Abstract
Background
The gut metabolome may be associated with the incidence and progression of numerous diseases. The composition of the gut metabolome can be captured by measuring metabolite levels in the feces. However, there is little data describing the effect of fecal sample collection methods on metabolomic measures.
Methods
We collected fecal samples from eighteen volunteers using four methods: no solution, 95% ethanol, fecal occult blood test (FOBT) cards, and fecal immunochemical test (FIT). One set of samples was frozen after collection (day 0), and for 95% ethanol, FOBT, and FIT, a second set was frozen after 96 hours at room temperature. We evaluated 1) technical reproducibility within sample replicates, 2) stability after 96 hours at room temperature for 95% ethanol, FOBT, and FIT, and 3) concordance of metabolite measures with the putative “gold standard,” day 0 samples without solution.
Results
Intraclass correlation coefficients (ICC) estimating technical reproducibility were high for replicate samples for each collection method. ICCs estimating stability at room temperature were high for 95% ethanol and FOBT (median ICC>0.87) but not FIT (median ICC=0.52). Similarly, Spearman correlation coefficients (rs) estimating metabolite concordance with the “gold standard” were higher for 95% ethanol (median rs=0.82) and FOBT (median rs=0.70) than for FIT (median rs=0.40).
Conclusions
Metabolomic measurements appear reproducible and stable in fecal samples collected with 95% ethanol or FOBT. Concordance with the “gold standard” is highest with 95% ethanol and acceptable with FOBT.
Impact
Future epidemiologic studies should collect feces using 95% ethanol or FOBT if interested in studying fecal metabolomics.
Keywords: metabolomics, feces, collection methods, reproducibility, stability
Introduction
Metabolomics is the systematic measurement of the low molecular weight compounds, often loosely termed “metabolites”, in biospecimens (1, 2). These metabolites typically come from the host, microbial symbionts, environmental exposures, or combinations of these sources (e.g. an environmental molecule modified by a bacterial enzyme) and can be measured using either a targeted or an untargeted approach. Targeted metabolomics platforms measure a limited number of specific metabolites of known identity making them well-suited for hypothesis-driven studies. In contrast, untargeted platforms measure as many metabolites as possible, of known and unknown identity, in a biospecimen allowing for the discovery of novel metabolic associations and disease pathways (3). Metabolomic phenotypes have been associated with diet (4–8), behaviors such as smoking (9, 10) and physical activity (11), and diseases such as diabetes (12), Crohn’s disease (13), prostate cancer (14, 15), and colorectal cancer (16–19). The majority of epidemiologic studies have measured metabolomic phenotypes (i.e. the full complement of measured metabolites) within blood (e.g. serum) (4–6, 9–12, 14–17); fewer have analyzed urine samples (7, 8, 14), and although only a few relatively small studies have analyzed fecal samples using untargeted platforms, they have found potential diagnostic markers of disease (13, 18). These studies highlight the need for future epidemiologic studies with large-scale fecal sample collection. However, methodological research to inform the collection protocols of these future studies is essential.
As epidemiologic studies increase collections of fecal samples for microbiome analysis (20, 21), we expect a considerable increase in the number of investigations on the relationship between fecal metabolites, which reflect complex interactions between dietary inputs, intestinal bacteria and host metabolism (22), and risk, or progression, of diseases such as colorectal cancer.
It is widely recognized that the microbiome alters the metabolome in ways that are associated with, and in mouse models, cause disease (23, 24). It is therefore advantageous to assess these phenotypes concurrently and, in epidemiological studies, use sample collection methods that are amenable to multiple molecular analyses (25). When selecting a suitable collection method for fecal samples in population-based research, key quality considerations include technical reproducibility (i.e. consistency of metabolites measurement for replicate samples collected and stored in the same manner), stability at ambient temperature for a period of time that mimics field conditions, and concordance of metabolite measurements with samples frozen soon after collection, which is currently considered the “gold standard” for microbiome and other “omic” analyses (26).
Therefore, we conducted a fecal metabolomics study to evaluate the technical reproducibility, stability, and concordance of three collection methods (95% ethanol, fecal occult blood test (FOBT) cards, and fecal immunochemical test (FIT) tubes) as compared with the “gold standard”. By design, this study was nested in a larger study of collection methods for fecal samples in microbiome analyses thereby permitting the identification of suitable collection methods for both fecal metabolome and microbiome studies. For microbiome analyses, we found that the fecal sample collection methods, considered herein, were relatively reproducible, stable and concordant with the “gold standard” sample that was frozen following collection (day 0) with no solution (Vogtmann et al, under re-review). In addition, a prior study of seven fecal sampling methods for microbiome analyses found that FOBT cards and samples stored in RNAlater had the highest stability, while FOBT cards and samples stored in 70% ethanol had the highest concordance with the “gold standard” sample that was frozen shortly after collection with no solution (27). This earlier study did not, however, consider the potential of each collection method for metabolomic analyses nor did it include FIT tubes, which are commonly used in colorectal cancer screening programs and thus a potential untapped resource in future diagnostic and etiologic studies of cancer if they were to be stored in −80°C freezers rather than discarded. Moreover, samples collected with RNAlater have a high sodium sulfate content making this collection incompatible with mass spectrometry-based metabolomics platforms (25) and highlighting the need for methods research to ascertain which fecal collection methods are most suitable for a variety of molecular analyses in future epidemiologic studies.
Materials and Methods
Study participants
A sample of 18 individuals (9 males and 9 females) was randomly selected from a larger microbiome methods study of 52 volunteers who were recruited from Mayo Clinic employees. Eligibility requirements for the main microbiome methods study included the following: minimum age of 18 years, no use of antibiotics or probiotics within the two weeks prior to enrollment, no history of pelvic radiation, and no current chemotherapy treatment. At study enrollment, participants self-reported demographic and health characteristics, tobacco use, alcohol consumption, oral health habits, and recent antibiotic exposure using a standardized questionnaire that was created for this study. All participants provided informed consent. The study was approved by the Mayo Clinic Institutional Review Board and by the National Cancer Institute Office of Human Subjects Research.
Fecal specimen collection
Following enrollment, participants were invited to return to the clinic at a later date to provide a fecal specimen. On the collection day, each participant was provided with an Exakt Pak canister (Inmark Packaging, Austell, GA, USA) for on-site fecal specimen collection. Immediately following collection, the study coordinator delivered each specimen to the laboratory for processing. Average time from collection to laboratory processing was 14 minutes.
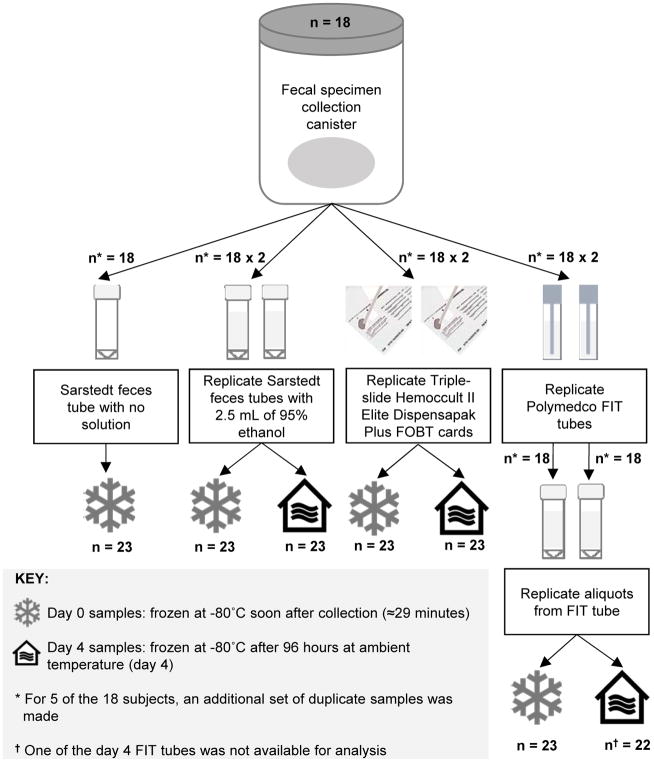
In the laboratory, each fecal specimen was mixed manually using a spatula, and aliquots for the different collection methods were generated in random order. For each participant, approximately 1–2 grams of feces, representing a full scoop of feces, was placed in a Sarstedt feces tube (Numbrecht, Germany) containing no solution or 2.5 mL of 95% ethanol (Sigma-Aldrich, St. Louis, Missouri). Triple-slide Hemoccult II Elite Dispensapak Plus for FOBT (Beckman Coulter, Brea, California) were smeared thinly with feces and the flap was closed. FIT tubes (Polymedco, Inc., Cortlandt Manor, New York) were created by dipping the fecal specimen with the FIT probe and the tube was shaken. Aliquots from each FIT tube were created and stored in cryovials. One set of replicates of the no solution, 95% ethanol, and FIT cryovials were frozen following processing at −80°C (day 0). One set of replicate FOBT cards was developed using two drops of Hemoccult Sensa Developer applied to the guaiac paper on the back of the card (i.e., the testing strategy for occult blood in colorectal cancer screening) and then frozen at −80°C (day 0). Average time from the beginning of processing to freezer for day 0 samples was 15 minutes. The remaining samples were left at ambient temperature for 96 hours. After 96 hours, the ambient temperature FOBT cards were developed and all remaining samples were frozen at −80°C (day 4). In total, 160 frozen samples from 18 participants, including blinded duplicate samples from 5, randomly selected, participants yielding 23 samples per collection method except for FIT tube/day 4, which had one sample missing (Figure 1), were shipped on dry ice to Metabolon (Durham, NC) for biochemical profiling. Following receipt, samples were stored at −80°C until processed.
FIGURE 1.
Flowchart of study design
Metabolomics analysis
Samples were prepared using an automated system. In brief, fecal samples collected with no solution and 95% ethanol were dried, weighed and then re-suspended at a 50:1 (50ul H2O for every 1mg of feces weight) ratio for homogenization and processing; thus, these samples were processed with equivalent amounts of material. For FOBT cards, all material was collected from the cards and re-suspended at a 30:1 ratio; thus, these samples were also processed at equivalent amounts. Finally, for the FIT tubes, a weight for the fecal component of these samples was not available; thus, 100ul of the suspension on a per sample basis was processed. To remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking followed by centrifugation. Following removal of the organic solvent, sample extracts were stored overnight under nitrogen before preparation for analysis.
All fecal samples were analyzed using ultra-performance liquid chromatography (UPLC) and high resolution/tandem mass spectrometry (MS/MS). The sample extract was dried then reconstituted in solvents compatible to each of the four collection methods. Each reconstitution solvent contained a series of standards at fixed concentrations to ensure injection and chromatographic consistency. One aliquot was analyzed using acidic positive ion conditions, chromatographically optimized for more hydrophilic compounds. In this method, the extract was gradient eluted from a C18 column using water and methanol with 0.05% perfluoropentanoic acid and 0.1% formic acid. Another aliquot was also analyzed using acidic positive ion conditions; however, it was chromatographically optimized for more hydrophobic compounds. In this method, the extract was gradient eluted from the same C18 column using water, methanol and acetonitrile with 0.05% perfluoropentanoic acid and 0.01% formic acid. A third aliquot was analyzed using basic negative ion optimized conditions following gradient elution with a separate C18 column using methanol, water, and 6.5 mM ammonium bicarbonate at pH 8. A fourth aliquot was analyzed using negative ionization following elution from an HILIC column using water, acetonitrile, and 10 mM Ammonium Formate, pH 10.8.
Raw data was extracted, peak-identified and processed by Metabolon using proprietary software as described elsewhere (28–30). In brief, compounds of exogenous, human, and microbial origin, were identified by comparison to library entries of purified standards or recurrent unknown entities. Metabolon maintains a dynamic and proprietary biochemical reference library of more than 4,500 known metabolites (based on authenticated standards) and more than 9,000 novel metabolites (without an identified chemical structure); each library entry contains the retention time/index (RI), mass to charge ratio (m/z), and chromatographic data (including MS/MS spectral data). Biochemical identifications are based on three criteria: retention index within a narrow RI window of the proposed identification, accurate mass match to the library +/− 10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards. The MS/MS scores are based on a comparison of the ions present in the experimental spectrum to the ions present in the library spectrum. Integrated ion peaks were quantified using area-under-the-curve. Each collection method was run separately; however, all samples for a given collection method were run on the same plate on the same day. For each collection method, metabolite peak values were rescaled to set the median equal to 1, and the missing values were imputed with the minimum value for a given metabolite.
In addition to our blinded duplicate samples, Metabolon included three types of controls that were analyzed in concert with the experimental samples. First, a pooled matrix sample, which was generated by taking a small volume of each experimental sample, served as a technical replicate throughout the data set. Second, extracted water samples served as process blanks. Finally, a cocktail of quality control standards that were carefully chosen not to interfere with the measurement of endogenous compounds were spiked into every analyzed sample to monitor instrument performance and aid chromatographic alignment. Experimental samples were randomized across the platform run with quality control samples spaced evenly among the injections. Metabolon determined instrument variability by calculating the median relative standard deviation (RSD) for the standards that were added to each sample prior to injection into the mass spectrometers; median instrument variability was 2%, 2%, 3%, and 4% for fecal samples collected with no solution, 95% ethanol, FOBT cards, and FIT tubes, respectively. In addition, Metabolon determined overall process variability by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 100% of the pooled matrix samples; median process variability was 6%, 8%, 8%, and 11% for fecal samples collected with no solution, 95% ethanol, FOBT cards, and FIT tubes, respectively. These values for instrument and process variability met Metabolon’s acceptance criteria (28).
Statistical analysis
For the analyses of technical reproducibility, stability, and concordance, we first limited the set of metabolites to those with ≥80% detectability (i.e. above the detection limit in a given batch) in day 0 samples collected with no solution, which was considered the “gold standard”. For analyses including samples collected with 95% ethanol, FOBT, or FIT, we further restricted the set of metabolites to those with ≥80% detectability in the collection method of interest. Limiting these analyses to only those metabolites with high detectability ensured an adequate number of samples for calculating ICCs in this small sample. For analyses, metabolite values were natural log-transformed to make them more normally distributed.
We defined “technical reproducibility” as a standard intraclass correlation coefficient, where is the between-individual variability and is the within-individual variability owing to sample handling or laboratory variability:
For each metabolite and each collection method, we calculated the values and using a linear mixed effects regression model with a random effect for subject. We calculated ICCs separately for day 0 and day 4 samples. Non-replicate samples were included in the ICC calculation since they contribute to the estimation of between-individual variability.
We similarly defined “stability” as an intraclass correlation coefficient, where is the between-individual variability and is the within-sample variability over time:
Our estimation of stability used data from replicate samples that were frozen at different time points (day 0 or day 4). For each metabolite, we calculated the values and using a linear mixed effects regression model with hierarchal random effects for subject and time of freezing to incorporate all available measurements for a given collection method. Note that measurement error (i.e. when evaluating technical reproducibility) does not factor into our definition of stability
We defined “concordance” as the Spearman correlation coefficient (rs) between metabolite values from samples frozen on day 0 for a given collection method and metabolite values from samples frozen on day 0 with no solution (i.e., “gold standard”). The Spearman correlation evaluates whether the rank order of metabolite values for a given collection method was preserved as compared with the “gold standard”. For the five individuals with duplicate day 0 samples we randomly selected a single sample for each of the collection methods so that there were two observations per participant, one for a given collection method (95% ethanol, FOBT, or FIT) and one for no solution.
RESULTS
Fecal samples from nine men and nine women, aged 22 to 56 years, were analyzed in this study. Two thirds of participants (66.7%) had at least a Bachelor’s degree, and a large majority (88.9%) was non-Hispanic white (Supplemental Table 1).
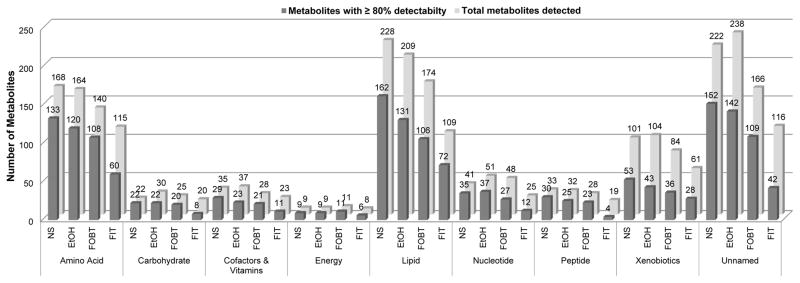
In total 859, 874, 704, and 496 metabolites were detected in fecal samples collected with no solution, 95% ethanol, FOBT cards, and FIT tubes, respectively (Supplemental Table 2). For the three collections methods with day 0 and day 4 samples, restricting to the subset of metabolites detected in day 0 samples only, there were 871, 703, and 466 metabolites in 95% ethanol, FOBT cards, and FIT tubes, respectively (Figure 2 and Supplemental Table 2). Further restricting to the subset of metabolites with ≥80% detectability, there were 625, 552, 461, and 243 metabolites in no solution, 95% ethanol, FOBT cards, and FIT tubes, respectively (Figure 2 and Supplemental Table 2).
FIGURE 2.
Number of metabolites in fecal samples, frozen on day 0, by collection method and metabolic super pathway
Abbreviations: NS, no solution; EtOH, 95% ethanol; FOBT, fecal occult blood test; FIT, fecal immunochemical test
In day 0 fecal samples with no solution, approximately half of all detected metabolites were classified as lipid (27%) or amino acid (20%) metabolites; a smaller percentage of detected metabolites were xenobiotic (12%), nucleotide (5%), peptide (4%), cofactor and vitamin (4%), carbohydrate (3%), or energy (1%) metabolites, and approximately 26% were unnamed metabolites. Although the number of detected metabolites varied considerably by collection method (Figure 2) the proportions of compounds classified as lipid (22% to 27%), amino acid (19% to 23%), xenobiotic (12%), nucleotide (5% to 7%), peptide (4%), cofactors and vitamins (4% to 5%), carbohydrate (3% to 4%), energy (1% to 2%), and unnamed (23% to 27%) metabolites were similar (Supplemental Figure and Supplemental Tables 3–6).
There were 508, 435, and 220 metabolites, with ≥80% detectability, in fecal samples collected with 95% ethanol, FOBT cards, and FIT tubes, respectively, that overlapped with the subset of 625 metabolites, with ≥80% detectability, in fecal samples collected with no solution (Table 1). Overall, 19%, 30%, and 65% of the metabolites with ≥80% detectability in no solution did not have ≥80% detectability in 95%, FOBT cards, and FIT tubes, respectively. The subset of overlapping metabolites was used to calculate technical reliability, stability, and concordance. The median ICC (interquartile range; IQR) for technical reproducibility for day 0 samples was 0.94 (0.86–0.98) for no solution, 0.86 (0.68–0.96) for 95% ethanol, 0.87 (0.75–0.95) for FOBT cards, and 0.83 (0.65–0.94) for FIT tubes (Figure 3 and Supplemental Table 7). The median ICCs for technical reproducibility for day 4 samples collected with 95% ethanol, FOBT, and FIT were similar to the day 0 ICCs (Supplemental Table 7). The median ICC (IQR) for stability was 0.87 (0.65–0.98) for 95% ethanol, 0.86 (0.69–0.99) for FOBT cards, and 0.52 (0.00–0.97) for FIT tubes (Figure 4 and Supplemental Table 7). For concordance, the median Spearman Correlation coefficient (IQR) with no solution was 0.82 (0.72–0.90) for 95% ethanol, 0.70 (0.51–0.85) for FOBT cards, and 0.40 (0.15–0.72) for FIT tubes (Figure 5 and Supplemental Table 7).
TABLE 1.
Comparison of the number of metabolites, with ≥80% detectability, in no solution (NS) to the number of metabolites, with ≥80% detectability, in fecal samples collected with 95% ethanol, FOBT cards, or FIT tubes
| Condition | Total no. Metabolites | No. of metabolites in condition and NS 1 | No. metabolites in NS only | No. metabolites in condition only |
|---|---|---|---|---|
| No solution (gold standard) | 625 | --- | --- | --- |
| 95% Ethanol | 561 | 508 | 117 | 53 |
| FOBT | 473 | 435 | 190 | 38 |
| FIT | 258 | 220 | 405 | 38 |
Subset of metabolites used in technical reliability, stability, and concordance calculations
Abbreviations: NS, no solution; FOBT, fecal occult blood test; FIT, fecal immunochemical test
FIGURE 3.
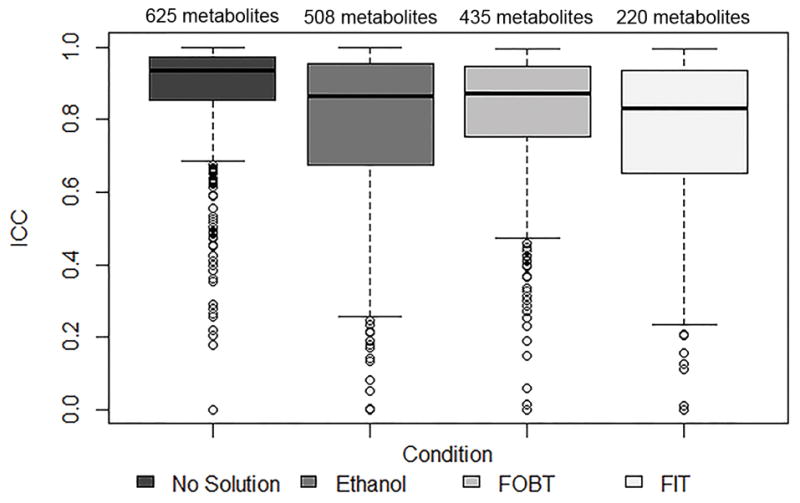
Technical reproducibility, estimated using intraclass correlation coefficients, of replicate fecal samples, frozen on day 0, for metabolites with ≥80% detectability in samples with no solution and in samples collected with 95% ethanol, FOBT, or FIT
Abbreviations: ICC, intraclass correlation coefficient; NS, no solution; FOBT, fecal occult blood test; FIT, fecal immunochemical test
FIGURE 4.
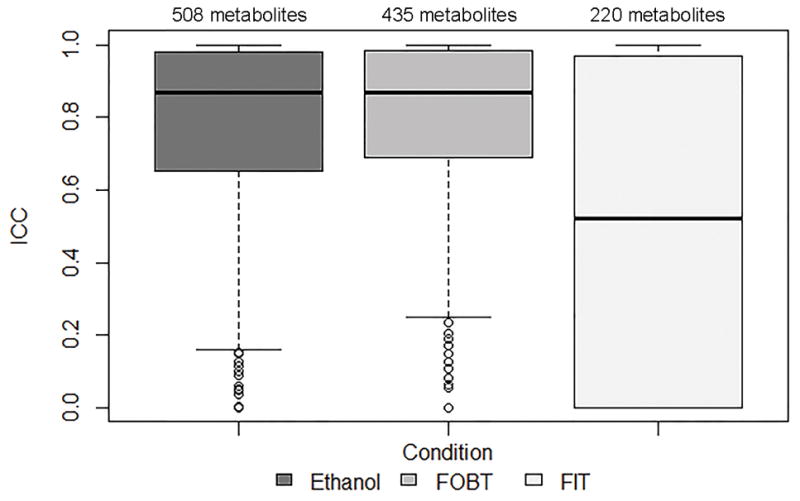
Stability, estimated using intraclass correlation coefficients estimated from fecal samples that were frozen on day 4 after 96 hours at ambient temperature and replicate fecal samples that were frozen on day 0, for metabolites with ≥80% detectability in samples with no solution and in samples collected with ethanol, FOBT, or FIT
Abbreviations: ICC, intraclass correlation coefficient; NS, no solution; FOBT, fecal occult blood test; FIT, fecal immunochemical test
FIGURE 5.
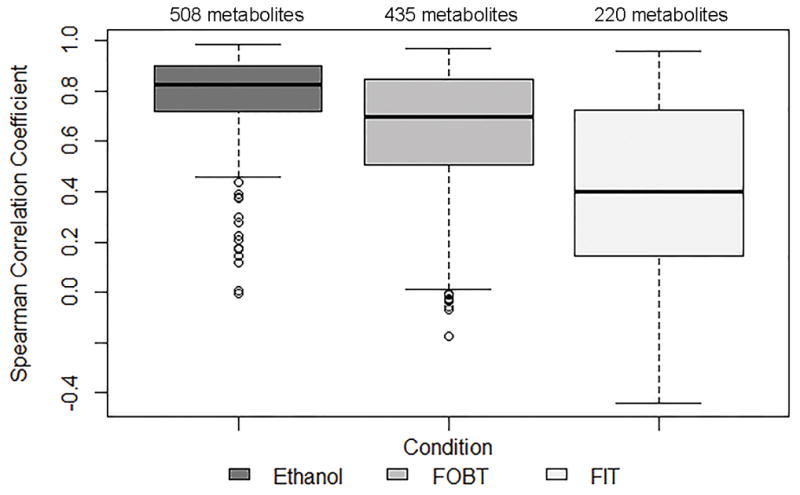
Concordance, estimated using Spearman correlation coefficients, of fecal samples that were collected with ethanol, FOBT, or FIT and frozen on day 0 with fecal samples that were frozen on day 0 with no solution for metabolites with ≥80% detectability in samples with no solution and in samples collected with ethanol, FOBT, or FIT
Abbreviations: NS, no solution; FOBT, fecal occult blood test; FIT, fecal immunochemical test
Known microbial metabolites were detected in fecal samples collected under varying conditions. For example, enterolactone, which is formed by the action of intestinal bacteria on lignan precursors, was detectable in all samples regardless of collection method (Supplemental Table 3–6) and freezing time (data not shown). Other microbial metabolites related to the degradation of polyphenols were detected under some but not all conditions. For example, in day 0 samples, caffeate was detected in 100% of samples that were collected with no solution, 95% ethanol, or FOBT cards but was not detected in fecal samples collected with FIT tubes; benzoate, on the other hand, was detected in 100% of samples collected with no solution or FOBT cards but in only 87% of samples that were collected with either 95% ethanol or FIT tubes, and 3-(3-Hydroxyphenyl)propionate was detected in 100% of samples collected with no solution or 95% ethanol but only in 91% of FOBT samples and 57% of FIT samples. Metabolon classified known metabolites by “super-pathway”, representing chemical classes, and “sub-pathway”, corresponding to the specific role of a compound in metabolism, on the basis of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (31). A complete list of metabolites, sorted by super- and sub-pathways, for each collection method frozen on day 0, can be found in Supplemental Tables 3 through 6.
DISCUSSION
This study found that metabolite measures obtained from an untargeted UPLC/MS-MS platform had high technical reproducibility for fecal samples collected with no solution, 95% ethanol, FOBT cards, or FIT tubes. In contrast, when comparing samples that were frozen after 4 days at room temperature to those that were frozen on day 0 for a given collection method, metabolite stability was high (median ICC>0.87) for fecal samples collected with 95% ethanol or FOBT cards, but low (median ICC=0.52) for the majority of metabolites measured in FIT tubes. Similarly, concordance with the putative “gold standard” was higher for fecal samples collected with 95% ethanol (median rs=0.82) or FOBT (median rs=0.70) and frozen on day 0 than for fecal samples collected with FIT tubes (median rs=0.40) and frozen on day 0.
To our knowledge, the impact of fecal collection method on metabolomic measurements, particularly fecal samples collected with no additive, 95% ethanol, FOBT cards, or FIT tubes, has not previously been assessed using an untargeted metabolomics platform. Moreover, the impact of incubating fecal samples collected with each of these methods for 96 hours at room temperature, a time period reflective of potential field conditions in a large population-based study, has not previously been studied. Our study shows that fecal samples collected with no additive are suitable for untargeted metabolomics analysis when frozen shortly after collection. Fecal samples that were collected with 95% ethanol or FOBT cards also appear suitable for untargeted metabolomics analysis and, more importantly, yield similar results when frozen after four days at ambient temperature or shortly after collection. FIT tubes that were frozen on day 0 are of intermediate quality, but given the existence of better methods, we recommend against this collection method. In contrast, FIT tubes that are left at room temperature for an extended period of time should not be used for untargeted metabolomics analysis. Although freezing samples right after collection with no solution yielded the greatest number of metabolites with high detectability and the highest estimates of technical reproducibility, this method of collection may not be practical for large epidemiological studies. Alternatively, fecal samples collected with 95% ethanol or FOBT cards appear well adapted to field conditions in which samples may be left at room temperature for an extended period of time.
In addition to the raw number of metabolites and these three measures of quality, researchers should consider which classes of compounds are of potential importance. For example, future studies with integrated measures of the metabolome and microbiome have the tremendous potential to increase our understanding of the mediating role of gut microbial metabolism in disease etiology. Accordingly, specific microbial metabolites may be of particular interest to researchers who are using fecal metabolomics to study interactions between dietary components and the microbiota. To aid in this effort, we have highlighted a small number of known microbial metabolites (enterolactone, caffeate, benzoate, and 3-(3-Hydroxyphenyl)propionate) with varying degrees of detectability by collection method and have included comprehensive lists of all detected metabolites by collection method in the supplemental material. Based on our results, fecal samples collected with no solution and frozen on day 0 as well as samples collected with 95% ethanol provide the largest number of metabolites with high detectability (≥80%) and the greatest coverage of metabolic super-pathways. In comparison, samples collected with FOBT cards as compared with no solution had a similar number (<20% difference) of metabolites with high detectability for metabolites related to the amino acid, carbohydrate, and energy super-pathways but substantially fewer metabolites related to (≥20% difference) cofactors and vitamins, lipid, nucleotide, peptide, and xenobiotic super-pathways. FIT tubes had many fewer detected metabolites for all metabolite classes as compared with other collection methods.
Metabolomics is a burgeoning technology and its potential to produce biomarkers of exposure and disease is considerable. Nevertheless, its integration into population-based research remains in its early stages (2). Rigorous methods studies identifying sources of variability in metabolite measurements for various types of biological specimens is critical (18, 32, 33). Presently, relatively few human studies have used untargeted metabolomics platforms to analyze fecal samples. In a small colorectal cancer case-control study approximately 1000 metabolites in lyophilized fecal samples by HPLC-GC/MS-MS were measured (18). Assay reproducibility in replicate samples exceeded 0.7 for 91% the 579 metabolites that were detected in at least 10% of the fecal specimens, and statistically significant differences between cancer cases and controls were found for 41 metabolites including some that appeared to reflect differences in gut microbial diversity. This study found potential markers to aid diagnosis and improve understanding of disease etiology thereby underscoring the need for future epidemiologic studies, including prospective cohort studies, with large-scale fecal sample collection and thus the need for research on how fecal collection methods and freezing conditions impact metabolite measurements.
A number of other small case-control (34, 35), cross-sectional (36, 37), and intervention (38–40) studies have used NMR spectroscopy to generate metabolic phenotypes of various types of fecal samples, including crude fecal samples (37), fecal water extracts (34–37, 40) and lyophilized feces (38, 39). Although these studies have shown reasonable reproducibility, they have been more targeted, capturing a smaller less diverse set of metabolites than untargeted approaches, and collectively have not used standardized sample collection, storage, or preparation methods. Interestingly, a number of NMR-based metabolomics studies have demonstrated associations between the fecal metabolome and microbiome (40, 41), further strengthening the case for the inclusion of fecal sample collection in population-based studies in combination with the development of standardized fecal sample collection methods suitable for multiple molecular analyses.
Limitations of this study include its small sample size and potentially limited generalizability as participants were relatively young, well-educated and primarily non-Hispanic white. It is possible that the quality metrics for fecal metabolomics considered herein may vary by collection methods among individuals with clinical or underlying disease or among individuals in different age or racial/ethnic groups. In addition, the samples in this study were collected at one-time point; consequently, we were unable to estimate within individual variability over time, which is an important consideration in population-based metabolomics studies. Using an untargeted metabolomics approach, one study found that, on average, approximately 40% of the biologic variability in serum metabolite measurements could be attributed to variation occurring within an individual over time; this estimate translates to a need for large sample sizes, upwards of 1000 individuals, to detect metabolite-disease associations in case-control studies (32). While sources of variability in urine, serum, and plasma metabolite measurements have been studied (32, 42, 43), data on sources of variability in fecal metabolite measurements is lacking.
Future methods work should be extended to the general population and should collect multiple fecal samples over a period of time to assess within individual variability over time. In addition, the feasibility of each collection method, including freezing time, should be carefully weighed when designing large population-based studies. For example, FOBT cards can be shipped in the mail, making them particularly suitable for large, geographically diverse cohorts. Finally, additional research is needed to ascertain whether these collection methods are suitable for other multiomic technologies, such as whole genome shotgun metagenomics and proteomics. In conclusion, metabolite measures, obtained from an untargeted UPLC/MS-MS platform, from fecal samples collected with 95% ethanol or FOBT cards are reflective of those from fecal samples with no solution that were frozen shortly after collection (i.e. “gold standard”) and appear reproducible and stable following incubation at room temperature.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health; a grant from the National Institutes of Health [1R01CA179243 to N. Chia], and a Howard Hughes Medical Institute award to R. Knight.
Footnotes
Conflicts of Interest: R. Knight has disclosed the following potential conflicts of interest: 1. Employment - CSO Entity: Biota Technology, Inc.; 2. Honoraria from Speakers Bureau - American Academy of Anti-Aging Medicine; 3. Ownership Interest (including patents) - Biota Technology, Inc.; 4. Consultant/Advisory Board - J&J/Janssen, CommenSe, Inc., and Prometheus Therapeutics & Diagnostics. All other authors have no conflicts to disclose.
References
- 1.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 2.Su LJ, Fiehn O, Maruvada P, Moore SC, O’Keefe SJ, Wishart DS, et al. The Use of Metabolomics in Population-Based Research. Adv Nutr. 2014;5:785–8. doi: 10.3945/an.114.006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzoulaki I, Ebbels TM, Valdes A, Elliott P, Ioannidis JP. Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol. 2014;180:129–39. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 4.Guertin KA, Moore SC, Sampson JN, Huang WY, Xiao Q, Stolzenberg-Solomon RZ, et al. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014;100:208–17. doi: 10.3945/ajcn.113.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Floegel A, von Ruesten A, Drogan D, Schulze MB, Prehn C, Adamski J, et al. Variation of serum metabolites related to habitual diet: a targeted metabolomic approach in EPIC-Potsdam. European journal of clinical nutrition. 2013;67:1100–8. doi: 10.1038/ejcn.2013.147. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt JA, Rinaldi S, Ferrari P, Carayol M, Achaintre D, Scalbert A, et al. Metabolic profiles of male meat eaters, fish eaters, vegetarians, and vegans from the EPIC-Oxford cohort. Am J Clin Nutr. 2015;102:1518–26. doi: 10.3945/ajcn.115.111989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vazquez-Fresno R, Llorach R, Urpi-Sarda M, Lupianez-Barbero A, Estruch R, Corella D, et al. Metabolomic pattern analysis after mediterranean diet intervention in a nondiabetic population: a 1- and 3-year follow-up in the PREDIMED study. J Proteome Res. 2015;14:531–40. doi: 10.1021/pr5007894. [DOI] [PubMed] [Google Scholar]
- 8.Edmands WMB, Ferrari P, Rothwell JA, Rinaldi S, Slimani N, Barupal DK, et al. Polyphenol metabolome in human urine and its association with intake of polyphenol-rich foods across European countries. Am J Clin Nutr. 2015;102:905–13. doi: 10.3945/ajcn.114.101881. [DOI] [PubMed] [Google Scholar]
- 9.Gu F, Derkach A, Freedman ND, Landi MT, Albanes D, Weinstein SJ, et al. Cigarette smoking behaviour and blood metabolomics. Int J Epidemiol. 2015:1–12. doi: 10.1093/ije/dyv330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross AJ, Boca S, Freedman ND, Caporaso NE, Huang WY, Sinha R, et al. Metabolites of tobacco smoking and colorectal cancer risk. Carcinogenesis. 2014;35:1516–22. doi: 10.1093/carcin/bgu071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao Q, Moore SC, Keadle SK, Xiang YB, Zheng W, Peters TM, et al. Objectively measured physical activity and plasma metabolomics in the Shanghai Physical Activity Study. Int J Epidemiol. 2016:1–12. doi: 10.1093/ije/dyw033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drogan D, Dunn WB, Lin WC, Buijsse B, Schulze MB, Langenberg C, et al. Untargeted Metabolic Profiling Identifies Altered Serum Metabolites of Type 2 Diabetes Mellitus in a Prospective, Nested Case Control Study. Clin Chem. 2015;61:487–97. doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- 13.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 15.Mondul AM, Moore SC, Weinstein SJ, Karoly ED, Sampson JN, Albanes D. Metabolomic analysis of prostate cancer risk in a prospective cohort: The alpha-tocolpherol, beta-carotene cancer prevention (ATBC) study. Int J Cancer. 2015;137:2124–32. doi: 10.1002/ijc.29576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross AJ, Moore SC, Boca S, Huang WY, Xiong X, Stolzenberg-Solomon R, et al. A prospective study of serum metabolites and colorectal cancer risk. Cancer. 2014;120:3049–57. doi: 10.1002/cncr.28799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liesenfeld DB, Grapov D, Fahrmann JF, Salou M, Scherer D, Toth R, et al. Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: the ColoCare study. Am J Clin Nutr. 2015;102:433–43. doi: 10.3945/ajcn.114.103804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, et al. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014;35:2089–96. doi: 10.1093/carcin/bgu131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, et al. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016;11:e0152126. doi: 10.1371/journal.pone.0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai V, Morris JG., Jr Need for prospective cohort studies to establish human gut microbiome contributions to disease risk. J Natl Cancer Inst. 2013;105:1850–1. doi: 10.1093/jnci/djt349. [DOI] [PubMed] [Google Scholar]
- 21.Fu BC, Randolph TW, Lim U, Monroe KR, Cheng I, Wilkens LR, et al. Characterization of the gut microbiome in epidemiologic studies: the multiethnic cohort experience. Ann Epidemiol. 2016;26:373–9. doi: 10.1016/j.annepidem.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–76. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes E, Wijeyesekera A, Taylor-Robinson SD, Nicholson JK. The promise of metabolic phenotyping in gastroenterology and hepatology. Nat Rev Gastroenterol Hepatol. 2015;12:458–71. doi: 10.1038/nrgastro.2015.114. [DOI] [PubMed] [Google Scholar]
- 24.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–63. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinha R, Vogtmann E, Chen J, Amir A, Shi J, Sampson JN, et al. Fecal microbiome in epidemiologic studies” - Response. Cancer Epidemiol Biomarkers Prev. 2016 doi: 10.1158/1055-9965.EPI-16-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song SJ, Amir A, Metcalf JL, Amato KR, Xu ZZ, Humphrey G, et al. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems. 2016;1:e00021. doi: 10.1128/mSystems.00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha R, Chen J, Amir A, Vogtmann E, Shi J, Inman KS, et al. Collecting Fecal Samples for Microbiome Analyses in Epidemiology Studies. Cancer Epidemiol Biomarkers Prev. 2016;25:407–16. doi: 10.1158/1055-9965.EPI-15-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 29.Evans AM, Mitchell MW, Dai H, DeHaven CD. Categorizing Ion–Features in Liquid Chromatography/Mass Spectrometry Metobolomics Data. Metabolomics. 2012;2:1–8. [Google Scholar]
- 30.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampson JN, Boca SM, Shu XO, Stolzenberg-Solomon RZ, Matthews CE, Hsing AW, et al. Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiol Biomarkers Prev. 2013;22:631–40. doi: 10.1158/1055-9965.EPI-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Q, Moore SC, Boca SM, Matthews CE, Rothman N, Stolzenberg-Solomon RZ, et al. Sources of variability in metabolite measurements from urinary samples. PLoS One. 2014;9:e95749. doi: 10.1371/journal.pone.0095749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monleon D, Morales JM, Barrasa A, Lopez JA, Vazquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22:342–8. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- 35.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–51. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 36.Saric J, Wang Y, Li J, Coen M, Utzinger J, Marchesi JR, et al. Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J Proteome Res. 2008;7:352–60. doi: 10.1021/pr070340k. [DOI] [PubMed] [Google Scholar]
- 37.Gratton J, Phetcharaburanin J, Mullish BH, Williams HR, Thursz M, Nicholson JK, et al. Optimized Sample Handling Strategy for Metabolic Profiling of Human Feces. Anal Chem. 2016;88:4661–8. doi: 10.1021/acs.analchem.5b04159. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs DM, Deltimple N, van Velzen E, van Dorsten FA, Bingham M, Vaughan EE, et al. H-1 NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomed. 2008;21:615–26. doi: 10.1002/nbm.1233. [DOI] [PubMed] [Google Scholar]
- 39.Zheng H, Yde CC, Clausen MR, Kristensen M, Lorenzen J, Astrup A, et al. Metabolomics Investigation To Shed Light on Cheese as a Possible Piece in the French Paradox Puzzle. J Agr Food Chem. 2015;63:2830–9. doi: 10.1021/jf505878a. [DOI] [PubMed] [Google Scholar]
- 40.Lamichhane S, Yde CC, Forssten S, Ouwehand AC, Saarinen M, Jensen HM, et al. Impact of Dietary Polydextrose Fiber on the Human Gut Metabolome. J Agr Food Chem. 2014;62:9944–51. doi: 10.1021/jf5031218. [DOI] [PubMed] [Google Scholar]
- 41.Yen S, McDonald JAK, Schroeter K, Oliphant K, Sokolenko S, Blondeel EJM, et al. Metabolomic Analysis of Human Fecal Microbiota: A Comparison of Feces-Derived Communities and Defined Mixed Communities. J Proteome Res. 2015;14:1472–82. doi: 10.1021/pr5011247. [DOI] [PubMed] [Google Scholar]
- 42.Floegel A, Drogan D, Wang-Sattler R, Prehn C, Illig T, Adamski J, et al. Reliability of Serum Metabolite Concentrations over a 4-Month Period Using a Targeted Metabolomic Approach. Plos One. 2011:6. doi: 10.1371/journal.pone.0021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholson G, Rantalainen M, Maher AD, Li JV, Malmodin D, Ahmadi KR, et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol Syst Biol. 2011;7:525. doi: 10.1038/msb.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.