Abstract
Genetic studies in fruit flies have implicated the chromatin remodeling complex NURF in immunity, but it has yet to be studied in mammals. Here we show that its targeting in mice enhances antitumor immunity in two syngeneic models of cancer. NURF was disabled by silencing of BPTF, the largest and essential subunit of NURF. We found that both CD8+ and CD4+ T cells were necessary for enhanced antitumor activity, with elevated numbers of activated CD8+ T cells observed in BPTF-deficient tumors. Enhanced cytolytic activity was observed for CD8+ T cells cocultured with BPTF silenced cells. Similar effects were not produced with TCR transgenic CD8+ T cells, implicating the involvement of novel antigens. Accordingly, enhanced activity was observed for individual CD8+ T cell clones from mice bearing BPTF silenced tumors. Mechanistic investigations revealed that NURF directly regulated the expression of genes encoding immunoproteasome subunits Psmb8 and Psmb9 and the antigen transporter genes Tap1 and Tap2. The PSMB8 inhibitor ONX-0914 reversed the effects of BPTF ablation, consistent with a critical role for the immunoproteasome in improving tumor immunogenicity. Thus, NURF normally suppresses tumor antigenicity and its depletion improves antigen processing, CD8 T cell cytotoxicity and antitumor immunity, identifying NURF as a candidate therapeutic target to enhance antitumor immunity.
Keywords: NURF, antigenicity, immunoproteasome, TAP, chromatin remodeling, antitumor immunity, CD8 T cell, immunotherapy
Introduction
Tumors which present in the clinic adapt to suppress antitumor immunity by reducing antigenicity, deregulating immune checkpoints, or amplifying immune suppressing cells (1). Immunotherapies block these adaptations and reestablish antitumor immunity for therapeutic benefit. In rare cases immunotherapies have positive therapeutic outcomes, but in a majority of cases they do not have lasting antitumor effects due to tumor adaptation (2). There is hope that combination therapy may be more effective, but to achieve these outcomes, we must discover novel pathways and targets which can be exploited for therapeutic advantage.
Decades of research in epigenetics has characterized many protein complexes with important roles in regulating nuclear processes. These complexes largely operate on the nucleosome, which is the fundamental unit of chromatin structure (3). One class of complexes which modifies the position and structure of nucleosomes are the ATP-dependent chromatin remodeling complexes (4). These chromatin remodeling processes are frequently essential for development, but are not generally necessary for cell survival (5-7). In addition to roles in normal development, nucleosome remodeling complexes have prominent roles in cancer specific biology, including putative driver functions for a variety of human cancers (8).
One largely uncharacterized chromatin remodeling complex is the nucleosome remodeling factor (NURF)(9). Mammalian NURF is composed of three subunits which include the largest, essential and unique subunit bromodomain PHD-finger containing transcription factor (BPTF), the ATPase SNF2L and a WD repeat containing protein pRBAP46/48 (10,11). These subunits recruit NURF to chromatin through interactions with sequence specific transcription factors or modified histones. Once recruited, NURF slides nucleosomes in cis to alter nucleosome positioning without histone eviction or exchange. These sliding reactions alter DNA accessibility to regulate transcription factor binding, and as a result gene expression (9).
Our previous studies demonstrate that BPTF preferentially regulates chromatin structure and the expression of the major histocompatibility locus (MHC) genes (5-7). These genes predominantly function to process and present antigens to immune cells. MHC genes with these functions include Psmb9 and Psmb8, which encode subunits of the immunoproteasome, and Tap1 and Tap2, which compose the transporter associated with antigen processing (TAP) complex (12). The immunoproteasome is a specialized proteasome which creates peptides with higher affinity to MHC I molecules, and are therefore more antigenic to CD8 T cells (12-14). Because genes located in the MHC are important for CD8 T cell activity, they are frequently repressed by tumor cells to avoid antitumor immunity (15). In an attempt to improve the immunogenicity of tumor cells, we depleted BPTF in the well-established B16F10 and 4T1 tumor models and monitored for changes in MHC expression and antitumor immunity.
Materials and Methods
Mice
BALB/c, C57BL/6, NOD/SCID, Ifrg2r −/− (NSG), B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J (Jackson Laboratories) and OT1 (Gift from Dr. Shawn Wang, VCU) female mice 6-8 weeks of age were housed under aseptic conditions. These studies have been approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Commonwealth University.
Cell Culture
4T1 (Fred Miller, Wayne State University in 2010), B16F10 (ATCC in 2011) and HEK 293T (ATCC in 2010) were cultured in complete media (CM) (DMEM (Life Sciences), 10% FBS (Hyclone), 1% NEAA, 2 mM glutamine and 1% Pen/Strep (Life Technologies)). T cells were cultured in CM with 10 mM HEPES, 5×10−5 M ß-mercaptoethanol (Sigma) and 50 U/ml IL-2 (R&D Systems). Hybridoma lines were cultured in RPMI-1640 (Life Sciences), 10% FBS, 1% NEAA, 2 mM glutamine and 1% Pen/Strep. Population doubling times were calculated by standard methods. Cell lines were authenticated by ATCC prior to shipment by short tandem repeat (STR) profiling.
Short-hairpin RNAs (shRNA) were introduced into 4T1 and B16F10 cells using Moloney murine leukemia virus (MMLV) using the pSIREN-RetroQ (Clonetech) system. Transduced cells were selected with 0.5 μg/ml puromycin (Life Technologies) after 48 hours. B16F10 cells were transfected with CMV-OVA along with a zeocin selectable marker and selected with 400 μg/ml zeocin (Invitrogen). Individual colonies were cloned and assayed for OVA expression, BPTF was then KD with a shRNA in OVA expressing lines. pSIREN plasmids Ctrl-sh1, Ctrl-sh2, Bptf-sh1 (4T1), Bptf-sh2 (4T1), Bptf-sh1 (B16F10), and Bptf-sh2 (B16F10) are available at Addgene as stock numbers 73665, 73666, 73669, 73668, 73667 and 73669, respectively.
Tumor Studies
1×104 4T1 cells were injected into the fourth mammary fat pad of BALB/c or NSG mice and tumors were analyzed at 21 days. 5×104 B16F10 cells were injected subcutaneously (S.C.) into the flank of C57BL/6 mice and tumors were analyzed at 18 days. For gemcitabine treated mice, 1×105 4T1 or 5×105 B16F10 cells were injected into BALB/c, NSG or C57BL/6 mice and 1.2 mg gemcitabine (Hospira) was injected intraperitoneously (I.P.) on day 5 and every 7 days following until tumors were analyzed on day 37 (4T1) or day 18 (B16F10).
mAb Depletions
GK1.5 (anti-CD8), 2.43 (anti-CD4) (Gift from Dr. Mosoud Manjili, VCU in 2012) and SH-34 (anti-asialo GM1) (ATCC in 2013) mAbs were purified from ascites fluid by ammonium sulfate fractionation (16). 225 μg/mouse mAb was injected I.P. into BALB/c or C57BL/6 mice on day -2 and day -1 and mice were inoculated with 1×105 4T1 or 5×104 B16F10 tumor cells on day 0. mAb was injected once every 5 days following tumor inoculation and 4T1 bearing mice were treated with gemcitabine as described previously. Hybridomas were authenticated by confirming antigen specific reactivity of produced antibodies by flow cytometry using commercial anti-CD8, anti-CD4, anti-asialo GM1 antibody standards. Depletion of CD8, CD4 T cells and NK cells from mice were comfirmed by flow cytometry.
Western Blot
Protein from cell cultures or homogenized tumors was extracted by TRI Reagent (Sigma). Primary antibodies: anti-BPTF (Millipore), anti-OVALBUMIN, anti-PMEL17 (Santa Cruz Biotechnology), anti-CYCLOPHILIN B (Abcam), anti-PSMB9, anti-TAP2 (Thermo Scientific), anti-PSMB8 and anti-TAP1 (Cell Signaling). Secondary antibody: horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse secondary antibody (Cell Signaling).
Metastasis Assay
Colony formation assay from the lungs of 4T1 tumor bearing mice were performed as previously described (17).
qRT-PCR
Total RNA was extracted from cultured cells with TRI Reagent and 5 μg RNA was reverse transcribed using Superscript II First-strand kit (Invitrogen). qPCR was performed using SYBR green PCR master mix (Biorad) with a 7900 HT fast real-time qPCR system (Applied Biosystems). The ddCt method was performed using normalization to Gapdh. Primer pairs are found in Supplementary DataSet S1.
Flow Cytometry
Cultured 4T1 and B16F10 cells were stained with H2K, H2D, Annexin V, OVA and 7AAD viability dye. Mean fluorescence intensity (MFI) was calculated using Cyflogic. To stain infiltrating lymphocytes, tumors were first digested with DNaseI and Collagenase and lymphocytes were purified using Percoll gradients as previously described (18). Purified lymphocytes were subsequently stained with CD8, CD69, TCRb, CD44, BTLA antibodies and 7AAD viability dye. All antibodies were purchased from BD Pharmingen.
Chromatin Immunoprecipitation
ChIP was performed as previously described (7). Briefly, cells were fixed in 1% paraformaldehyde for 15 min and chromatin was sheared to an average size of 300 bp by sonication. Lysates were incubated with 0.5 μg anti-BPTF (Millipore) or control rabbit IgG bound to Protein G Dyna beads (Life Technologies) overnight, subsequently washed with low and high salt buffers and eluted with 0.1 M NaHCO3 in 1% SDS. Primer pairs are found in Supplementary DataSet S1.
CTL Cytotoxicity Assay
CD8a+ cells were purified by negative selection using MACS Separation Columns. The purified CTLs were restimulated with mitomycin C-treated 4T1 or B16F10 tumor cells. CTLs were harvested, purified with by percoll gradient and cocultured with mitomycin C-treated tumor cells in 96-well plates. For hyperactivation, purified splenic CD8a+ T cells were treated with 0.8 μM PMA, 0.35 μM Ionomycin and 500 U/ml IL-2 on mitomycin C-treated tumor cells. For total splenocyte and OVA analysis, splenocytes from tumor bearing mice or naïve OT1 mice were restimulated in vitro as described. For ONX-0914 treatment, ONX-0914 was added to targets for 24 hours before incubation with splenocytes at a 50:1 E:T ratio. For all assays, T cells/splenocytes were incubated on targets for 48 hours and cell death was measured using the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega).
ELISPOT Assay
2×106 B16F10 tumor infiltrating lymphocytes were cocultured with 1×106 splenocytes from naïve mice and stimulated with 1 μg/ml Gp10025–33 (KVPRNQDWL), TRP2180-188 (SVYDFFVWL) or p15E604-611 (KSPWFTTL) and 50 U/ml IL-2 for 96 hours at 37°C in wells precoated with IFN-γ capture antibody. ELISPOT was performed as previously described (19).
Limited Dilution
T cell limited dilution was performed as previously described (20). Clones were assayed for activity against control and BPTF KD targets with the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega).
Formaldehyde Assisted Isolation of Regulatory Elements (FAIRE)
FAIRE was performed essentially as described (21). FAIRE results were normalized to a control locus which does not have BPTF dependent FAIRE when normalized to equal DNA mass. Primer pairs are found in Supplementary DataSet S1.
Microarray
RNA extraction, microarray analysis and statistical analysis were performed as described (22,23). Briefly, necrotic tissue was selected out by macrodissection and total RNA was extracted from frozen tumor tissues with TRI reagent MagMAX-96 Total RNA Isolation Kit. Biotinylated cRNA was generated with the GeneChip 3’IVT Express kit and 10μg was applied to the GeneChip®Mouse Genome 430A 2.0 Array. Arrays were scanned using an Affymetrix GeneChip Scanner 3000 and data was processed as described as described in GEO accession GSE71864.
Results
BPTF Depletion Increases T Cell Antitumor Immunity to 4T1 Tumors
Consistent with previous work showing that subunits of the NURF complex (Fig. 1A) are frequently overexpressed in cancer cells (8), we found each of the NURF subunits is expressed in a variety of mouse and human breast cancer cell lines (Supplementary Fig. S1A). To discover functions of NURF in cancer cell biology, we repressed BPTF expression in mouse 4T1 cells by retroviral introduced shRNA knock-down (KD) (Fig. 1B). In vitro, BPTF KD cells show no significant differences in doubling time, cellular morphology or levels of apoptosis (Supplementary Fig. S1B-D).
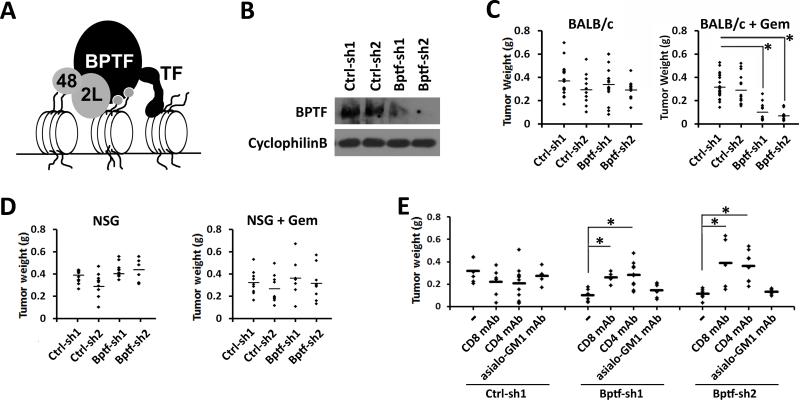
Figure 1.
Depletion of BPTF reduces 4T1 tumor weights in mice with CD8+ and CD4+ T cells. A, cartoon of NURF (BPTF, 2L, 48) bound to chromatin. Grey circles: histone modifications, TF: transcription factor. B, BPTF Western blot analysis from control (Ctrl-sh1, Ctrl-sh2) and BPTF KD (Bptf-sh1, Bptf-sh2) 4T1 total cell extracts. Cyclophilin B loading control. C-D, weights of primary control and BPTF KD 4T1 tumors after growth in C, BALB/c mice (n ≥ 12, * = ttest pvalue < 9.6×10−7) or D, NSG mice (n = 9). E, weights of 4T1 tumors after growth in undepleted, CD8+, CD4+ or asialo-GM1+ mAb depleted BALB/c mice. (n ≥ 5, * = ttest pvalue < 0.02).
To uncover functions for BPTF in tumor biology, we introduced BPTF KD 4T1 cells into the 4th mammary fat pad of syngeneic BALB/c mice. As part of our studies, we inoculated the cells into both untreated and gemcitabine treated mice. Gemcitabine is a routinely used chemotherapeutic which inhibits DNA replication and induces apoptosis in tumors (16). We observed significantly reduced weights for BPTF KD tumors only in gemcitabine treated mice (Fig. 1C). We chose weight and not volume to monitor tumor growth because BPTF KD tumors are morphologically flat compared to controls, confounding a volume based measurement (Supplementary Fig. S2A,B).
In addition to inducing cancer cell apoptosis, gemcitabine also inhibits the proliferation of several cells of the immune system, most prominently myeloid derived suppressor cells (MDSC) (16,24-27). MDSCs are dramatically amplified in mice bearing 4T1 tumors, which suppresses the antitumor immune response (25). To determine if the effect of gemcitabine on BPTF KD tumors is a result of enhanced antitumor immunity, we repeated our tumor studies in immune compromised NOD/SCID, Ifrg2r −/− (NSG) mice (28). In NSG mice, we observed similar growth of control and BPTF KD tumors with gemcitabine treatment (Fig. 1D), demonstrating that the reduced tumor growth in BALB/c mice is not due to gemcitabine alone, but rather in combination with antitumor immunity.
To determine if BPTF KD cells are eliminated from tumors grown in immune competent mice, we measured BPTF levels in the primary tumors. We show that BPTF expression increases in tumors from BALB/c (Supplementary Fig. S2C) but not NSG mice (Supplementary Fig. S2D), suggesting that a functional immune system either directly or indirectly selects for cells which re-express BPTF.
To understand the immune response to gemcitabine treated BPTF KD 4T1 tumors, we selectively depleted CD8+ T cells, CD4+ T cells or NK cells by antibody dependent cellular cytotoxicity (ADCC). We observed a rescue of BPTF KD tumor growth when CD8+ or CD4+ cells were depleted (Fig. 1E), demonstrating that the antitumor response to BPTF KD 4T1 tumors requires T cells.
MDSC Amplification and Tumor Metastasis are BPTF-Independent
4T1 tumors dramatically amplify MDSCs, resulting in splenomegaly with spleen size proportional to the degree of MDSC amplification (25). As previously reported, we observed a significant increase in spleen weight with introduction of 4T1 tumors into BALB/c mice (Supplementary Fig. S2E). The observed increase in spleen weight was proportional to tumor weight and was not affected by BPTF KD, demonstrating that splenomegaly is BPTF-independent (Supplementary Fig. S2F).
The 4T1 model is also widely used to study breast cancer cell metastasis (17). To determine if BPTF regulates metastasis, we analyzed metastases to the lung of both untreated and gemcitabine treated BALB/c mice using a colony formation assay (17). We show that tumor size, but not BPTF KD, was significantly correlated with the number of colonies, suggesting that metastasis of 4T1 is BPTF-independent (Supplementary Fig. S2G,H).
BPTF Depletion Increases T Cell Antitumor Immunity to B16F10 Tumors
To further verify our findings, we used the B16F10 melanoma model which is syngeneic to C57BL/6 (29). First, we confirmed that each NURF subunit is expressed in B16F10 cells (Supplementary Fig. S3A). Additionally, BPTF KD (Fig. 2A) had no significant effect on doubling time, cellular morphology or apoptosis of B16F10 cells (Supplementary Fig. S3B-D).
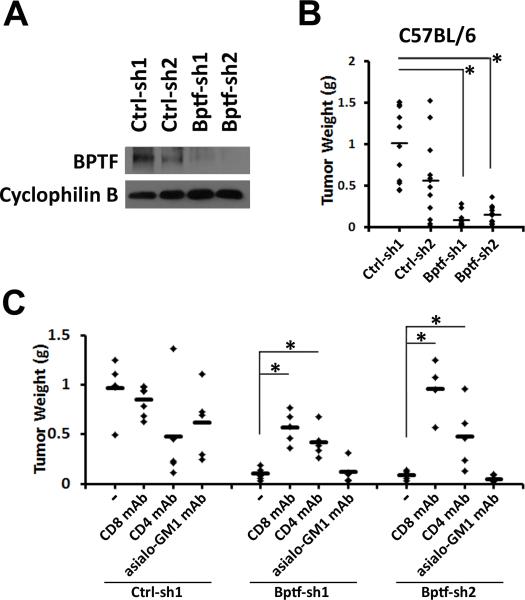
Figure 2.
BPTF depletion reduces B16F10 tumor weights in mice with CD8+ and CD4+ T cells. A, BPTF Western blot analysis of control (Ctrl-sh1, Ctrl-sh2) and BPTF KD (Bptf-sh1, Bptf-sh2) B16F10 total cell extracts. Cyclophilin B loading control. B, weights of primary control and BPTF KD B16F10 tumors after growth in C57BL/6 mice (n ≥ 9, * = ttest pvalue < 1.4×10−7). C, weights of B16F10 tumors after growth in undepleted, CD8+, CD4+ or asialo-GM1+ mAb depleted C57BL/6 mice. (n ≥ 5, * = ttest pvalue < 0.02).
Consistent with 4T1, B16F10 KD tumors had a flattened morphology, preventing caliper measurements from accurately comparing tumor growth (Supplementary Fig. S3E,F). As with 4T1, we observed significant reductions in BPTF KD B16F10 tumor weights (Fig. 2B). However, unlike in the 4T1 model, gemcitabine treatment did not selectively reduce BPTF KD B16F10 tumor growth (Supplementary Fig. S3G).
To determine if CD4 and CD8 T cells are required for reduced B16F10 BPTF KD tumor growth, we repeated our ADCC experiments. Consistent with an enhanced T cell response, we observed a rescue of BPTF KD tumor growth with CD4+ and CD8+ cell depletion, but not NK cell depletion (Fig. 2C). These results in combination support a model of an enhanced CD8 T cell cytotoxic response to BPTF KD tumors, which requires CD4 T helper cell activity.
CD8 T Cells are More Abundant and Activated in BPTF KD Tumors
We then measured the infiltration and activation status of intratumoral CD8 T cells by flow cytometry. As expected, we observed a greater number of CD8 T cells (CD8+, TCRb+) in the BPTF KD tumor microenvironment (Fig. 3A,B)(Supplementary Fig. S4). We next measured the abundance of activation markers CD69 and CD44 and the anergy marker BTLA on intratumoral CD8 T cells. We observed a greater percentage of CD69high CD8+ cells in both 4T1 and B16F10 BPTF KD tumors (Fig. 3C,D). In contrast, CD44 and BTLA expression differed significantly on CD8 T cells between control and BPTF KD B16F10, but not 4T1, tumors (Fig. 3E-H). This could be the result of differences in the tumor microenvironment between 4T1 and B16F10 (see Discussion). Together, these results indicate that the BPTF KD tumor microenvironment has a greater number of active CD8 T cells.
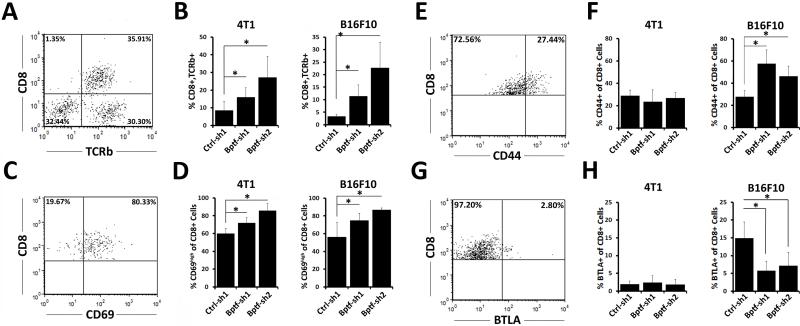
Figure 3.
Enhanced presence and activity of CD8 T cells in BPTF KD tumors. A,C,E,G representative dot plots of live 4T1 tumor infiltrating lymphocytes stained for CD8 and TCRb, CD69, CD44 or BTLA, respectively. B, percentages of live intratumor CD8+,TCRb+ lymphocytes as a percent of all live lymphocytes from 4T1 and B16F10 tumors (n ≥ 7, * = ttest pvalue < 0.05). D,F,H, percentages of intratumor CD8+ lymphocytes that are CD69high, CD44+ or BTLA+ from 4T1 and B16F10 tumors, respectively (n = 6, * = ttest pvalue < 0.05).
CD8 T Cells have Enhanced Cytotoxic Activity Toward BPTF KD Cells In Vitro
To investigate if BPTF KD 4T1 and B16F10 tumor cells are more efficiently targeted by CD8 T cells, we performed in vitro cytotoxicity assays. Coculture of purified splenic CD8 T cells isolated from BPTF KD tumor bearing mice with BPTF KD targets results in enhanced cytolytic activity compared to similar experiments using controls (Fig. 4A). We next determined if BPTF KD cells are more susceptible to T cell induced cell death. Toward this end, we used PMA + ionomycin activated naïve CD8 T cells and observed similar cytolytic activity between control and BPTF KD targets (Fig. 4B). These results demonstrate that enhanced CD8 T cell killing of BPTF KD target cells was not due to increased sensitivity to CD8 T cell mediated cell death.
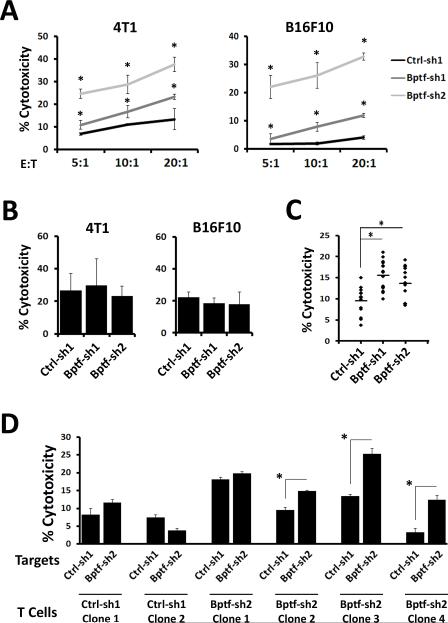
Figure 4.
BPTF depletion sensitizes tumor cells to CD8 T cell cytotoxicity in vitro. A-D, percent target cell cytotoxicity determined by LDH release. A, purified splenic CD8 T cells from control or BPTF KD tumor bearing mice were cocultured on control or BPTF KD targets, respectively, at the indicated effector to target (E:T) ratios. (n = 3, * = ttest pvalue < 0.05). B, coculture of naïve purified CD8 T cells treated with PMA + Ionomycin with 4T1 or B16F10 targets at a 10:1 E:T ratio. C, CD8 T cell clones isolated from spleens of 4T1 control or BPTF KD tumor bearing mice were cocultured with 4T1 control or BPTF KD targets, respectively, at a 10:1 E:T ratio. Each dot represents one clone and is an average of 3 biological replicates (* = ttest pvalue < 5.0×10−3). D, six representative CD8 T cell clones from panel C were cocultured with either control or BPTF KD 4T1 targets at a 10:1 E:T ratio. Results are representative of 3 biological replicates for each clone (* = ttest pvalue < 0.04).
To investigate if BPTF KD cells are more antigenic than control cells, we used the OT1 and pmel CD8 T cell TCR transgenic models (30,31). The pmel TCR recognizes peptides from the endogenously expressed Pmel, whereas the OT1 TCR recognizes peptides from chicken Ovalbumin (OVA) presented by H2-Kb, which is expressed on B16F10, but not 4T1 (30,31).
Coculture experiments show that neither pmel nor OT1 CD8 T cells had enhanced reactivity to BPTF KD B16F10 cells (Supplementary Fig. S5A,B) even though they expressed equivalent OVA and increased PMEL protein (Supplementary Fig. S5C,D). Consistent with BPTF-independent OVA antigen presentation, an antibody which recognizes the OVA peptide (SIINFEKL) in context with H2-Kb (32) equivalently stains control and BPTF KD B16F10 cells (Supplementary Fig. S5E). To further characterize the CD8 T cell response to known antigens, we quantified gp100, TRP2, and p15E reactive intratumoral CD8 T cells by ELISPOT. From this we observed similar numbers of peptide reactive CD8 T cells between control and BPTF KD tumors (Supplementary Fig. S5F). Together, these results suggest that enhanced CD8 T cell activity to BPTF KD tumors occurs to novel antigens.
To investigate if BPTF KD cells present novel antigens, we cloned CD8 T cells from both BPTF KD and control tumor bearing mice. Coculture assays showed enhanced activity of CD8 T cell clones from BPTF KD tumor bearing mice to BPTF KD target cells compared to similar experiments using controls (Fig. 4C). Furthermore, this enhanced activity usually occurs only when cocultured with BPTF KD, but not control, target cells (5 of 9 assayed clones)(Fig. 4D). In contrast, we do not observe enhanced cytolytic activity when CD8 T cells isolated from control tumor bearing mice are cocultured with BPTF KD target cells (0 of 4 assayed clones)(Fig. 4D). These results support the hypothesis that BPTF KD tumors express novel antigens.
BPTF Directly Regulates Antigen Processing
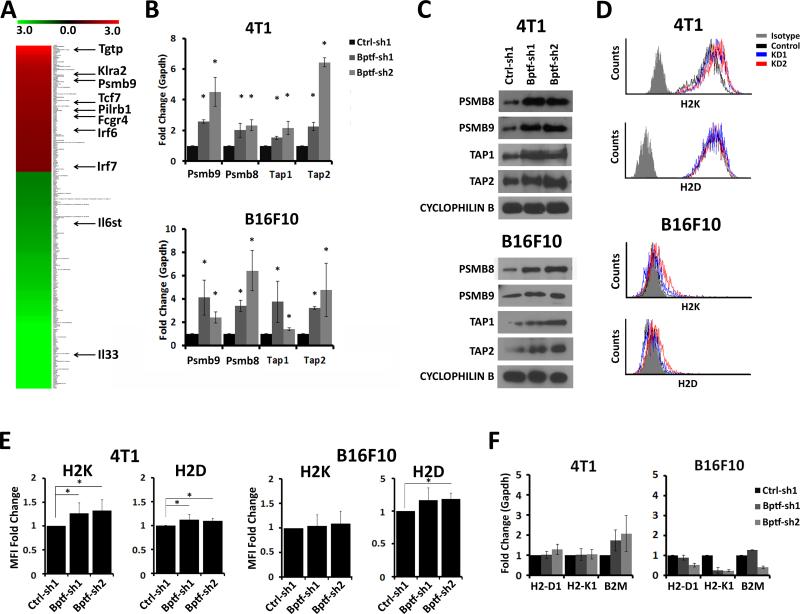
To identify BPTF-dependent genes which could alter tumor cell antigenicity, we performed genome wide expression arrays on 4T1 tumors harvested from NSG mice (Fig. 5A). We observed 115 upregulated genes and 199 down regulated genes (>1.5 fold change in expression, FDR<0.05)(Fig. 5A)(Supplementary DataSet S1, Supplementary Table S1). Of genes identified, we focused on Psmb9 because it regulates antigenicity as a subunit of the immunoproteasome (33). We confirmed Psmb9 upregulation in vitro, and observed elevated expression of the neighboring antigen processing genes Psmb8, Tap1 and Tap2 in both tumor models (Fig. 5B,C). The upregulation of these genes correlates with equivalent to slight increases in cell surface expression of H2K or H2D in BPTF KD cells as measured by MFI, which were not the result of increased gene expression (Fig. 5D-F). In addition, the expression of the interferon inducible genes Oas1a and Oas2 were not upregulated with BPTF KD, indicating that upregulation of Psmb8, Psmb9, Tap1 and Tap2 are not a result of a general interferon response (Supplementary Fig. S6A).
Figure 5.
BPTF regulates immunoproteasome and TAP subunit expression. A, microarray heat map of genes significantly deregulated in BPTF KD 4T1 tumors from NSG mice (n = 3). Scale represents +/− 3 fold expression change. Genes related to the immune response are highlighted. B, qRT-PCR analysis of Psmb8, Psmb9, Tap1 and Tap2 expression from control or BPTF KD 4T1 and B16F10 cells. (n = 3 biological replicates, * = ttest pvalue < 0.05). C, PSMB8, PSMB9, TAP1 and TAP2 Western blot analysis of 4T1 (top) and B16F10 (bottom) total cell extracts. D, representative flow cytometry histograms of 4T1 and B16F10 cells stained for the MHC class I molecules H2K and H2D. E, fold change in MFI of H2K and H2D (n ≥ 3 biological replicates, * = ttest pvalue < 0.05). F, qRT-PCR analysis of MHC class I gene expression from 4T1 and B16F10 cells (n = 3 biological replicates).
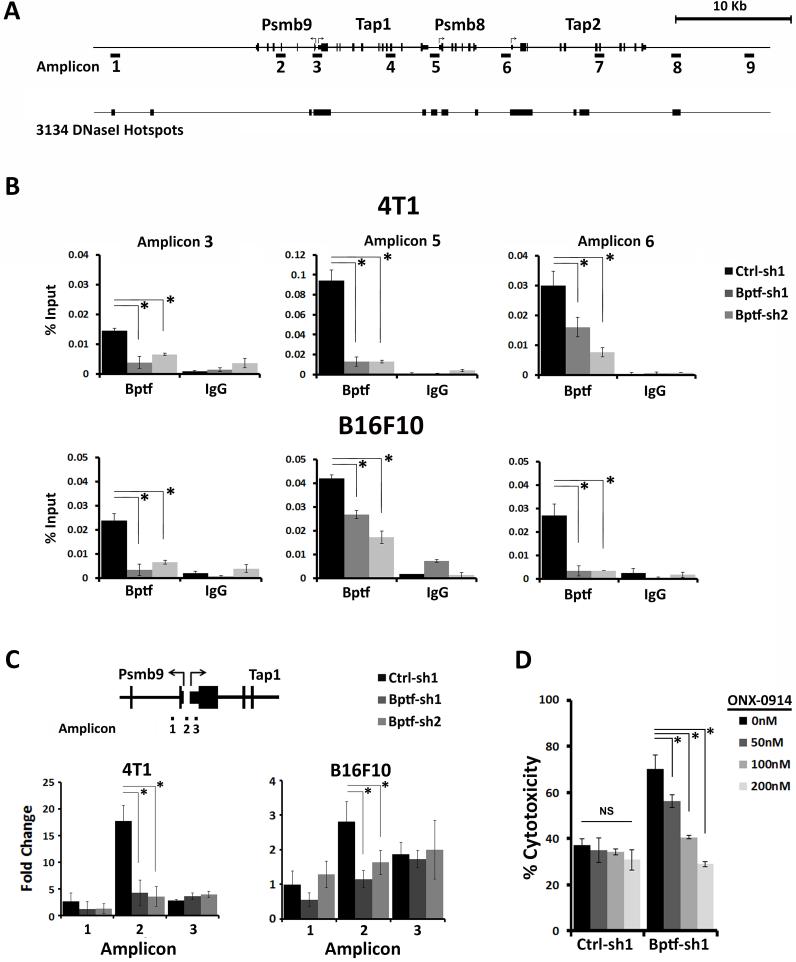
To determine if BPTF directly regulates the expression of Psmb8, Psmb9, Tap1 and Tap2, we measured BPTF occupancy by chromatin immunoprecipitation (ChIP) (34). Sites chosen for ChIP were guided by previously identified DNaseI hypersensitivity hotspots, and therefore possible regulatory elements, from genome wide studies done in the 3134 mouse mammary epithelial cell line (35) (Fig. 6A). The most consistent enrichment of BPTF between 4T1 and B16F10 was detected at the promoters, consistent with BPTF directly repressing these genes (Fig. 6B, Supplementary Fig. S6B,C). To determine if BPTF chromatin remodeling activity could be relevant to changes in gene expression, we focused on the well characterized Psmb9-Tap1 divergent promoter (36), using formaldehyde assisted isolation of regulatory elements (FAIRE) (21). With this technique, open chromatin is isolated from fixed cells using phenol extractions and quantified by qPCR relative to a BPTF independent reference site. FAIRE shows that BPTF maintains chromatin structure of the Psmb9-Tap1 promoter in both cell lines (Fig. 6C).
Figure 6.
BPTF occupies and regulates chromatin structure at Psmbs and TAPs. A, cartoon showing the position of PCR amplicons used for ChIP analysis and DNaseI hotspots from the mouse mammary epithelial line 3134 (35). B, BPTF ChIP at Psmb9, Psmb8, Tap1 and Tap2 promoters (n = 3 biological replicates, * = ttest pvalue < 0.05). C, FAIRE at the divergent Psmb9-Tap1 promoter. Values are normalized to a BPTF-independent control site (n = 3 biological replicates, * = ttest pvalue < 0.05). D, percent target cell cytotoxicity determined by LDH release for 4T1 splenocytes from control or BPTF KD tumor bearing mice stimulated at a 50:1 E:T ratio on control or BPTF KD targets, respectively, after treatment with 50-200nM ONX-0914 (n = 3 biological replicates, * = ttest pvalue < 0.04).
To verify that increased expression of the immunoproteasome subunits is responsible for the enhanced antigenicity of BPTF KD cells, we utilized the PSMB8 selective inhibitor ONX-0914 (37). Cytotoxicity assays show that enhanced CD8 T cell cytotoxicity to BPTF KD targets is ablated after treatment with ONX-0914 (Fig. 6D). These results allow us to propose a model where BPTF depletion upregulates the antigen processing genes Psmb8, Psmb9, Tap1 and Tap2, which results in enhanced antigenicity and improved T cell antitumor immunity. As a corollary, we propose the BPTF normally suppresses antitumor immunity by repressing antigen processing in cancer cells.
Discussion
Using KD of BPTF as a means to deplete the NURF complex, we show that it suppresses T cell antitumor immunity in two mouse tumor models with different genetic backgrounds. Intratumor CD8 T cells are more active in BPTF KD tumors. Differences between the tumor models include CD44 and BTLA expression. This is partially the result of MDSC activity in 4T1 tumors, which is known to suppress CD44, but not CD69, abundance on CD8 T cells (38). Differences in BTLA could result because the 4T1 tumor microenvironment has elevated costimulatory signals compared to B16F10, preventing T cell anergy (39). It is unlikely that BPTF regulates costimulatory molecules on the tumor because not all CD8 T cell clones are preferentially reactive to BPTF KD targets. Our in vitro assays reveal that BPTF protects tumor cells from direct CD8 T cell cytotoxic activity. Enhanced antitumor activity to BPTF KD cells is observed only when CD8 T cells are primed and activated with BPTF KD tumor cells, suggesting that BPTF suppresses antigen presentation. It is not likely that BPTF is a general regulator of antigen presentation because we observe approximately equivalent levels of cell surface H2K and H2D with BPTF KD. The degree to which an antigen stimulates T cell activity depends on both the number of an antigen:MHC complex presented and the quality of the antigen (40,41). We utilized OT1 and pmel transgenic CD8 T cells to determine if BPTF suppresses the presentation of known antigens (30,31). Despite equal or enhanced expression of OVA or PMEL antigens, coculture experiments show that BPTF does not lend protection against OVA and pmel reactive CD8 T cells. These results indicate that BPTF represses the presentation of select antigens to CD8 T cells. Consistent with this, assay of CD8 T cell clones isolated from 4T1 BPTF KD tumor bearing mice identified many clones with enhanced reactivity specifically toward BPTF KD targets. The identity of these antigens is currently unknown, but they likely result from BPTF regulating presentation of tumor specific antigens (TSA) or tumor associated antigens (TAA)(42). Our microarray analysis of 4T1 tumors did not reveal any known TSA, but these results would not detect differences in expression of alloantigens. The 4T1 tumor line has a BALB/cfC3H hybrid genotype, and therefore presents alleles from the C3H background, which would be recognized as alloantigens in BALB/c mice (17).
In addition to the presentation of TAA and TSA, novel antigens can be created by changes in antigen processing. From our experiments, we observe that BPTF represses the expression of Psmb8, Psmb9, Tap1, and Tap2. Upregulation of the immunoproteasome subunits Psmb8 and Psmb9 with BPTF depletion would result in increased immunoproteasome activity, generating more antigenic peptides (14,33,43). Peptides with greater antigenicity can more favorably bind to MHC molecules or promote higher affinity interactions with the TCR (40). In addition, upregulation of the TAPs alters the repertoire of peptides presented by changing the types of peptides transported into the ER for loading into MHC molecules (13). BPTF could directly regulate these genes because it is localized to the Psmb and Tap promoters in both cell lines. BPTF also occupies distal elements, most significantly in 4T1, which could also influence gene expression. BPTF, presumably through NURF, could remodel chromatin structure at either of these locations to repress gene expression. This is supported by an observed change in chromatin structure by FAIRE at the Psmb9-Tap1 promoter with BPTF KD. Psmb8, Psmb9, Tap1 and Tap2 promoters are regulated by interferon gamma through the STAT1 and IRF-1 transcription factors (44,45), and are repressed in tumor cells by DNA methylation and histone deacetylation (46-48). Our finding that BPTF represses their expression, presumably through the NURF complex, presents a novel epigenetic mechanism to suppress tumor cell antigenicity.
The NURF subunits are rarely deleted and frequently amplified or overexpressed in cancer cells, suggesting that a majority of tumor cells will have a NURF complex to inhibit (8,49). BPTF is not necessary for the viability of any primary cell type examined (5-7), suggesting that NURF inhibition may be tolerated in adults. Therefore, it is plausible that inhibiting NURF could be a viable strategy to improve tumor cell antigenicity. Further, NURF is an enzyme with several substrate binding sites (DNA, histones and ATP) and critical interaction surfaces with chromatin (transcription factors, DNA and modified histones), each of which could be targeted by small molecules (9).
As with most therapies, a NURF inhibitor would be more effective when used in combination with other chemo or immunotherapies. BPTF depletion improves antigenicity, but these effects are only relevant for antitumor immunity when MDSC abundance is low. Hence the need to use gemcitabine to deplete MDSCs in the 4T1 model. In contrast the B16F10 model has low levels of MDSC, not requiring the use of gemcitabine (50). The contrast between these two tumor models provides proof of principle for the utility of a NURF inhibitor in combinatorial therapeutic regimens.
Supplementary Material
Acknowledgements
The authors would like to thank Joyce Lloyd, Masoud Manjili, Tomas Kordula and Chunqing Guo for critically reading the manuscript. These experiments were funded by start-up funds from the Department of Human and Molecular Genetics (JWL), the Massey Cancer Center (JWL), and an early stage investigator grant from the V Foundation for Cancer Research (JWL).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Mittal D, Gubin MM, Schreiber RD, Smyth MJ. New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol. 2014;27:16–25. doi: 10.1016/j.coi.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake CG, Lipson EJ, Brahmer JR. Breathing new life into immunotherapy: review of melanoma, lung and kidney cancer. Nat Rev Clin Oncol. 2014;11(1):24–37. doi: 10.1038/nrclinonc.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol. 2010;2(5):a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker PB, Workman JL. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol. 2013;5(9) doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4(10):e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 2011;25(3):275–86. doi: 10.1101/gad.2007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu Z, Song C, Malakouti N, Murray D, Hariz A, Zimmerman M, et al. Functional interactions between NURF and Ctcf regulate gene expression. Mol Cell Biol. 2015;35(1):224–37. doi: 10.1128/MCB.00553-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayes K, Qiu Z, Alhazmi A, Landry JW. ATP-dependent chromatin remodeling complexes as novel targets for cancer therapy. Adv Cancer Res. 2014;121:183–233. doi: 10.1016/B978-0-12-800249-0.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alkhatib SG, Landry JW. The nucleosome remodeling factor. FEBS Lett. 2011;585(20):3197–207. doi: 10.1016/j.febslet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 2003;22(22):6089–100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, et al. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8(3):531–43. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- 12.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–36. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 13.Durgeau A, El Hage F, Vergnon I, Validire P, de Montpreville V, Besse B, et al. Different expression levels of the TAP peptide transporter lead to recognition of different antigenic peptides by tumor-specific CTL. J Immunol. 2011;187(11):5532–9. doi: 10.4049/jimmunol.1102060. [DOI] [PubMed] [Google Scholar]
- 14.Angeles A, Fung G, Luo H. Immune and non-immune functions of the immunoproteasome. Front Biosci (Landmark Ed) 2012;17:1904–16. doi: 10.2741/4027. [DOI] [PubMed] [Google Scholar]
- 15.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 16.Le HK, Graham L, Cha E, Morales JK, Manjili MH, Bear HD. Gemcitabine directly inhibits myeloid derived suppressor cells in BALB/c mice bearing 4T1 mammary carcinoma and augments expansion of T cells from tumor-bearing mice. Int Immunopharmacol. 2009;9(7-8):900–9. doi: 10.1016/j.intimp.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–405. [PubMed] [Google Scholar]
- 18.Yu XW, Sha C, Guo YL, Xiao R, Xu Y. High-level expression and characterization of a chimeric lipase from Rhizopus oryzae for biodiesel production. Biotechnol Biofuels. 2013;6(1):29. doi: 10.1186/1754-6834-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wang XY, Subjeck JR, Kim HL. Covalent crosslinking of tumor antigens stimulates an antitumor immune response. Vaccine. 2010;28(40):6613–20. doi: 10.1016/j.vaccine.2010.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassanin H, Serba S, Schmidt J, Marten A. Ex vivo expanded telomerase-specific T cells are effective in an orthotopic mouse model for pancreatic adenocarcinoma. Clin Exp Immunol. 2009;158(1):125–32. doi: 10.1111/j.1365-2249.2009.03935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci U S A. 2003;100(11):6364–9. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh SK, Bhardwaj R, Wilczynska KM, Dumur CI, Kordula T. A complex of nuclear factor I-X3 and STAT3 regulates astrocyte and glioma migration through the secreted glycoprotein YKL-40. J Biol Chem. 2011;286(46):39893–903. doi: 10.1074/jbc.M111.257451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumur CI, Sana S, Ladd AC, Ferreira-Gonzalez A, Wilkinson DS, Powers CN, et al. Assessing the impact of tissue devitalization time on genome-wide gene expression analysis in ovarian tumor samples. Diagn Mol Pathol. 2008;17(4):200–6. doi: 10.1097/PDM.0b013e318169bfaf. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin Cancer Res. 2005;11(18):6713–21. doi: 10.1158/1078-0432.CCR-05-0883. [DOI] [PubMed] [Google Scholar]
- 25.Gallina G, Dolcetti L, Serafini P, De Santo C, Marigo I, Colombo MP, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116(10):2777–90. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazzoni A, Bronte V, Visintin A, Spitzer JH, Apolloni E, Serafini P, et al. Myeloid suppressor lines inhibit T cell responses by an NO-dependent mechanism. J Immunol. 2002;168(2):689–95. doi: 10.4049/jimmunol.168.2.689. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol. 2009;182(1):240–9. doi: 10.4049/jimmunol.182.1.240. [DOI] [PubMed] [Google Scholar]
- 28.Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174(10):6477–89. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 29.Elvin P, Evans CW. Cell adhesion and experimental metastasis: a study using the B16 malignant melanoma model system. Eur J Cancer Clin Oncol. 1984;20(1):107–14. doi: 10.1016/0277-5379(84)90041-5. [DOI] [PubMed] [Google Scholar]
- 30.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 31.Overwijk WW, Theoret MR, Finkelstein SE, Surman DR, de Jong LA, Vyth-Dreese FA, et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med. 2003;198(4):569–80. doi: 10.1084/jem.20030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6(6):715–26. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 33.Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med. 2001;193(11):1319–26. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19(3):425–33. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 35.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, et al. Chromatin accessibility predetermines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright KL, White LC, Kelly A, Beck S, Trowsdale J, Ting JP. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J Exp Med. 1995;181(4):1459–71. doi: 10.1084/jem.181.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muchamuel T, Basler M, Aujay MA, Suzuki E, Kalim KW, Lauer C, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat Med. 2009;15(7):781–7. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- 38.Schouppe E, Mommer C, Movahedi K, Laoui D, Morias Y, Gysemans C, et al. Tumor-induced myeloid-derived suppressor cell subsets exert either inhibitory or stimulatory effects on distinct CD8+ T-cell activation events. Eur J Immunol. 2013;43(11):2930–42. doi: 10.1002/eji.201343349. [DOI] [PubMed] [Google Scholar]
- 39.Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, et al. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J Immunother. 2013;36(9):477–89. doi: 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemmer B, Pinilla C, Gran B, Vergelli M, Ling N, Conlon P, et al. Contribution of individual amino acids within MHC molecule or antigenic peptide to TCR ligand potency. J Immunol. 2000;164(2):861–71. doi: 10.4049/jimmunol.164.2.861. [DOI] [PubMed] [Google Scholar]
- 41.Kageyama S, Tsomides TJ, Sykulev Y, Eisen HN. Variations in the number of peptide-MHC class I complexes required to activate cytotoxic T cell responses. J Immunol. 1995;154(2):567–76. [PubMed] [Google Scholar]
- 42.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–71. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 43.Hutchinson S, Sims S, O'Hara G, Silk J, Gileadi U, Cerundolo V, et al. A dominant role for the immunoproteasome in CD8+ T cell responses to murine cytomegalovirus. PLoS One. 2011;6(2):e14646. doi: 10.1371/journal.pone.0014646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min W, Pober JS, Johnson DR. Kinetically coordinated induction of TAP1 and HLA class I by IFN-gamma: the rapid induction of TAP1 by IFN-gamma is mediated by Stat1 alpha. J Immunol. 1996;156(9):3174–83. [PubMed] [Google Scholar]
- 45.Chatterjee-Kishore M, Kishore R, Hicklin DJ, Marincola FM, Ferrone S. Different requirements for signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J Biol Chem. 1998;273(26):16177–83. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- 46.Hasim A, Abudula M, Aimiduo R, Ma JQ, Jiao Z, Akula G, et al. Post-transcriptional and epigenetic regulation of antigen processing machinery (APM) components and HLA-I in cervical cancers from Uighur women. PLoS One. 2012;7(9):e44952. doi: 10.1371/journal.pone.0044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 48.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 49.Buganim Y, Goldstein I, Lipson D, Milyavsky M, Polak-Charcon S, Mardoukh C, et al. A novel translocation breakpoint within the BPTF gene is associated with a pre-malignant phenotype. PLoS One. 2010;5(3):e9657. doi: 10.1371/journal.pone.0009657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181(8):5791–802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.