Abstract
The antimetabolite 5-fluorouracil (5-FU) is one of the most widely used chemotherapy drugs. Dihydropyrimidine dehydrogenase (DPD) is a major determinant of 5-FU response and toxicity. While DPYD variants may affect 5-FU metabolism, they do not completely explain the reported variability in DPD function or the resultant differences in treatment response. Here, we report that H3K27 tri-methylation (H3K27me3) at the DPYD promoter regulated by Ezh2 and UTX suppresses DPYD expression by inhibiting transcription factor PU.1 binding, leading to increased resistance to 5-FU. Enrichment of H3K27me3 at the DPYD promoter was negatively correlated with both DPYD expression and DPD enzyme activity in peripheral blood specimens from healthy volunteers. Lastly, tumor expression data suggests that DPYD repression by Ezh2 predicts poor survival in 5-FU-treated cancers. Collectively, the findings of the present manuscript suggest that a previously uncharacterized mechanism regulates DPD expression and may contribute to tumor resistance to 5-FU.
Keywords: Histone H3K27me3, Ezh2, UTX, dihydropyrimidine dehydrogenase, 5-fluorouracil response
Introduction
Two major obstacles that limit the effectiveness of chemotherapy are low response rates and the high degree of treatment-associated toxicities. As one of the most widely used chemotherapeutics, the antimetabolite 5-fluorouracil (5-FU) confronts the same issues. When administered as a single agent, the response rate for 5-FU is usually less than 20% (1). The addition of other drugs (i.e., combination chemotherapy) can improve response rates to as high as 50%; however, this improvement in efficacy is often accompanied by a dramatic increase in adverse toxicity (2, 3). Therefore, a better understanding of the mechanisms that contribute to drug resistance and toxicity has the potential to improve treatment efficacy.
Dihydropyrimidine dehydrogenase (DPD, encoded by DPYD) catabolizes 85% of administered 5-FU to the non-toxic metabolic intermediate 5-dihydrofluorouracil (4). Specific DPYD variants have been shown to impair DPD enzymatic activity in vitro (5) and significantly increase the risk of 5-FU toxicity in vivo (6). However, known SNPs only account for approximately 30% of cases of severe toxicity to 5-FU (7, 8), and the majority of patients who develop 5-FU toxicity do not carry known toxicity-associated DPYD SNPs (6). In addition to genetic variation in DPYD, differences in DPYD expression have been reported in cancer patients (9, 10). A limited retrospective study demonstrated that patients with low tumor expression of DPYD experienced higher response rates to 5-FU compared to patients with high tumor expression of DPYD (11). Collectively, these observations suggest that the regulation of DPYD expression in both normal and tumor tissues controls 5-FU catabolism and response. Therefore, DPYD transcriptional control is paramount to the sensitivity and resistance to 5-FU.
Gene expression is known to be epigenetically regulated by the methylation of DNA and/or by the modification of histones. Tri-methylation of lysine 27 on histone H3 (H3K27me3) acts to repress gene expression (12). Changes in H3K27me3 are frequently observed in cancer (13). Additionally, inactivating somatic mutations in the H3K27me3 demethylase UTX have been identified in diverse cancers, including multiple myeloma, esophageal, renal, and bladder cancers (14). Mutations in, or overexpression of, the H3K27me3 methyl-transferase EZH2 have also been observed in various cancers, including leukemia, prostate, breast, lung, and liver cancers (15).
In this manuscript we present a previously unrecognized mechanism contributing to 5-FU toxicity and sensitivity. We demonstrate that DPYD expression is directly regulated by the histone H3K27me3 methyl-transferase Ezh2 and the demethylase UTX. H3K27me3 inhibits PU.1 binding, decreasing DPYD expression and increasing 5-FU sensitivity. Tumors with high DPYD and low EZH2 expression are significantly more resistant to 5-FU–based therapy, strongly suggesting that H3K27me3 at the DPYD promoter may be a robust predictive biomarker for 5-FU response.
Material and Methods
Cell lines
Low-passage HEK293T/c17 (ATCC CRL-11268), Hela (ATCC CCL-2), HepG2 (ATCC HB-8065), HCT116 (ATCC CCL-247), RKO (ATCC CRL-2577), SW480 (ATCC CCL-228), SW620 (ATCC CCL-227) and HT-29 (ATCC HTB-38) cells were obtained from the American Type Culture Collection (ATCC) in 2013 and maintained at 37°C in a humidified incubator with an atmosphere of 5% CO2. Cells were cultured using DMEM (Mediatech) supplemented with 10% FBS (Denville Scientific), penicillin and streptomycin (Mediatech).
Antibodies and primers
Antibodies were used as following: Histone H3 (WB 1:5000) was a generous gift from Dr. Zhiguo Zhang; anti-DPD (ab6556, WB 1:2000), TS (ab58287, WB 1:2000) and Tubulin (ab186407, WB 1:5000) were purchased from Abcam; H3K27me3 (C36B11, WB 1:5000) and Ezh2 (D2C9, WB 1:2000) were purchased from Cell Signaling Technology; PU.1 (sc-352 X, WB 1:1000) was purchased from Santa Cruz. Primers used for real-time PCR are list in Table S1.
Vector construction
To generate gRNA expression vectors, annealed oligonucleotides (Integrated DNA Technologies, listed in Table S2) were cloned into the lentiGuide-Puro vector, a gift from Dr. Feng Zhang, which was obtained from Addgene (plasmid #52963). The expression vector for dCas (plasmid #46910) was a gift from Stanley Qi & Jonathan Weissman. The lentiviral packaging vectors pMD2.G (plasmid #12259) and psPAX (plasmid #12260), were obtained from Addgene. Lentivirus vectors expressing shRNAs targeting PU.1 (sh1: TRCN0000426240; sh2: TRCN0000417534) and EZH2 (sh1: TRCN0000353069; sh2: TRCN0000286290) were obtained from Sigma-Aldrich. Wildtype and mutant histone H3 expression vectors were generous gifts from Dr. Zhiguo Zhang.
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as described (16) with minor changes. Cells were fixed with 1% formaldehyde for 10 min at room temperature and quenched with 125 mM glycine. Chromatin was sheared by sonication (Bioruptor Pico) to average lengths of 500 bp. Chromatin was immunoprecipitated using 10 µg relevant antibodies and protein A beads. DNA was recovered from the protein A complex using elution buffer (10 mM Tris, 10 mM EDTA, 1% SDS, 150 mM NaCl, 5 mM DTT, pH 8.0) and purified using Macherey-Nagel PCR Clean-up kit. The amount of specific DNA sequences recovered during each ChIP assay was quantified by real-time PCR.
Cell viability and Real-time cellular analysis
Cells were seeded at standardized concentrations to ensure sub-confluence at 48 hours after treatment. Drug treatments were applied 24 hours post-seeding. Viable was estimated using CellTiter-Blue (Promega) 48 hours after treatment. Real-time cellular analysis using the xCELLigence RTCA system (Acea Biosciences) was performed as described (5).
Healthy volunteer specimens
Healthy volunteers that did not carry the known deleterious DPYD variants rs3918290, rs55886062, or rs67376798 (6) were identified using the Mayo Clinic Center for Individualized Medicine Biobank (17). DPD enzyme activity was measured in PBMC lysates as previously described (18). RNA was extracted from PAXgene Blood RNA Tubes (Becton Dickinson) using the PAXgene Blood RNA kit (Qiagen) using standard protocols. All participants provided informed consent for these studies, which were approved by our institutional review board.
Transcription factor binding site prediction
The promoter region of DPYD (1.5 kb upstream of transcription start site) was analyzed by PROMO (19) using version 8.3 of TRANSFAC transcription factor database with random expectation value threshold set to <0.005.
Statistical analyses
RNA-seq data for individual TCGA cancer types (Illumina HiSeqV2, version 2015-02-24, RSEM normalized) and all clinical information were retrieved using the UCSC Cancer Browser. A cohort of colorectal cancer patients with known treatment information was obtained from publically available data set GSE40967. Standard scores (z-scores) of log2 transformed normalized read counts were calculated for expression analyses. Pearson’s correlations, Kaplan-Meier plots, and Cox proportional-hazards regression analyses were performed using R version 3.2.3. IC50 and two-tailed Student’s t-tests were calculated using GraphPad Prism6.
Results
Identifying EZH2 as a potential regulator of DPYD expression
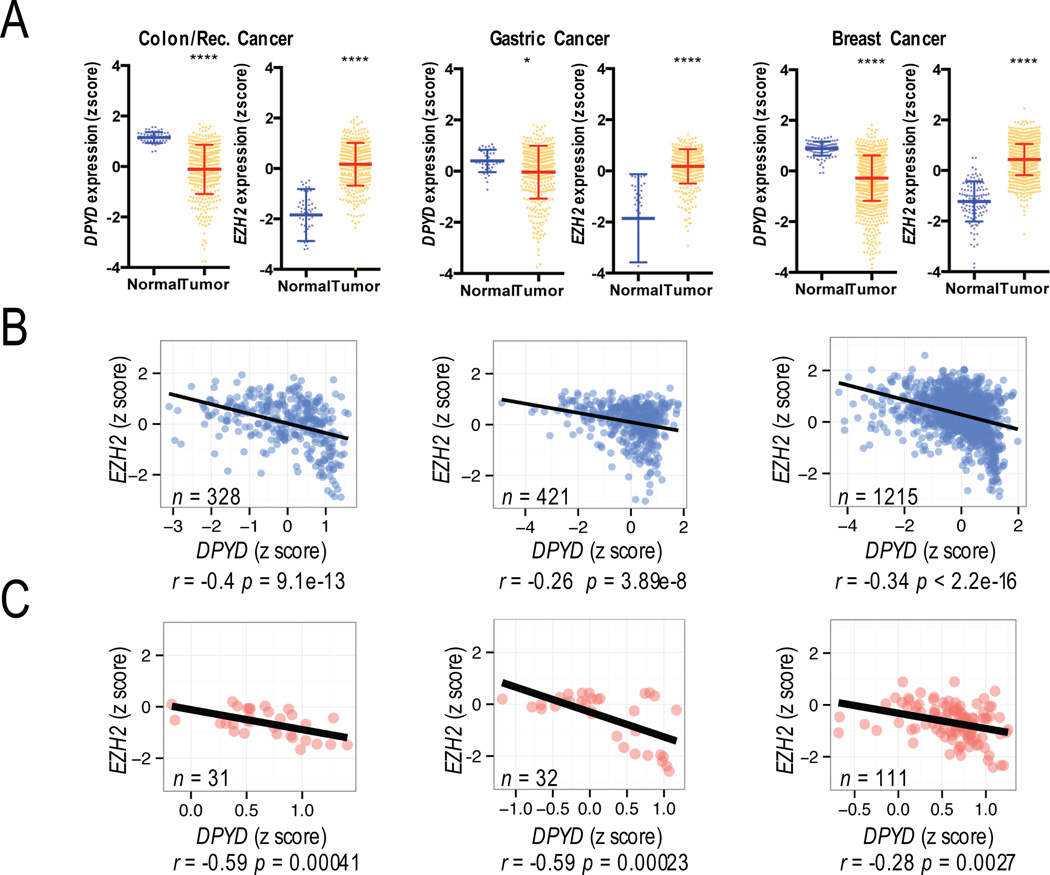
To identify potential pathways through which DPYD could be regulated, we screened expression data from The Cancer Genome Atlas (TCGA) for correlations between epigenetic regulatory genes and DPYD gene expression. Overall, DPYD expression was significantly reduced in tumor samples compared to normal tissues for various cancers, including colon, gastric, and breast cancers (Figure 1A; DPYD expression data for additional cancer types is shown in Figure S1A). In contrast, expression of the H3K27 methyl-transferase EZH2 was significantly elevated in colon, gastric, and breast tumors (Figures 1A; data for additional cancers is shown in Figure S1A). The expression of EZH2 showed negative correlation with DPYD expression in both tumor (Figures 1B and S1B) and normal (Figure 1C) tissues. To further address the link between Ezh2 and DPYD, we treated HEK293T cells, which we previously showed express undetectable levels of DPD (5), with various chemical inhibitors directed against the major histone modification enzymes that control transcription. The greatest increase in DPYD expression was noted for cells treated with GSK-126 (Figure S1C), a highly selective small molecule inhibitor of Ezh2 that has been shown to inhibit global H3K27 tri-methylation (20).
Figure 1. Relationship between DPYD and EZH2 expression.
A. Expression of DPYD and EZH2 in tumor and normal tissues was compared in TCGA datasets for colon/rectum, gastric, and breast cancer. Plots represent the z-scores for log2-transformed normalized RNA-seq read counts. The calculated means +/− SD are represented by bars and whiskers. B. The correlations between DPYD and EZH2 are presented for colon/rectum, gastric, and breast cancers. C. The correlations between DPYD and EZH2 expression in normal tissue adjacent to colon/rectum, gastric, and breast cancers are shown. * p<0.05; **** p<0.0001
It is widely recognized that the epigenetic landscape in tumor cells differs greatly from that of normal cells. Therefore, we tested for correlations between expression in tumor and matched normal tissues for DPYD and EZH2. The expression of neither gene showed evidence for correlation between tumor and normal tissues (Figure S1D), suggesting that the regulation of both genes in tumors is independent from their regulation in normal tissues.
Inhibition of Ezh2 increases DPYD gene expression
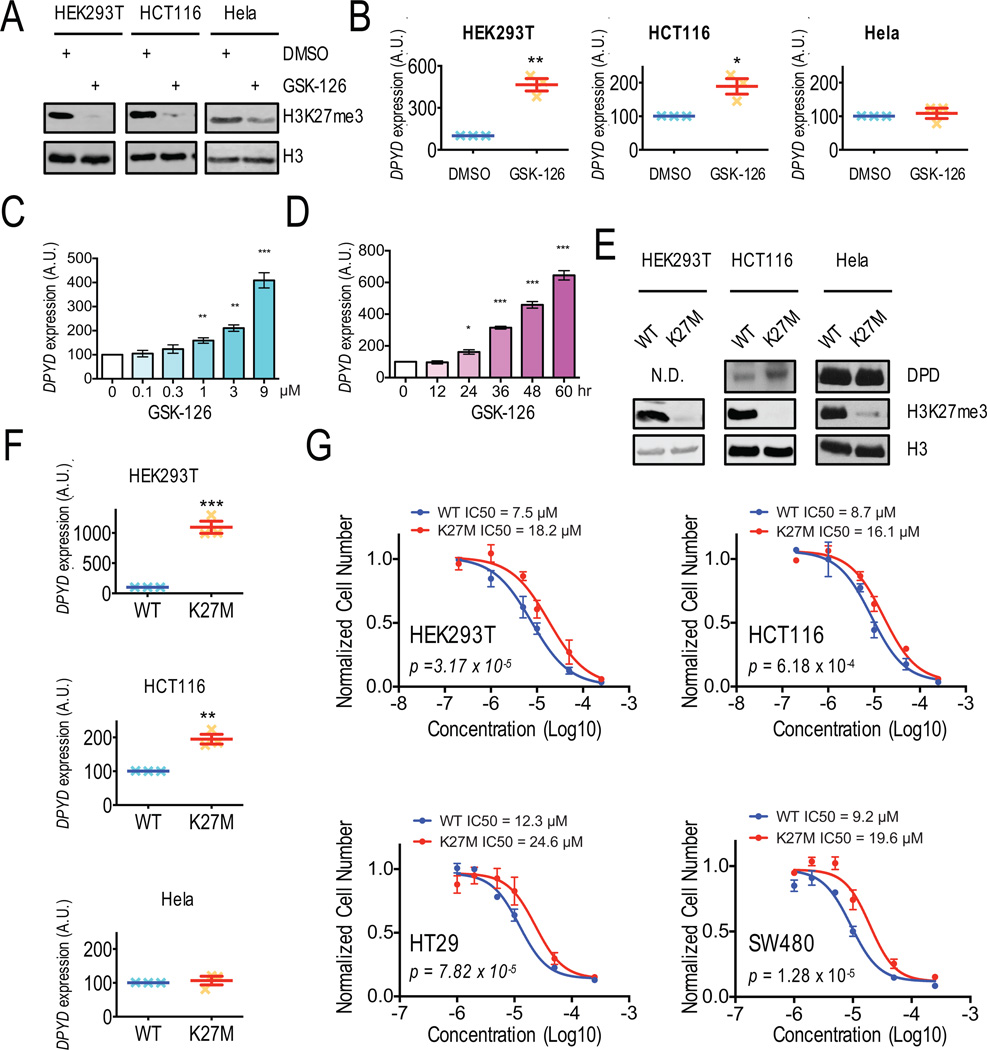
Based on the correlations observed in TCGA data, we hypothesized that H3K27 tri-methylation by Ezh2 would repress DPYD expression. Therefore, inhibition of Ezh2 would be expected to increase DPYD levels. To test this hypothesis, various cell lines were treated with GSK-126. As previously reported, global levels of H3K27me3 were reduced in all cell types treated with GSK-126 (Figure 2A and S2A). Additionally, GSK-126 significantly increased DPYD expression in HEK293T, HCT116, SW480, SW620 and RKO cells, but not Hela cells (Figure 2B and S2B). Knocking down EZH2 by specific shRNA showed similar effect in HEK293T, HCT116, HT29 and Hela cells. (Figures S2C and S2D). Dose- and time-dependent increases of DPYD expression following GSK-126 treatment were noted using HEK293T (Figures 2C and 2D) and HCT116 cells (Figures S2E and S2F).
Figure 2. Inhibition of Ezh2 results in increased DPYD expression and desensitizes cells to 5-FU.
A. Western blotting was used to measure histone H3 and H3K27me3 levels in HEK293T, HCT116, and Hela cells treated with DMSO or 1 µM GSK-126. B. DPYD expression in cells from (A) was measured by q-RT-PCR. C. DPYD expression was measured in HEK293T cells treated with the indicated concentrations of GSK-126. D. DPYD expression in HEK293T cells was measured at indicated time points following treatment with 1 µM GSK-126. E. DPD, H3K27me3, and total H3 expression in HEK293T, HCT116 and Hela cells stably expressing wild-type H3 and H3K27M were measured by western blotting. F. DPYD expression was measured in cells from (E). G. IC50 concentrations for 5-FU were determined using HEK293T, HCT116, HT29 and SW480 cells stably expressing wild-type H3 (WT) or H3K27M (K27M). In all panels means +/− SD are represented by bars and whiskers. N.D., not detectable; * p<0.05; ** p<0.01; *** p<0.001; N.D. not detectable.
To confirm the results obtained by using the chemical inhibitor GSK-126, we generated cell lines that express a lysine 27 to methionine mutant version of histone H3 designated herein as H3K27M that originally been detected in pediatric glioblastomas (K27M) (21, 22), which inhibited global tri-methylation levels of H3K27 in heterozygous carriers in a strongly dominant-negative manner (16, 23, 24). As expected, cells that stably expressed H3K27M showed significant reductions in global H3K27me3 levels, but no overall change in H3 expression (Figure 2E and S2G). For the colorectal tumor lines HCT116, HT29, and SW480, but not Hela cells, DPD protein expression was elevated in H3K27M-expressing cells compared to matched wildtype cells (Figure 2E and S2G). With the exception of Hela cells, DPYD expression was significantly higher in cells expressing H3K27M (Figure 2F and S2H). Additionally, the IC50 for 5-FU was significantly increased in H3K27M expressing cells compared to cells expressing wildtype H3 for all cell lines tested (Figure 2G). Taken together, these results corroborated the observations from the chemical inhibition of Ezh2 with GSK-126 and strongly suggest that DPYD expression is suppressed by Ezh2.
Thymidylate synthetase (TS, encoded by TYMS) is the other major gene that has been suggested to contribute to 5-FU resistance and response (25). No significant changes in TYMS expression were noted for cells treated with GSK-126 (Figure S2I) or for cells stably expressing H3K27M (Figure S2J).
Promoter enrichment of Ezh2 and H3K27me3 correlates with DPYD expression
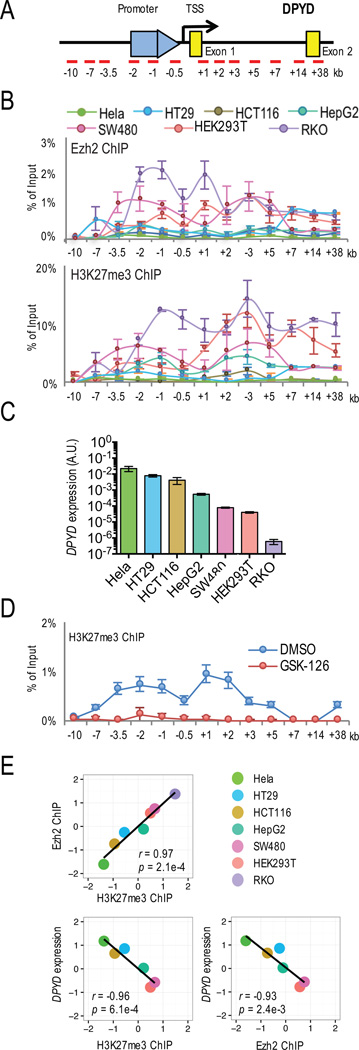
To determine if Ezh2 and H3K27me3 were enriched on the DPYD promoter of cells expressing low levels of DPYD, chromatin immunoprecipitation (ChIP) was performed using antibodies against Ezh2 and H3K27me3. The region that was examined included the promoter, the transcription start site (TSS), and the first two exons (Figure 3A). Ezh2 and H3K27me3 were highly enriched on the DPYD promoter and exon 1 in RKO, HEK293T and SW480 cell lines (Figure 3B), which expressed the lowest levels of DPYD (Figure 3C). Ezh2 and H3K27me3 were moderately enriched on the DPYD promoter in HepG2 and HCT116 cells; almost no enrichment was noted in Hela and HT29 cell lines (Figure 3B). The localized enrichment of H3K27me3 could be reversed using GSK-126 (Figure 3D).
Figure 3. Enrichment of H3K27me3 and Ezh2 on the DPYD promoter and effect on DPYD expression.
A. Schematic of the qPCR primer placement relative to the DPYD promoter, TSS, and first two exons is presented. B. Ezh2- (upper panel) and H3K27me3- (lower panel) associated genomic DNA from the indicated cell lines was obtained by chromatin immuno-precipitation (ChIP) and the enrichment at specific genomic regions was quantified by qPCR using primers indicated in (A). C. DPYD expression in Hela, HT29, HCT116, HepG2, SW480, HEK293T and RKO cells was measured. D. H3K27me3 enrichment was measured by qPCR using ChIP DNA from HCT116 cells treated with 1 µM GSK-126 or DMSO. E. Pearson’s correlation coefficients were calculated for DPYD, Ezh2 enrichment and H3K27me3 enrichment at the DPYD promoter using the sum of relative ChIP enrichment at −2 kb, −1 kb, −0.5 kb and +1 kb. DPYD mRNA and ChIP enrichment level were log2 transformed and standardized to z score.
We estimated the cumulative Ezh2 and H3K27me3 ChIP enrichment at the DPYD promoter and Exon1 (i.e., from −2 kb to +1 kb from the TSS; Figure 3A) to determine correlation with DPYD expression (Figure 3E). ChIP enrichment of Ezh2 and H3K27me3 were highly correlated (r=0.97, p=0.0003). DPYD expression was negatively correlated with both Ezh2 and H3K27me3 accumulation (Ezh2: r=−0.93, p=0.0024; H3K27me3: r=−0.96, p=0.0006). These data support our hypothesis that the level of Ezh2 and H3K27me3 could explain the difference in DPYD gene expression between various different cell lines.
Inhibiting the demethylases UTX and Jmjd3 decreases DPYD gene expression
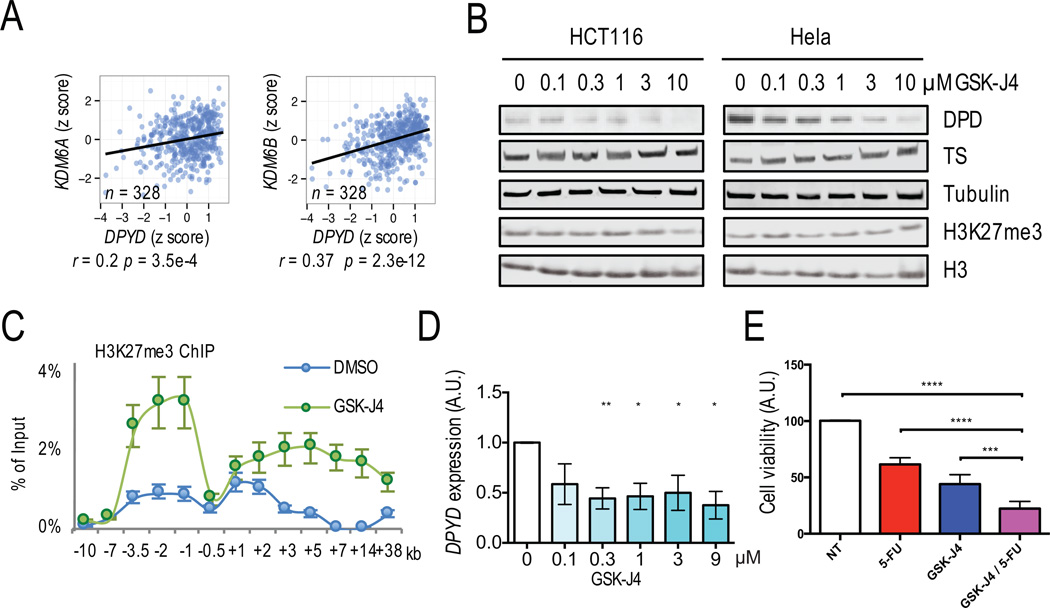
We hypothesized that the H3K27-specific demethylases UTX and Jmjd3 (26) promote DPYD expression through removal of inhibitory H3K27me3 from the DPYD promoter. In TCGA RNA-seq data, UTX and JMJD3 were positivity correlated with DPYD expression in colon cancer (Figure 4A), which is consistent with the negative correlation observed between DPYD and EZH2 (Figures 1B and 1C). Expression of UTX and JMJD3 are both reduced in tumor compare to normal tissues (Figure S3), whereas expression of EZH2 was increased in tumors (Figure 1A).
Figure 4. H3K27me3 demethylases UTX and Jmjd3 regulated DPYD expression.
A. Correlations between DPYD and UTX (left panel) or JMJD3 (right panel) expression were determined using normalized RNA read counts. B. Whole cell lysates from HCT116 and Hela cells treated with the indicated concentrations of GSK-J4 were immunoblotted using the indicated antibodies. C. H3K27me3 was immunoprecipitated from HCT116 cells treated with 1 µM GSK-J4 or DMSO control and analyzed by qPCR. D. Expression of DPYD was measured in cells from (B). E. The viability of HCT116 cultures treated with 5-FU, GSK-J4, 5-FU/GSK-J4 combination, or non-treated control (NT) was measured by realtime cell analysis. In all panels means +/− SD are represented by bars and whiskers. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001
To experimentally corroborate these findings, we suppressed demethylation of H3K27me3 using GSK-J4, an inhibitor of the catalytic sites of UTX and Jmjd3. Consistent with previous findings (27), we did not observe an overall change of H3K27me3 level in Hela and HCT116 cell lines (Figure 4B). GSK-J4 treatment significantly increased H3K27me3 ChIP enrichment at the DPYD promoter in HCT116 cells (Figure 4C). Dose-dependent reductions in DPD and DPYD expression were observed for both cell lines (Figures 4B and 4D).
To determine if induction of H3K27me3 enhances 5-FU anti-tumor activity in vitro, HCT116 cells were treated with 5-FU, GSK-J4, or a combination of 5-FU and GSK-J4. Cell viability was significantly impaired with treatment of GSK-J4 or 5-FU alone (Figure 4E). The decrease in viability was more pronounced in cells treated with both GSK-J4 and 5-FU (Figure 4E), suggesting that H3K27 demethylation enhances 5-FU efficacy by up-regulating DPD.
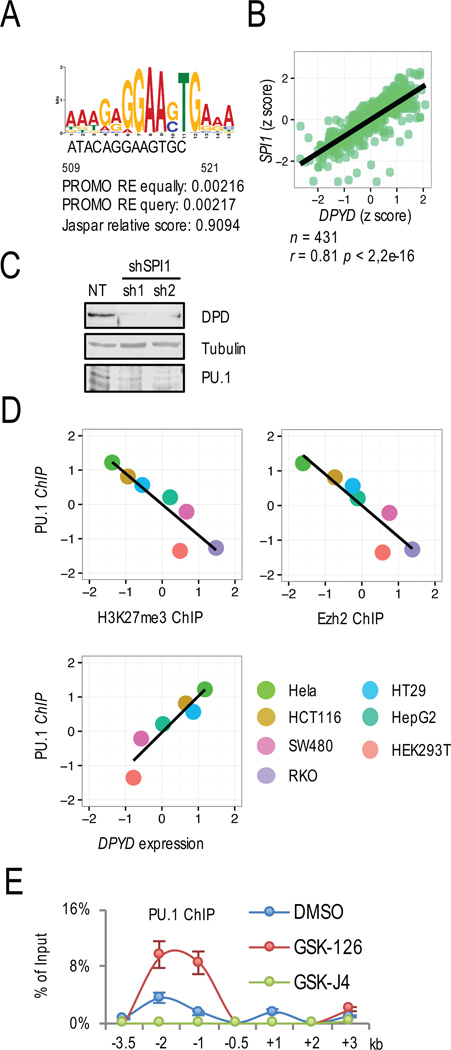
H3K27me3 at the DPYD promoter inhibits transcription factor PU.1 binding
Six candidate transcription factor binding sites in the DPYD promoter were identified using PROMO (19). SPI1, which encodes for PU.1, showed the strongest correlation with DPYD expression in colorectal (Figures 5A, 5B, and S4A) and other (Figure S4B) tumor types in the TCGA dataset. In cellular studies, PU.1 knockdown significantly reduced DPD expression (Figure 5C). It is noted that DPYD promoter-specific PU.1 enrichment varied between cell lines (Figure S4C). Both PU.1 enrichment and Ezh2 or H3K27me3 enrichment on the DPYD promoter were negatively correlated, whereas PU.1 ChIP enrichment on the DPYD promoter positively correlated with DPYD expression (Figure 5D).
Figure 5. H3K27 trimethylation prevented PU.1 binding at the DPYD promoter.
A. The PU.1 binding site was identified on the DPYD promoter located 509–521 nucleotide upstream from the TSS using PROMO. B. Correlation between DPYD and SPI1 expression was determined in colon/rectum cancer TCGA data. C. DPD and PU.1 expression were measured by western blotting of HCT116 cells treated with two different shRNAs directed at PU.1 (sh1 and sh2) or non-target control shRNA (NT). D. Correlations between PU.1 and H3K27me3 enrichment at the DPYD promoter (top left), PU.1 and Ezh2 enrichment at the DPYD promoter (top right), and PU.1 and DPYD mRNA (bottom left) were determined for cell lines indicated (bottom right). Cumulative ChIP enrichment was calculated as in Figure 3E. E. Changes in PU.1 enrichment at the indicated positions (relative to DPYD TSS as indicated) were determined by ChIP-qPCR from HCT116 cells treated with 1 µM GSK-126, 1 µM GSK-J4, or DMSO control.
Given the negative correlation between PU.1 and H3K27me3 ChIP enrichment (Figure 5D), we hypothesized that changing the H3K27 tri-methylation level would alter PU.1 binding affinity. GSK-126 significantly increased PU.1 enrichment on the DPYD promoter, whereas GSK-J4 showed the opposite effect (Figure 5E). These data, combined with the decrease and increase in H3K27me3 levels measured after GSK-126 and GSK-J4 treatments (Figures 3D and 4C), respectively, strongly suggest that tri-methylation of H3K27 at the DPYD promoter regulates transcription by inhibiting PU.1 binding.
Promoter-specific targeting of Ezh2 or UTX alters DPYD expression and cellular sensitivity to 5-FU
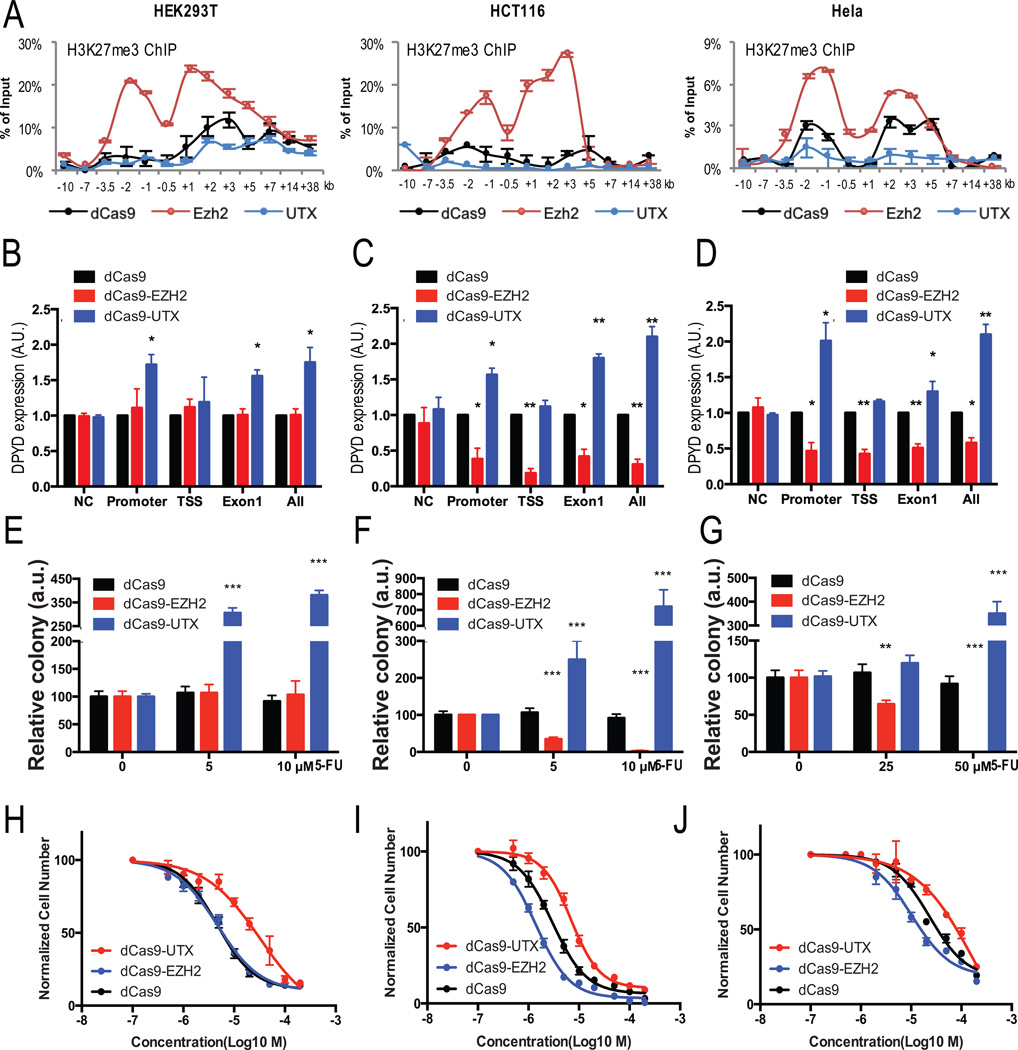
Modified CRISPR methodology was utilized to manipulate H3K27me3 levels specifically at the DPYD promoter. Briefly, a nuclease-deficient Cas9 (dCas9) protein, was fused to either Ezh2 or UTX and expressed in various cell lines together with guide RNAs (gRNAs) that were designed to target different regions of the DPYD gene, including the promoter, TSS, or Exon1 (Figure S5A). To minimize potential artifacts that could be introduced from overexpressing Ezh2 or UTX, low levels of the fusion proteins were expressed that did not change the overall H3K27me3 level (Figure S5B).
Ezh2 and UTX targeting was confirmed using H3K27me3 ChIP-qPCR on cells co-expressing all six gRNAs. DPYD promoter H3K27me3 levels significantly increased in all dCas9-Ezh2–expressing cell lines compared to cells expressing dCas9 control (Figure 6A). H3K27me3 enrichment on the DPYD promoter decreased in all UTX-targeting cell lines (Figure 6A). Changes were restricted to the region of −7 kb to +7 kb of the DPYD TSS (Figure 6A and data not shown), in agreement with unchanged global H3K27me3 levels (Figure S5B).
Figure 6. Regulating H3K27me3 specifically at the DPYD promoter changed DPYD expression and affected 5-FU sensitivity.
A. H3K27me3 enrichment at the indicated regions (relative to DPYD TSS) was measured by ChIP-qPCR in HEK293T (left), HCT116 (center), and Hela (right) cells expressing dCas9 (control), dCas9-Ezh2, or dCas9-UTX and gRNAs targeting the promoter, TSS and 1st exon (see Figure S5A for the location of gRNA targets). B–D. DPYD expression was measured in HEK293T (B), HCT116 (C), and Hela (D) cells expressing dCas9, dCas9-Ezh2 or dCas9-UTX and gRNA targeting indicated positions. E–G. HEK293T (E), HCT116 (F), and Hela (G) cells co-expressing gRNAs targeting promoter and the indicated dCas9 constructs were treated with the indicated concentration of 5-FU. Colonies were counted after four weeks. H–J. IC50 values for 5-FU were calculated for HEK293T (H), HCT116 (I), and Hela (J) cells co-expressing gRNAs targeting promoter. In all panels means +/− SD are represented by bars and whiskers. * p<0.05; ** p<0.01; *** p<0.001.
In HEK293T cells, which possessed the highest level of H3K27me3 on the DPYD promoter (Figure 3B), targeting dCas9-Ezh2 to the DPYD promoter showed no effect on DPYD expression (Figure 6B), suggesting that the natural level of tri-methylation in this cell line may be adequate to effectively inhibit PU.1 binding. On the other hand, targeting dCas9-UTX to either the promoter or Exon1 greatly increased DPYD expression (Figure 6B). For HCT116 cells, which expressed moderate levels of DPYD, targeting dCas9-Ezh2 suppressed DPYD expression, while guiding dCas9-UTX to the promoter or Exon 1 increased DPYD expression (Figure 6C). In Hela cells, dCas9-Ezh2 inhibited DPYD expression when targeted to any of the regions surrounding the promoter, whereas dCas9-UTX increased DPYD expression only when targeted to the promoter and Exon1 (Figure 6D).
To determine the effect of DPYD-specific H3K27 tri-methylation and de-methylation on 5-FU sensitivity, we treated cell lines co-expressing promoter-targeted gRNAs and dCas9, dCas9-Ezh2, or dCas9-UTX with various concentrations of 5-FU. Targeting of dCas9-Ezh2 to the DPYD promoter increased 5-FU resistance in Hela, HCT116, and HEK293T cells (summarized in Figure 6E–G; representative images are shown in Figure S5C). Guiding dCas9-Ezh2 to the DPYD promoter caused Hela and HCT116 cells, but not HEK293T cells, to be more sensitive to 5-FU treatment (Figure 6E–G and S5C). These findings are consistent with the changes in DPYD expression noted for the various cell lines expressing promoter-guided dCas9-Ezh2 or dCas9-UTX (Figures 6B–D).
To further characterize the effects on 5-FU sensitivity, the IC50 for 5-FU was measured (Figure 6H–J). Targeting of dCas9-Ezh2 to the DPYD promoter did not significantly affect the IC50 for 5-FU in HEK293T cells (4.8 µmol/L for dCas9-UTX compared to 4.6 µmol/L for dCas9 control; p =0.97); however, targeting of dCas9-UTX significantly increased the IC50 (32.2 µmol/L; p=1.5×10−3). The lack of effect noted when Ezh2 is targeted to the DPYD promoter can be explained by the high level of H3K27me3 on the DPYD promoter in HEK293T cells (Figure 3A). The IC50 for 5-FU in Hela cells expressing dCas9 was 38.9 µmol/L. Targeting of dCas9-UTX to the promoter significantly increased the IC50 (174 µmol/L; p=6.7×10−6), and targeting dCas9-Ezh2 significantly reduced the IC50 (20 µmol/L; p =1.7×10−4). Similar changes were noted in HCT116 cells, for which the IC50 for dCas9 was 2.9 µmol/L, dCas9-UTX was 8.9 µmol/L (p=3.6×10−4), and dCas9-Ezh2 was 1.3 µmol/L (p =1.1×10−4). Collectively, these results demonstrate that anchoring Ezh2 or UTX to the DPYD promoter specifically alters local H3K27me3 levels, DPYD gene expression, and 5-FU sensitivity/resistance.
Use of Ezh2 and H3K27me3 levels as predictive biomarkers of 5-FU sensitivity/resistance
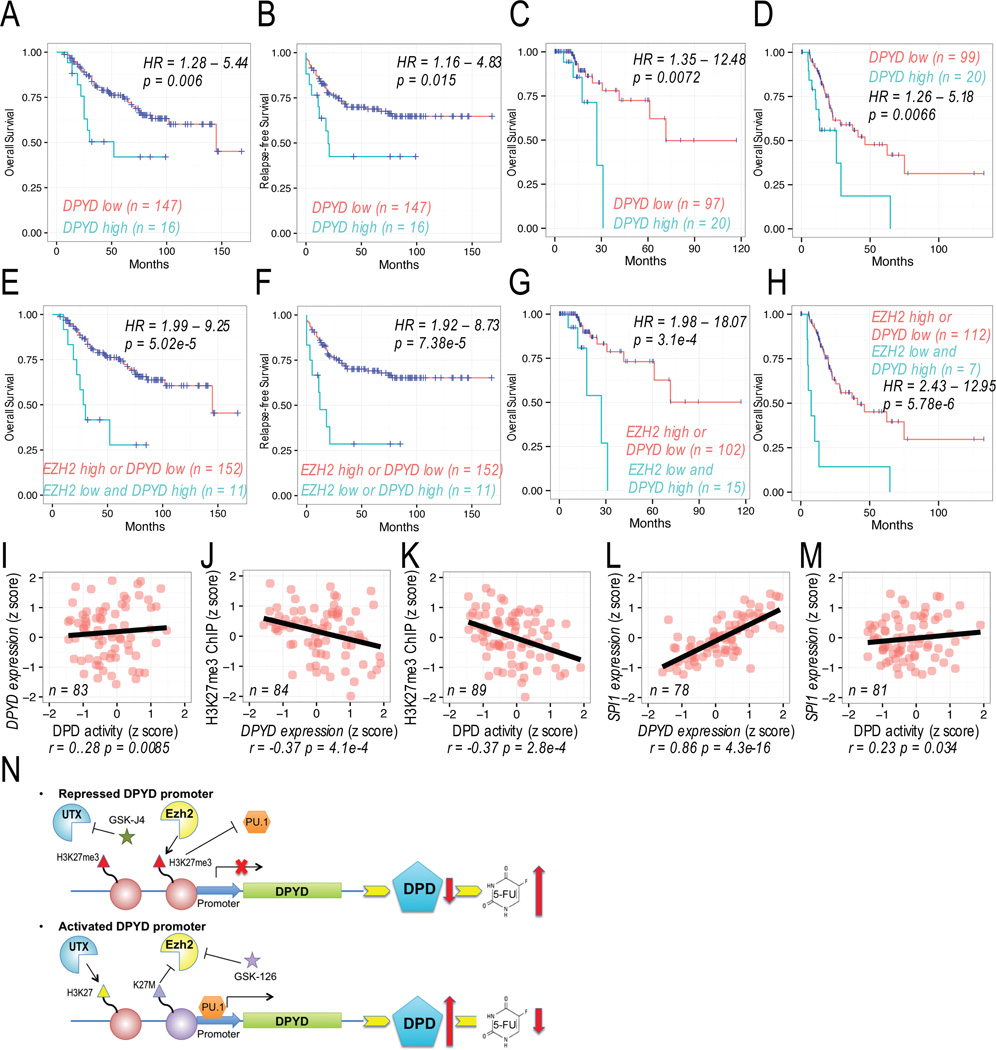
To assess the relationship between DPYD and H3K27me3 levels and tumor resistance to 5-FU, we analyzed overall survival and relapse-free survival data following treatment with a 5-FU-containing regimen in a publically available colorectal cancer dataset GSE40967 (28) and in the TCGA data set for colorectal and gastric cancer patients. For the 20% of patients who had tumors with the highest DPYD expression, overall survival and relapse-free survival were significantly reduced compared to patients whose tumors had lower DPYD expression (Figure 7A–D). COX regression analysis showed that high DPYD expression associated with significantly shorter survival. The median survival for colorectal cancer patients in the GSE40967 dataset with tumors expressing low DPYD levels was 4060 days, compared to 840 days for patients with high tumor DPYD levels (HR=2.64, log-rank p=0.006; Figure 7A). The median relapse-free survival for patients in the GSE40967 dataset with tumors expressing high tumor DPYD levels was 560 (note that fewer than 50% of low tumor DPYD level patients had died during the follow-up period; HR=2.36, log-rank p=0.015; Figure 7B). The median overall survival for colorectal cancer patients in the TCGA dataset with tumors expressing low DPYD levels was 2003 days, compared to 758 days for patients with high tumor DPYD levels (HR=4.26, log-rank p=0.0072; Figure 7C). The median overall survival for gastric cancer patients with tumors expressing low DPYD levels was 1294 days, while median survival for patients with high tumor DPYD expression was 712 days (HR=2.56, log-rank p=0.0066; Figure 7D).
Figure 7. Role of Ezh2 in predicting DPYD expression and 5-FU response.
A–B. Overall survival (A) and relapse-free survival (B) were determined for 5-FU treated colon/rectal cancer patients in GSE40967 dataset stratified by DPYD expression. C–D Overall survival was determined for 5-FU treated colon/rectal cancer (C) and gastric cancer (D) in TCGA dataset. E–F. Overall survival (E) and relapse-free survival (F) were determined for 5-FU treated colon/rectal cancer patients stratified by DPYD and EZH2 expression level as indicated. G–H Overall survival was determined for 5-FU treated colon/rectal cancer (G) and gastric cancer (H) patients stratified by DPYD and EZH2 expression levels as indicated. I–M. Pearson’s correlations were determined for DPD activity, DPYD expression, H3K27me3 enrichment on the DPYD promoter, and SPI1 expression in peripheral mononuclear cells from healthy volunteers. N. A model of epigenetic regulation of the DPYD promoter and PU.1 accessibility is shown together with the effect on DPD expression and resultant 5-FU catabolism.
Because we noted an inverse correlation between tumor expression of DPYD and EZH2 (Figure 1B), we sought to determine if the addition of EZH2 expression improved prediction of survival. In 5-FU-treated patients, a combination of high DPYD and low EZH2 expression was significantly associated with shorter survival (Figure 7E–H) and served as a better predictor of survival than DPYD expression alone (Figure 7A–D). In the GSE40967 dataset, median overall survival improved from 784 to 4060 days (HR=4.3, log-rank p=5.02×10−5; Figure 7E). Similarly, median relapse-free survival was also significantly different between the two groups (HR=4.10, log-rank p=7.38×10−5; Figure 7F). Comparable results were noted in TCGA datasets for colorectal cancer (HR=5.99; log-rank p=3.1×10−4; Figure 7G) and gastric cancer (HR=5.61; log-rank p=5.78×10−5, Figure 7H). It is notable that stratification by EZH2 expression alone did not yield significant differences in survival in any of the datasets analyzed (Figure S6 A–D).
To determine if H3K27me3 could also serve as a biomarker for DPYD expression in normal tissues, and thus as a potential predictor for severe 5-FU toxicity risk, we measured H3K27me3 on the DPYD promoter, DPYD expression, DPD enzyme activity, and SPI1 expression in peripheral blood mononuclear cells (PBMCs) from healthy volunteers. DPYD expression was positively correlated with DPD enzyme activity (r=0.28, p=0.0085; Figure 7I). Negative correlations with H3K27me3 ChIP enrichment on the DPYD promoter were noted for both DPYD expression (r=-0.37, p=4.1×10−4 Figure 7J) and DPD activity (r=-0.37, p=2.8×10−4 Figure 7K). Consistent with observations from the TCGA datasets (Figure 5B and S4B), SPI1 positively correlated with both DPYD expression (r=0.86, p=4.3×10−16; Figure 7L) and DPD activity (r=0.23, p=0.34 Figure 7M).
Discussion
Our data demonstrate that promoter H3K27me3 regulates DPYD expression and likely represents a clinically relevant determinant of tumor response to 5-FU. Our results support a model (Figure 7N) wherein Ezh2 and UTX have opposing roles at the DPYD promoter by regulating the binding of transcription factor PU.1. Modifying global H3K27me3 using either chemical (GSK-126 or GSK-J4) or genetic (H3K27M) approaches effectively modulated PU.1 binding to the DPYD promoter, resulting in altered DPYD expression and 5-FU sensitivity. Additionally, using CRISPR/dCas9-mediated targeting of Ezh2 or UTX, we confirmed that these results are due to tri-methylation of H3K27 specifically at the DPYD promoter. The combination of high DPYD and low EZH2 expression in tumor cells was associated with poor outcome in 5-FU-treated colorectal and gastric cancer patients. Finally, the enrichment of H3K27me3 on the DPYD promoter was negatively correlated with cellular DPD activity and DPYD gene expression.
Our results strongly suggest that epigenetic repression of DPYD by H3K27me3 is a critical determinant of 5-FU resistance. Previous data have suggested that high DPYD expression associated with poor outcome following 5-FU treatment (9). Our findings further indicated that addition of tumor EZH2 expression may help to further stratify patients based on the risk of tumor recurrence. This approach may help to identify patients that could benefit from aggressive adjuvant 5-FU chemotherapy. Specifically treatment options for stage II colorectal cancer patients represent a therapeutic challenge since limited benefit has been observed with adjuvant therapy in this group (29), despite a high risk of recurrence (30). Additional targeted studies of this association in stage II colorectal cancer may help direct therapeutic strategies justifying aggressive treatment in certain patients.
Ezh2 has been previously suggested to contribute to colorectal cancer development (31), but the role of Ezh2 in colorectal cancer recurrence or therapeutic response is largely unknown. Our data suggest that EZH2 alone may have limited potential to predict outcome following 5-FU treatment in colorectal cancer (Figure S6), which is consistent with a previous report that showed a small to modest difference in outcome when patients were stratified by Ezh2 levels (31). Our data demonstrate that tumor DPYD expression may be a better predictor of outcome (Figure 7A–D), especially when combined with EZH2 expression (Figure 7E–H). Collectively, these findings suggest that defects in Ezh2 (e.g., mutations) may disrupt the Ezh2-DPYD regulatory axis and affect 5-FU sensitivity. Inhibitors of Ezh2 or the KDM6 family (UTX and Jmjd3) have proven to be effective therapies for various cancers (27, 32–34). Ezh2 inhibitors have been shown to reverse drug-resistance in liver, brain, ovarian, and lung cancers (35–38), suggesting that the H3K27me3 epigenomic landscape may contribute to chemotherapy response. In the present study, our finding that Ezh2 regulates cellular response to 5-FU by inhibiting DPYD expression is perhaps unexpected based on a previous report that suggested that Ezh2 inhibition may sensitize tongue cancer cells to 5-FU (39). However, we confirmed our findings using multiple approaches including chemically inhibiting H3K27me3 demethylation using GSK-J4, as well as using genetic approaches by expressing the dominant negative H3K27M mutant. Additionally, we demonstrate that the 5-FU response was due to direct modulation of H3K27me3 at the DPYD promoter using dCas9-mediated targeting of Ezh2 or UTX to the DPYD promoter (Figure 6).
In this manuscript, we identified the PU.1 transcription factor as an activator of DPYD expression (Figure 5). PU.1 is generally thought to regulate gene expression in circulating cells (40), but has also been identified as a metastatic suppressor in hepatocellular carcinoma (41). PU.1 has also been suggested to be involved in osteoclast development, adipogenesis, and aberrant Notch1 signaling linked to infertility (42–44). In the present study, we observed a positive correlation between DPYD and SPI1 expression in RNA-seq data from TCGA datasets (Figure 5B) and in volunteer PBMCs (Figure 7L and 7M). These correlations, together with the observed reduction in DPYD after PU.1 knockdown (Figure 5C) and PU.1 enrichment on the DPYD promoter (Figure S4B) strongly indicate that PU.1 may be a key transcription factor that promotes DPYD expression in tumor and normal tissues.
In conclusion, understanding the mechanisms that contribute to drug sensitivity has the potential to significantly improve treatment planning decisions. Changes in the expression of genes related to drug metabolism could greatly affect tumor sensitivity to chemotherapy. In this manuscript we present a novel epigenetic mechanism of DPYD regulation and identify potential biomarkers that may be clinically useful in identifying patients who may benefit from aggressive adjuvant chemotherapy.
Supplementary Material
Acknowledgments
We thank Dr. Zhiguo Zhang for the histone H3 antibody, wildtype and K27M mutant plasmids, and for insightful discussions concerning dCas9-Ezh2 plasmid construction. The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. We thank the Mayo Clinic Center for Individualized Medicine for the use of healthy volunteer samples from the Biobank and the Mayo Clinic Cancer Center for supporting additional shared resources (NIH CA 15083).
Financial support: NIH 5P30CA015083-42 (R.B.D.)
Footnotes
The authors have no conflict of interest to disclose.
Reference
- 1.Carethers JM. Systemic treatment of advanced colorectal cancer: tailoring therapy to the tumor. Therap Adv Gastroenterol. 2008;1:33–42. doi: 10.1177/1756283X08093607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham JS, Cassidy J. Adjuvant therapy in colon cancer. Expert Rev Anticancer Ther. 2012;12:99–109. doi: 10.1586/era.11.189. [DOI] [PubMed] [Google Scholar]
- 3.Lee AM, Shi Q, Pavey E, Alberts SR, Sargent DJ, Sinicrope FA, et al. DPYD variants as predictors of 5-fluorouracil toxicity in adjuvant colon cancer treatment (NCCTG N0147) J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heggie GD, Sommadossi JP, Cross DS, Huster WJ, Diasio RB. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987;47:2203–2206. [PubMed] [Google Scholar]
- 5.Offer SM, Wegner NJ, Fossum C, Wang K, Diasio RB. Phenotypic profiling of DPYD variations relevant to 5-fluorouracil sensitivity using real-time cellular analysis and in vitro measurement of enzyme activity. Cancer Res. 2013;73:1958–1968. doi: 10.1158/0008-5472.CAN-12-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, Diasio RB, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol Ther. 2013;94:640–645. doi: 10.1038/clpt.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee AM, Shi Q, Alberts SR, Sargent DJ, Sinicrope FA, Berenberg JL, et al. Association between DPYD c.1129–5923 C>G/hapB3 and severe toxicity to 5-fluorouracil-based chemotherapy in stage III colon cancer patients: NCCTG N0147 (Alliance) Pharmacogenet Genomics. 2015 doi: 10.1097/FPC.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosmarin D, Palles C, Pagnamenta A, Kaur K, Pita G, Martin M, et al. A candidate gene study of capecitabine-related toxicity in colorectal cancer identifies new toxicity variants at DPYD and a putative role for ENOSF1 rather than TYMS. Gut. 2015;64:111–120. doi: 10.1136/gutjnl-2013-306571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salonga D, Danenberg KD, Johnson M, Metzger R, Groshen S, Tsao-Wei DD, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 10.Fujiwara H, Terashima M, Irinoda T, Takagane A, Abe K, Kashiwaba M, et al. Quantitative measurement of thymidylate synthase and dihydropyrimidine dehydrogenase mRNA level in gastric cancer by real-time RT-PCR. Jpn J Cancer Res. 2002;93:1342–1350. doi: 10.1111/j.1349-7006.2002.tb01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichikawa W, Uetake H, Shirota Y, Yamada H, Nishi N, Nihei Z, et al. Combination of Dihydropyrimidine Dehydrogenase and Thymidylate Synthase Gene Expressions in Primary Tumors as Predictive Parameters for the Efficacy of Fluoropyrimidine-based Chemotherapy for Metastatic Colorectal Cancer. Clinical Cancer Research. 2003;9:786–791. [PubMed] [Google Scholar]
- 12.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 13.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 14.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson JE, Ryu E, Johnson KJ, Koenig BA, Maschke KJ, Morrisette JA, et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88:952–962. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offer SM, Diasio RB. Response to "A case of 5-FU-related severe toxicity associated with the P.Y186C DPYD variant". Clin Pharmacol Ther. 2014;95:137. doi: 10.1038/clpt.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 21.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 22.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Lecomte T, Ferraz JM, Zinzindohoue F, Loriot MA, Tregouet DA, Landi B, et al. Thymidylate synthase gene polymorphism predicts toxicity in colorectal cancer patients receiving 5-fluorouracil-based chemotherapy. Clin Cancer Res. 2004;10:5880–5888. doi: 10.1158/1078-0432.CCR-04-0169. [DOI] [PubMed] [Google Scholar]
- 26.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 27.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 30.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 31.Fluge O, Gravdal K, Carlsen E, Vonen B, Kjellevold K, Refsum S, et al. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br J Cancer. 2009;101:1282–1289. doi: 10.1038/sj.bjc.6605333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514:513–517. doi: 10.1038/nature13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125:346–357. doi: 10.1182/blood-2014-06-581082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Bi C, Cheong LL, Mahara S, Liu SC, Tay KG, et al. The histone methyltransferase inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and targets leukemia cells in AML. Blood. 2011;118:2830–2839. doi: 10.1182/blood-2010-07-294827. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Liu G, Lin C, Liao G, Tang B. Silencing the EZH2 gene by RNA interference reverses the drug resistance of human hepatic multidrug-resistant cancer cells to 5-Fu. Life Sci. 2013;92:896–902. doi: 10.1016/j.lfs.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Fan TY, Wang H, Xiang P, Liu YW, Li HZ, Lei BX, et al. Inhibition of EZH2 reverses chemotherapeutic drug TMZ chemosensitivity in glioblastoma. Int J Clin Exp Pathol. 2014;7:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzo S, Hersey JM, Mellor P, Dai W, Santos-Silva A, Liber D, et al. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol Cancer Ther. 2011;10:325–335. doi: 10.1158/1535-7163.MCT-10-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Wang Y, Qiu J, Li Q, Yuan C, Zhang W, et al. The polycomb group protein EZH2 is a novel therapeutic target in tongue cancer. Oncotarget. 2013;4:2532–2549. doi: 10.18632/oncotarget.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 41.Song LJ, Zhang WJ, Chang ZW, Pan YF, Zong H, Fan QX, et al. PU.1 Is Identified as a Novel Metastasis Suppressor in Hepatocellular Carcinoma Regulating the miR-615-5p/IGF2 Axis. Asian Pac J Cancer Prev. 2015;16:3667–3671. doi: 10.7314/apjcp.2015.16.9.3667. [DOI] [PubMed] [Google Scholar]
- 42.Su RW, Strug MR, Jeong JW, Miele L, Fazleabas AT. Aberrant activation of canonical Notch1 signaling in the mouse uterus decreases progesterone receptor by hypermethylation and leads to infertility. Proc Natl Acad Sci U S A. 2016;113:2300–2305. doi: 10.1073/pnas.1520441113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei N, Wang Y, Xu RX, Wang GQ, Xiong Y, Yu TY, et al. PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. Anim Genet. 2015;46:133–140. doi: 10.1111/age.12275. [DOI] [PubMed] [Google Scholar]
- 44.Ishiyama K, Yashiro T, Nakano N, Kasakura K, Miura R, Hara M, et al. Involvement of PU.1 in NFATc1 promoter function in osteoclast development. Allergol Int. 2015;64:241–247. doi: 10.1016/j.alit.2015.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.