- Posadas EM, Al-Ahmadie H, Robinson VL, Jagadeeswaran R, Otto K, Kasza KE, Tretiakov M, Siddiqui J, Pienta KJ, Stadler WM, et al. FYN is overexpressed in human prostate cancer. BJU Int. 2009;103:171–177. doi: 10.1111/j.1464-410X.2008.08009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CJ, Gao Q, Mitsopoulous C, Zvelebil M, Pearl LH, Pearl FM. MoKCa database--mutations of kinases in cancer. Nucleic Acids Res. 2009;37:D824–D831. doi: 10.1093/nar/gkn832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger MA, Nagar B, Frank F, Cao X, Henderson MN, Kuriyan J. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal G, Davis B, Kristensen SM, Sankar A, Linacre J, Stein RC, Panayotou G, Waterfield MD, Driscoll PC. Solution structure of the C-terminal SH2 domain of the p85 alpha regulatory subunit of phosphoinositide 3-kinase. Journal of Molecular Biology. 1998;276:461–478. doi: 10.1006/jmbi.1997.1562. [DOI] [PubMed] [Google Scholar]
- Singh MM, Howard A, Irwin ME, Gao Y, Lu X, Multani A, Chandra J. Expression and activity of Fyn mediate proliferation and blastic features of chronic myelogenous leukemia. PLoS One. 2012;7:e51611. doi: 10.1371/journal.pone.0051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrynnikov NR, Millet O, Kay LE. Deuterium spin probes of side-chain dynamics in proteins. 2. Spectral density mapping and identification of nanosecond time-scale side-chain motions. J Am Chem Soc. 2002;124:6449–6460. doi: 10.1021/ja012498q. [DOI] [PubMed] [Google Scholar]
- Superti-Furga G, Fumagalli S, Koegl M, Courtneidge SA, Draetta G. Csk inhibition of c-Src activity requires both the SH2 and SH3 domains of Src. Embo J. 1993;12:2625–2634. doi: 10.1002/j.1460-2075.1993.tb05923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessari M, Gentile LN, Taylor SJ, Shalloway DI, Nicholson LK, Vuister GW. Heteronuclear NMR Studies of the Combined Src Homology Domains 2 and 3 of pp60 c-Src. Biochemsitry. 1997;36:14561–14571. doi: 10.1021/bi9712044. [DOI] [PubMed] [Google Scholar]
- Tsai C, Delsol A, Nussinov R. Allostery: Absence of a Change in Shape Does Not Imply that Allostery Is Not at Play. Journal of Molecular Biology. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer T, Werner J, Campbell I. SH3-SH2 domain orientation in Src Kinases NMR studies of Fyn. Structure (London, England : 1993) 2002;10:901–991. doi: 10.1016/s0969-2126(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Whitley MJ, Lee AL. Frameworks for understanding long-range intra-protein communication. Curr Protein Pept Sci. 2009;10:116–127. doi: 10.2174/138920309787847563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Belrose J, Trepanier CH, Lei G, Jackson MF, MacDonald JF. Fyn, a potential target for Alzheimer’s disease. J Alzheimers Dis. 2011;27:243–252. doi: 10.3233/JAD-2011-110353. [DOI] [PubMed] [Google Scholar]
- Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic Coupling between the SH2 and SH3 Domains of c-Src and Hck Underlies Their Inactivation by C-Terminal Tyrosine Phosphorylation. Cell. 2001;105:115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- Zafra Ruano A, Cilia E, Couceiro JR, Ruiz Sanz J, Schymkowitz J, Rousseau F, Luque I, Lenaerts T. From Binding-Induced Dynamic Effects in SH3 Structures to Evolutionary Conserved Sectors. PLoS Comput Biol. 2016;12:e1004938. doi: 10.1371/journal.pcbi.1004938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Smithgall TE, Gmeiner WH. Self-association and backbone dynamics of the hck SH2 domain in the free and phosphopeptide-complexed forms. Biochemistry. 1998;37:7119–7126. doi: 10.1021/bi972077e. [DOI] [PubMed] [Google Scholar]