Abstract
Purpose
As humans, we are constantly exposed to ionizing radiation from natural, man-made and cosmic sources which can damage DNA, leading to deleterious effects including cancer incidence. In this work we introduce a method to monitor strand breaks resulting from damage due to the direct effect of ionizing radiation and provide evidence for sequence-dependent effects leading to strand breaks.
Materials and methods
To analyze only DNA strand breaks caused by radiation damage due to the direct effect of ionizing radiation, we combined an established technique to generate dehydrated DNA samples with a technique to analyze single strand breaks on short oligonucleotide sequences via denaturing gel electrophoresis.
Results
We find that direct damage primarily results in a reduced number of strand breaks in guanine triplet regions (GGG) when compared to isolated guanine (G) bases with identical flanking base context. In addition, we observe strand break behavior possibly indicative of protection of guanine bases when flanked by pyrimidines, and sensitization of guanine to strand break when flanked by adenine (A) bases in both isolated G and GGG cases.
Conclusions
These observations provide insight into the strand break behavior in GGG regions damaged via the direct effect of ionizing radiation. In addition, this could be indicative of DNA sequences that are naturally more susceptible to strand break due to the direct effect of ionizing radiation.
Keywords: DNA damage, Guanine, Radiation Chemistry, Radiosensitivity, Gel electrophoresis, Direct Effect, Ionizing Radiation
Introduction
The effect of ionizing radiation (IR) on humans at high to lethal absorbed dose levels has been well documented. Studies have demonstrated a higher incidence of leukemia as well as breast, bladder, lung, colon and stomach cancers and a number of non-cancer health effects in populations exposed to between 5 and 150 mGy IR (Cullings et al. 2006; Douple et al. 2011). Global gene expression and cell signaling pathways are also altered after exposure to IR (Ding et al. 2005). This is likely linked to the observed increased frequency of clustered and deleterious DNA damage when exposed to IR, i.e. closely juxtaposed strand breaks, formation of abasic sites, and oxidized bases (Sutherland et al. 2001). Investigation into the effect of IR damage to DNA has implications for all individuals, due to natural background levels of radiation. Moreover, these studies are particularly relevant to those who receive higher than normal levels of IR, including people undergoing radiation therapy treatments, diagnostic imaging procedures that use IR and the health of radiation workers who receive larger than average doses of IR due to the nature of their vocation (Ayouaz et al. 2008).
Damage to DNA by IR occurs via two mechanisms in vivo. The first is due to the indirect effect of IR. In this case, IR creates free radicals and other reactive species (hydroxyl radicals (•OH), solvated electrons (e−aq), hydrogen peroxide (H2O2) etc.) that can damage DNA in a diffusion-limited manner (von Sonntag 1987). However, in the nuclei of human cells, the tight packaging of DNA with histone proteins into chromatin and the concentration of other molecules that react with diffusible species reduce the proportional DNA damage via the indirect effect. As a result, damage via the direct effect of IR to DNA represents a significant fraction of the total damage (Krisch et al. 1991; Becker and Sevilla 1993; von Sonntag 2006; Close 2008; Bernhard 2009; Becker et al. 2010). Radiation damage to DNA via the direct effect involves direct deposition of energy into DNA or water molecules directly bound to DNA. This produces similar DNA base damage products including 8-oxo-guanine (8-oxo-Gua), dihydrothymine (DHThy), and dihydrouracil (DHUra) (Bernhard 2009). The direct effect of IR also produces sugar radicals on the DNA backbone, which can lead to DNA strand breaks. These damage products, however, are produced in different proportions via the direct effect compared to the products of the indirect effect of IR (Bernhard 2009). Krish et al. separated these types of damage in cells through freezing and hydroxyl scavenging techniques, determining that damage by the direct effect of IR accounts for at least 50% of the total IR-dependent strand breaks (Krisch et al. 1991). DNA damage by IR results in both DNA base and sugar lesions, some of which result in physical scission of covalent bonds in the DNA backbone. However, it is unknown to what extent specific DNA sequences or chromosome fragile sites are affected by the direct effect of IR. The effect of radiosensitivity by permutations in specific DNA bases has been analyzed by quantifying free base release, but such studies only provide an average probability of strand break associated with each of the four DNA bases (Sharma et al. 2008). Here, we have developed an assay that not only provides information on the identity of a DNA base present at a strand break site, but also the specific sequence at the site of damage. Specifically, films of dry duplex DNA pre-radiolabeled on the 5′ end with phosphorus-32 (32P) were irradiated in order to quantify strand breaks formed via the direct effect of IR. These strand breaks were quantified through the use of denaturing sequencing gels. This has enabled us to identify two specific trends surrounding strand break occurrence at guanine, and its dependence on base context. In addition to the indication of direct influence of base context on strand break probability at guanine bases, we find that guanine triplets are less sensitive to prompt strand break than single guanine bases with analogous base context.
Materials and Methods
DNA Substrates
Double stranded, 16- and 15-base pair DNA oligodeoxynucleotides (ODN(s)), containing 16-base pair single-G and 15-base pair triple-G base pair sequences (see Table 1), were purchased from Integrated DNA Technologies (IDT, Coralville, Iowa, USA). The ODNs were analyzed using the OligoAnalyzer program to avoid secondary structures and self-dimerization. ODNs were chosen as such to maintain equal number of adenines on the 5′ and 3′ end of the 16-mer.
Table I.
Oligonucleotide Sequences. Outline of the labeled ODNs used in this study and the complementary strand used in each case for producing labeled, duplexed DNA samples. In both cases the G-rich strand is labeled in the single stranded state before duplexing with its complement sequence indicated below.
| Oligonucleotide | Length | Listed 5′ – 3′ |
|---|---|---|
| G-rich Strand | ||
| Single-G | 16 | AAAATGTCGCAGAAAA |
| Triple-G | 15 | TGGGTCGGGCAGGGA |
| Compliment | ||
| Single-G Compliment | 16 | TTTTCTGCGACATTTT |
| Triple-G Compliment | 15 | TCCCTGCCCGACCCA |
Radioactive Labeling and Purification of Oligonucleotides
ODNs were radioactively labeled at their 5′ termini by phosphoryl transfer with phosphorous-32 adenosine tri-phosphate [γ-32P]ATP. Briefly, an ODN (20 nmoles) was incubated with 2 microliters (μl) T4 Polynucleotide Kinase (PNK) (10 units/μl, New England Biolabs, Ipswich, MA, USA), 8 μl [γ-32P]ATP (10 μCi/μl), 8 μl PNK Buffer A (New England Biolabs, Ipswich, MA, USA) and 25 μl double-distilled water (ddH2O) for 1 hour at 37°C. One μl of the reaction was separated by 12% denaturing polyacrylamide gel electrophoresis (PAGE) for 1 hour at 150 volts (V). One of the glass plates was removed to expose the gel. Plastic-wrap was placed on the gel and light-sensitive markers (Glogos II Markers, Agilent Technologies, Santa Clara, CA, USA) were placed in the corners for orientation. The gel was exposed to X-ray film (Kodak, Rochester, NY, USA) for 20 seconds and the film subsequently developed (XOMAT, Carestream Health, Rochester, NY, USA). The band containing the labeled ODN was cut out of the gel and the specific activity was determined by placing the acrylamide gel slice into a scintillation tube and subjected to direct Cherenkov counting on a Beckman LS600SC (Global Medical Instrumentation, Ramsey, MN, USA) using the 32P channel. The remainder of the ODN was mixed with 20 μl of 100% formamide. The solution was incubated for 1 minute at 90°C to remove any homo-dimerization or secondary structures, and loaded onto an 18% urea-polyacrylamide gel and subjected to electrophoresis at 75 watts for 2 hours. The band containing the radiolabeled ODN was located and excised as described above and the gel slice placed into an Eppendorf tube (1.5 ml Eppendorf LoBind Microcentrifuge Tubes, ThermoFisher Scientific, Waltham, MA, USA), crushed to a near powder with a plastic pestle. Next, 500 μl of 1X TE (10 mM Tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl), pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA)) was added and the tube was placed on a rotator overnight at 25°C. The contents of the tube were then transferred to a Corning Costar Spin-X microcentrifuge filter tube (cellulose acetate membrane, pore size 0.22 μm) (Sigma-Aldrich, St. Louis, MO, USA) and centrifuged for 5 minutes at 2.3 g to separate the gel pieces from the solution. Recovery of DNA was determined by spotting 0.5 μl of solution on Whatman filter paper (GE Healthcare Life Sciences, Pittsburgh, PA, USA) and subjecting to direct counting as above. To the tube, 2 μl glycogen (Roche Applied Science, Indianapolis, IN, USA), 20 μl 7.5M ammonium acetate, and 1200 μl ice cold 99% ethanol was added, the tube was inverted twice, then incubated in a −80°C freezer for 1 hour. After incubation, the sample was centrifuged at 14,500 g for 20 minutes at room temperature. The supernatant was carefully aspirated off the precipitated DNA and 500 μl of 70% ethanol solution was added with gentle agitation to wash the pellet. The tube was then subjected to centrifugation for 10 minutes at 14,500 g. Most of the supernatant was removed by pipette and the tube was placed in a 37°C heat block to remove the remaining ethanol. Finally, 50 μl of 1X TE was added to the near-dry pellet to solubilize the DNA.
To generate double-stranded (DS) ODNs, the labeled ODNs were mixed with an equimolar amount of the unlabeled complementary strand in 1X annealing buffer (10 mM Tris, pH 8.0, 50 mM NaCl, 1 mM EDTA). The solution was heated to 90°C for 10 minutes then the heating block switched off and allowed to cool for 4 hours.
DNA Film Production and Irradiation
High purity Suprasil (Heraeus, Hanau, Germany) quartz tubes (1 mm O.D.) were cut into 3 cm segments, open at both ends, and then treated with Aquasil siliconizing solution (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Forty μl of the 5′ end labeled DS-ODN, suspended in the annealing buffer indicated above, were pipetted into the quartz tubes and the tubes placed in a desiccation chamber in the presence of 50 grams of diphosphorus pentoxide (P2O5) (Sigma-Aldrich, St. Louis, MO, USA) with an applied vacuum of 30 mmHg for 3 days to produce films of DNA with a nominal hydration level, Γ (2.5 mol water per mol nucleotide) (Swarts et al. 1992). At this hydration level, films are stable at ambient humidity and will not hydrate during the time it takes to process the samples. DNA films were irradiated at room temperature using a Varian OEG-76 tungsten target X-ray source (Varian, Palo Alto, CA, USA) operated at 70 kilovolts (kV) and 20 milliamps (mA), at a distance of 20.0 millimeters (mm) between the sample and the X-ray window. At these conditions, the dose rate at the sample was 1.7 kGy/minute, which was confirmed through radiochromic film dosimetry. Specifically, radiochromic films were irradiated at the sample position for a known amount of time using these operational conditions. Based on the film calibration curve established using a 60Co source, we were able to determine the dose rate at this position. Doses delivered were 15, 30, 60, 120, and 240 kGy in addition to the control sample, which was unirradiated.
Sample Analysis
After irradiation, samples were dissolved in 100 μl of 1X TE buffer containing 5% glycerol to scavenge any secondary production of solvent radicals. 1 μl of each sample was removed and 10 μl 2X denaturing sample loading buffer (90% formamide, 1 ul 10X TBE buffer, 4 mg bromphenol blue) was added to the final mixture. The samples were immediately analyzed by polyacrylamide gel electrophoresis (PAGE) on 18% sequencing gels (19:1 acrylamide:bisacrylamide, 1X TBE (90 mM Tris(hydroxymethyl)aminomethane hydrochloride (Tris)-borate, pH 8.3, 1 mM EDTA), 8M urea). Markers were produced via Maxam-Gilbert sequencing reactions for G, G+A, C and C+T performed on the parent sequences and applied to the gels to identify specific bands in the experimental lanes. The gels were electrophoresed for 2.5 hours at 75 watts, dried under vacuum at 80°C for 1 hour and exposed to phosphor screens overnight. Images were visualized on a GE Healthcare Typhoon phosphorimager and the total volume of each band as a function of dose determined using ImageQuant software (GE Healthcare Life Sciences, Pittsburg, PA, USA). Plots of volume as a function of absorbed dose were fit to a linear regression curve using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA). The slope of the linear regression was used as a metric for strand break frequency at that site. For each sequence, three sample sets were analyzed, providing an effective N of 3. Error bars to the slopes were based on standard deviation calculation of the average slope for each fragment.
Results
When analyzed in standard laboratory (dilute) solutions, naked DNA exposed to IR will be primarily damaged via the indirect effect of IR because of the overwhelming cross-sectional target of water compared to DNA. To analyze only DNA strand breaks caused by the direct effect of IR, we combined an established technique to generate dehydrated DNA samples with a technique to analyze single strand breaks on short oligonucleotide (ODN) sequences via denaturing gel electrophoresis.
A 16- or 15-nt DNA ODN was radiolabeled at the 5′ end and annealed with its complement. The double stranded ODNs were placed in siliconized quartz tubes and dehydrated to Γ=2.5 (see Materials and Methods) by incubation in the presence of P2O5. We note that quartz siliconization was necessary to produce a small target for irradiation and to allow consistent recovery of the DNA after dehydration. The dehydrated samples were irradiated at room temperature and atmospheric pressure with X-ray doses ranging from 15 to 240 kGy. Samples were dissolved in buffer containing 5% glycerol to scavenge any residual diffusible radicals and DNA strand breaks analyzed by running the samples on 18% denaturing PAGE sequencing gels as described in the Materials and Methods section. Each band on the gel corresponds to damage events resulting in strand breaks at an individual nucleotide in the polymer chain. An example of a PAGE data set is provided in Figure 1. Maxam-Gilbert base-specific scission of the deoxyribose backbone (cleavage reactions) were performed with the unirradiated double-stranded ODNs to provide a reference set of cleavage products with known chemical structures containing 3′ monophosphoester termini. The unirradiated ODN exhibits a background of bands that could be due to incomplete synthesis and/or aberrant cleavage by autoradiolysis. Although free 32P was cleared initially after labeling using a P-6 filter (Bio-Rad Laboratories, Hercules, CA), we suspect that the drying method may have relieved additional 32P and once exposed to water in solution could have undergone additional autoradiolysis events as exhibited in the unirradiated (‘0’) lane (reviewed in Gates 2009). Additional free radical quenchers or a second filter step could be necessary to minimize the effect of autoradiolysis. However, as the X-ray dose increases from the unirradiated sample in Lane 1 to 240 kGy in Lane 6 (Fig 1), the amount of visible cleavage products increases for all positions within the ODN, indicating that damage by the direct effect of IR results in prompt strand breaks throughout the double-stranded ODN. Importantly, the vast majority of radiation-induced products align with products of Maxam-Gilbert cleavage at specific bases, indicating that damage largely results in 3′ monophosphoester termini. We have therefore labeled these bands according to the identity of the base at which 3′ cleavage occurs (Fig 1).
Figure 1.
Visualization of site-specific DNA strand breaks due the direct effect of IR damage by X-rays. Image of bands produced through denaturing PAGE analysis of samples of single-G double stranded irradiated to the indicated absorbed dose. Lane 1 contains the unirradiated single-G oligonucleotide (ODN). Lanes 2–6 received doses of 15, 30, 60, 120 and 240 kGy, respectively. The four lanes on the right contain cleavage products produced by Maxam-Gilbert sequencing used to identify bands produced through radiolytic cleavage. Bands labeled with an arrow exhibit apparent precursor/product relationships, as a function of dose.
We also observe that some bands are present in between the prominent bands corresponding to Maxam-Gilbert cleavage products. Most of these have precursors that are present in the unirradiated ODN and appear to exhibit a precursor/product relationship as a function of radiation dose (some examples are indicated with and arrow in Fig 1). These products may have termini distinct from 3′-monophosphoester termini that specifically degrade upon irradiation or altered non-native base content remaining from the ODN synthesis. Nevertheless, the coincident migration of the vast majority of irradiation products indicates that that majority of strand breaks originating from the canonical DNA structure result in 3′ monophosphoester termini.
We analyzed the cleavage by quantifying and plotting the volume of corresponding bands as a function of dose to elucidate a dose response for DNA strand cleavage at each base position in the ODN, thus revealing relative strand break probabilities within the same sequence. For each data set, the volume of the band assigned to a specific fragment was plotted as a function of dose after normalization to the parent band and subtraction of the corresponding band in the unirradiated control lane. The dose response of each fragment was fit by a linear regression function and the slope used as a metric for relative site-specific strand break probability. From the volume analysis for the gel provided in Figure 1, it can be seen that the dose response of the fragments is highly linear with significant differences in slope existing between fragments (Fig 2a).
Figure 2.
A plot showing the volume analysis of 3 example fragments of the single-G ODN at the indicated dose. Site-specific DNA strand breaks induced via the direct effect of IR increase with dose. For each fragment on both single- and triple-G ODN a linear regression was applied and slopes were calculated to provide a metric for strand break probability at that site. Parentheses around the final base indicated in the legend signify the base lost due to cleavage as a result of the irradiation. Dose response of radiation-generated fragments for the (B) single-G and (C) triple-G sequences. Relative cleavage probabilities as a function of dose were collected. Error bars represent one standard deviation.
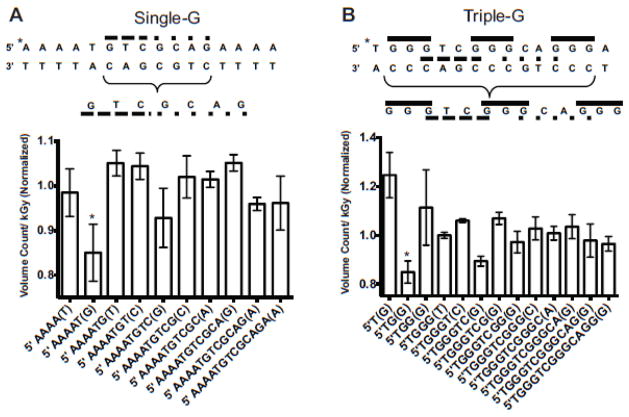
The probability of cleavage as indicated by the slope of the linear fit to the volume analysis was used to generate the bar graphs (Fig 2b and c). In all volume analysis results we observe a systematic increase in the apparent volume as the fragments get larger, related to the accumulation of more closely-spaced bands and a smoothly increasing background from the bottom to the top of the gel. In order to isolate qualitative trends of local strand break behavior, we applied a normalization algorithm (indicated below) to all data sets and provide the normalized, average slope of the damage response of each fragment in Figure 3.
Figure 3.
Normalized dose response for radiation-generated fragments. Normalization of data shown in Figure 2 based on rolling average within each data set to emphasize relative trends. Both single- and triple-G substrates are represented. Error bars represent the standard deviation for each group of data.
m′N represents the new normalized slope of strand break response at the 3′ terminus of a base N, mN corresponds to the original slope at site N before normalization, and the summation relations describes the average of the site being normalized, as well as the slope at sites 5′ and 3′ adjacent. In order to normalize the slope of strand break at either end of the ODN, we applied a linear regression to the 3 terminal bases, which was used to provide a predictive rolling average to the terminal site of damage. This normalization technique leads to a general reduced apparent error due to reduction in the variation of gel background and a higher contrast in cleavage trends.
The analysis indicates that cleavage levels likely depend on DNA sequence and/or position within the double-stranded ODN, as there are subtle but reproducible differences in cleavage at each base position. Given the high potential of guanine for oxidative damage, we chose to focus on strand break behavior at, and directly adjacent to, guanine base regions within the sequence. For example, the lowest-frequency cleavage occurs at guanine residues in both sequences studied, while strand breaks occur at other guanine residues at higher frequencies. Within both sequences, a guanine positioned adjacent to an adenine residue is sensitized to strand break damage. In the single-G sequence (Fig 3a) guanines positioned between two cytosine or thymine residues appear to be protected from cleavage. Additionally, in both cases the highest strand break frequency occurs in a guanine region when it is flanked by an adenine. This indicates that for both sequences a neighboring adenine sensitizes guanine to damage, thereby increasing frequency of stand breaks. In contrast, the presence of a neighboring pyrimidine protects guanine, lowering the frequency of strand breaks, with the possibility of thymine providing a stronger protective effect than cytosine. This effect is also evident in a comparison of the three guanine (GGG) sequences in the triple-G ODN (marked by solid lines in Figure 3b), which show very different cleavage profiles. The 5′-most GGG is bordered by a 3′ adenine residue and incurs strand breaks with high frequency, while a second GGG region harbors a central guanine residue cut with low frequency and is bordered by a 3′ cytosine. A third GGG sequence at the 3′ end of the ODN shows reduced cleavage at the central guanine residue, and is bordered by a 5′ thymine.
Another indication of the effect of sequence is seen for the sequence 5′-GCAG-3′, which occurs in both ODN (marked by dotted lines in Figures 3a and 3b), and appears to have a very similar cleavage profile in both. In contrast, the sequence 5′-GTC-3′ occurs at the same position in both ODNs but the guanine is cleaved at a much higher frequency when this sequence is bordered by a thymine rather than a guanine (marked by dashed lines in Figs 3a and 3b). Clearly, neighboring bases influence the probability of damage accumulation in addition to the identity of the base at the site of damage and strand breakage.
In addition to the effects of local base context on strand break formation, we also compared the relative strand break probability of guanine triplets within the triple-G sequences to their single-G analogues. In all cases, the relative probability of strand break of each guanine within a GGG is lower than the corresponding probability of strand break at the analogue single G site. This analysis is provided in Table II. These probabilities were calculated using the following equation:
Table II.
Relative Strand Break Probabilities. Analysis of the frequency of strand break of GGG regions (normalized against the frequency of all strand breaks within the substrate) compared with the strand break frequency of analogous triplets normalized using the same treatment. Analysis indicates a reduction in strand break occurrence for GGG regions when compared to their analogues with identical flanking base identity.
| Oligonucleotide | Triplet Sequence | Strand Break Probability |
|---|---|---|
| AAGAC | 0.147 ± 0.0056 | |
| Single-G | ACGCT | 0.117 ± 0.0071 |
| CTGTA | 0.086 ± 0.0109 | |
| AGGGA | 0.105 ± 0.0151 | |
| Triple-G | CGGGC | 0.096 ± 0.0117 |
| TGGGT | 0.063 ± 0.0064 |
Where RFPx is the Relative Fragmentation Probability of Fragment x, mx is the slope of fragment formation of Fragment x in volume count per kGy (See Fig 2), and mtot is the slope of fragment formation of all fragments for either single-G or triple-G sequences. Comparison of fragment formation, normalized against fragmentation of the entire ODN, indicates that guanine triplets are less susceptible to prompt strand breaks due to the direct effect of ionizing radiation compared to genomic regions lacking serial guanine bases.
Discussion
PAGE analysis of irradiated DNA films of a known sequence revealed an important correlation between radiation-produced strand cleavages and base context. It has been established that guanine is the most effective sink for oxidative damage in DNA (Steenken and Jovanovic 1997; Morikawa et al. 2014). As such, we focused on the cleavage probabilities at isolated guanine bases and guanine triplet repeats within two double-stranded sequences of interest, designated single-G and triple-G (Table 1) annealed to their complements, respectively. Importantly, we described the first application of this analysis method to determine site-specific strand break frequencies in dry DNA. To date, similar techniques have been used to analyze products formed via the indirect effect of IR damage, yielding a large amplification of base damage in guanine triplet regions (Oikawa et al. 2001; Spotheim-Maurizot and Davidkova 2011; Saito et al. 1998; Yoshioka et al. 1999). Our method relies on X-ray irradiation of DNA samples in which the amount of water has been reduced to a level well below what would be required to minimize cleavage due to the indirect effect of IR damage. To ensure that residual radicals do not result in damage due to the indirect effect upon re-solvation of the sample, we include an efficient radical scavenger (glycerol) in the buffer at a level far above what would be required to eliminate damage from diffusible hydroxyl radical generated either by chemical means (Dixon et al. 1991) or by IR dependent photolysis of water (results not shown).
We used this method to analyze DNA strand breaks that arise purely from direct effect of IR damage within single-G and triple-G sequences irradiated at a dose range from 15 to 240 kGy. This dose range is required for proper resolution of radiolytic cleavage products and previous studies have shown that strand break formation remains linear with respect to dose from low (mGy) to high (kGy) dose ranges (Sharma K et al. 2010; Sharma K et al. 2011). This linear relationship ensures that relative strand break probabilities observed at low dose will persist in the physiological dose range.
We demonstrate that strand breaks caused by the direct effect of IR can be visualized and quantified from denaturing 18% PAGE gels, subsequently dried and exposed to phosphor screens. As migration through such gels is sensitive to the identity of the ODN termini (Balasubramanian et al. 1998) our data indicate that the strand break mechanism produces 3′ monophosphoester termini, which migrate identically to fragments produced by Maxam-Gilbert sequencing reactions. We observed direct effect IR damage-induced strand breaks distributed throughout the ODN and occurring at all four bases. However, we find that strand breaks resulting from the direct effect of IR do not occur randomly along the DNA backbone.
Our analysis of cleavage data indicates that base context is a critical factor in the likelihood of strand break formation for a given sequence. As free base release analysis, quantification of products like C (8-oxo-G), and the ESR studies of mechanisms of 8-oxo-G cation radical formation from guanine cation radical has pointed to guanine as a sink for DNA damage induced by an oxidative pathway (Shukla et al. 2004; Steenken and Jovanovic 1997; Morikawa et al. 2014) we analyzed the relative probability of strand break occurrence at a guanine triplet compared to a single guanine base with identical flanking bases. This investigation builds on previous work in which ultraviolet A (UVA, wavelength 400-315 nanometers) irradiation studies identified GGG as a sink for base damage, showing dramatic increases in the production of 8-oxo-G through one-electron oxidation of the G within GGG regions, which resulted in strand breaks after hot piperidine treatment (Chung et al. 1992). It is known that strand breaks produced through the direct effect of IR are the result of formation of free radicals on deoxyribose moieties within the DNA backbone. Free radical formation on DNA bases, in comparison, does not produce strand breaks (von Sonntag 1987).
One could assume from this, that GGG regions within the genome would be less susceptible to strand breaks induced via the direct effect, as they are more susceptible to base damage. We tested this hypothesis through comparison of strand break formation in short ODNs of two sequences, one containing GGG and the other containing guanine bases isolated from other guanine within the sequence. Indeed we find a reduction of strand break probability of GGG when compared to their heterogeneous base triplets in the single-G sequence. For example, comparison of direct effect damage strand breaks at AGA within single-G to that at GGG flanked by adenine in triple-G indicates a significant reduction in relative strand break probability from 14.7% to 10.5%, in guanine regions compared to their triple-G analogue (Table II). This behavior persists for all single-G to triple-G strand break probability comparisons, indicating a reduction of susceptibility to prompt strand break from direct-type damage, regardless of base context.
Mechanisms of DNA Prompt Strand Break Formation Due to the Direct Effect of Ionizing Radiation
There are several observed mechanisms of prompt strand break formation after damage caused by the direct effect of IR found in the literature. First there is the neutral radical precursor mechanism via deprotonation following hole trapping at the deoxyribose moiety on the DNA backbone (Bernhard 2009; Close 2008; Sagstuen and Hole 2009; Adhikary et al. 2012). This reaction is in direct competition with hole transfer from the site of original hole formation on the sugar to the base stack and is driven by the oxidation potential of the DNA base neighboring the originally oxidized sugar. Once deprotonation occurs at any of the deoxyribose carbon atoms following one-electron oxidation, strand break occurs and damage transfer to the base stack can no longer take place (Adhikary et al. 2012, 2013). Studies performed have determined the free radical distribution at low temperatures using electron paramagnetic resonance (EPR). One such study determined a hole distribution of 78% trapped on the base stack, primarily at guanine bases, and 22% trapped on the sugar-phosphate backbone (Purkayastha et al. 2006). In addition, it was determined that only ⅓ of electron holes initially formed on the DNA backbone end up trapped at deoxyribose moieties at low temperature. This mechanism of strand break can be affected by DNA sequence context, with increased rates of hole transfer from the sugar to the base stack occurring with a nearby guanine base present (von Sonntag 2006; Becker et al. 2010). This could indicate an increased likelihood of strand break occurrence in DNA regions of high pyrimidine content, due to a less favorable oxidation potential of these bases and therefore a weaker driving force for hole transfer into the base stack.
Later work focusing on strand break patterns in DNA elucidated through the quantification of free base release products resulted in the discovery of sequence dependent factors in strand break probability (Sharma et al. 2010). This group found that guanine has the ability to sensitize thymine and protect cytosine from strand break. While this is interesting, we must consider the radiation chemistry behind each of these effects. The sensitization of thymine, as proposed by the authors, is likely due to two one-electron oxidation events producing a carbocation, resulting in strand break. Since the strand breaks we observe in this study are formed linearly with dose, we must consider this mechanism and other mechanisms of strand break employing sequential one-electron oxidation events (Bernhard 2009) to less frequently contribute to strand breaks than single-hit mechanisms outlined in this work. The second base-context dependent effect is more relevant as it is postulated to be due to transfer of a radical cation formed on cytosine to the sugar-phosphate backbone. It is proposed that this could occur only due to the unfavorable oxidation potential of the cytosine base. This mechanism only requires a single ionizing event and therefore could explain the less prominent protective effect observed in this work at the 5′ end of guanine flanked by cytosine bases when compared to guanine flanked by thymine bases.
Another mechanism that has been proposed by others is the transfer of an excited state cation from the base stack to the sugar-phosphate backbone. It is possible, due to secondary electron interactions with a one-electron oxidized group, to produce a base cation in the excited state. This radical species can then transfer to the deoxyribose, thereby resulting in a strand break. Of particular interest is work performed using ultraviolet (UV) excitation of trapped one-electron oxidized radicals in DNA (Becker et al. 2010). This group observed the formation of sugar radicals after UV excitation of base radicals at low temperature and a base context effect that could explain the results we outline in this work. Specifically, this group discovered that a neighboring thymine base can out-compete the transfer of the excited state electron hole from the guanine base to its sugar. This could be contributing to the protective effect we observe in which incidence of strand scission at the 5′ sugar of a guanine base is reduced when flanked on the 3′ side by a thymine or cytosine group. Moreover, it was observed that transfer rates of the excited state cation radicals increased with temperature, making this mechanism even more plausible considering our irradiations were performed at room temperature. Finally, this mechanism would only require one ionization per strand break, consistent with our finding that strand breaks occur linearly with dose.
A more recently discovered mechanism of strand break due to direct-type ionizing radiation damage is the dissociative electron attachment (DEA) of low energy electrons (LEE). In this case, LEEs are defined by an energy range of 0.5–50 eV. Under this mechanism of damage, free electrons at the end of an ionization track have the resonance energy required to break covalent bonds upon attachment to the DNA sugar-phosphate backbone. This is supported in the literature through both experimental results and density functional theory calculations (Caron and Sanche 2011; Kumar and Sevilla 2012). In the case of experimental evidence, investigators have documented both single strand breaks and double strand breaks and connected the probability of strand break to electron energy (Arumainayagam et al. 2010). While some measurement of LEE induced strand break was performed on DNA nucleotides in the gas phase, (Abdoul-Carime et al. 2004) the most compelling evidence was gathered on single layers of DNA ODNs in the condensed phase (Naaman and Sanche 2007; Ray et al. 2007; Zheng et al. 2005). These investigations were focused on single strand break formation in single stranded DNA (ssDNA), but some data was gathered on double stranded DNA (dsDNA). One interesting finding from these efforts was the increased free base release incidence at terminal bases. Moreover, base context dependence was observed with the highest free base release of A and T bases with T having been at the 3′ terminus of a tetramer. This could correspond to our results in which we observed a dramatic increase in strand breaks at the 3′ terminal T within our ‘single-G’ sequence (Figure 3). Further work in tetramers in which abasic sites were introduced (Zheng et al. 2006) showed a dramatic reduction in strand break incidence upon the absence of purine bases within the DNA strand. This again exhibits the importance of base context in the induction of strand breaks due to LEE. Other LEE investigations on DNA in the condensed phase observed sequence dependencies on strand break induction. Namely, it was found that electron capture probability increased with the presence of clustered guanine bases. This study also indicated decreased capture of LEEs in dsDNA compared to ssDNA (Ray et al. 2006). It can be argued that all free electrons produced along a radiation track eventually fall into this energy range, but it is unclear what the relative contribution LEE play in total strand breaks produced by ionizing radiation in vivo. In the context of our study we believe that stand breaks produced through LEE must be occurring and could be responsible for the base-context specific finding we have presented, namely behavior at the terminal T in our single-G sequence. However, DEA by LEE is not sufficient to explain our finding that guanine triplets are less susceptible to strand break given findings indicating increased LEE attachment to clustered guanine bases.
Future Studies
Our study observes DNA ODNs containing GGG regions to address the initial questions of strand break behavior under influence of base context. Previously, it has been observed that the probability of a strand break occurring at a particular site due to chemically generated reduced oxygen species is dependent on the DNA base sequence at that site. A pioneering study by Henle et al. showed that hyperoxia specifically created strand breaks more frequently to adjacent guanines (especially in GG or GGG formation) via site-specific Fenton reactions catalyzed by Fe2+. As a result, G-rich sequences exhibit an increased susceptibility to base damage and strand breaks, which can lead to end-to-end fusions, DNA mutations, cell death and other detrimental consequences (Henle et al. 1999; Honda et al. 2001; Ilyenko et al. 2011; Kawanishi et al. 1999; Kawanishi and Oikawa 2004; Lindahl 1993; Lu and Liu 2010; von Zglinicki 2002; von Zglinicki and Martin-Ruiz 2005; von Zglinicki et al. 1995, 2003). Notably, shielding of triple-G rich telomeric DNA by chromatinization to diffusible radicals and the reduced proportion of free water molecules within the nucleus greatly reduces the contribution of IR damage caused by the indirect effect of IR (Becker and Sevilla 1993). Additionally, the relationship between telomere dysfunction and IR is well reported (Ayouaz et al. 2008; Fumagalli et al. 2012; Genesca et al. 2006; Salin et al. 2008; Wong et al. 2000). These studies combined make telomere sequences prime subjects for direct-effect IR damage which is why in future experiments we would like to generate DNA substrates that simulate in vivo telomeric DNA. These would include double-stranded telomeric DNA and the highly stable G-quadruplex structures within the single-stranded G-rich overhang (Biffi et al. 2013; Gomez et al. 2006; Hänsel et al. 2013; Kumar et al. 2008; Lam et al. 2013; Sen et al. 1988; Wu et al. 2008; Paeschke 2005; Paeschke et al. 2008).
Our findings on sequence-dependent prompt strand break point to a single-event mechanism, but the specific mechanism of strand break observed here could not be explicitly determined given our experimental design. Future studies will be geared towards investigating the mechanism of sequence context on strand break incidence. Of particular interest is a comparison of sequences tested here to those containing abasic sites. This will determine not only the direct influence of a neighboring base on single-strand break amplification or protection, but can also control for any 3′ or 5′ damage preference for a given sequence. Through the use of enzymatic cleavage of specific base damage products, we can further reveal the interaction between base context and base damage as well as base context and prompt strand break formation.
In regards to our observed preference for large fragment formation, future work will also include 3′ end radiolabeling techniques as this would determine if this preference for strand break on the 3′ end of our sequences is an artifact of our assay or authentic. It must be noted, however, that our analysis accounted for this preference for large fragments by employing our rolling average correction, reinforcing the base context dependent behavior provided in Figure 4. Finally, it is possible that long irradiation times could have revealed heat labile damage sites or potential oxygen effects since irradiations occurred in open air. To address these possibilities, future experiments will include irradiations done on samples at 77 K by submersion in liquid nitrogen. This will address both temperature and oxygen effects that could be present.
With regard to discussed limitations of the performed study, we cannot make a firm conclusion about a quantification of base sequence effect. We demonstrate here the possible application of our technique to investigate this behavior in more detail with a larger number of sequences to fully describe the effect of base context on prompt strand break due to the direct effect of IR. We hope that this work will encourage other groups to adopt this technique to expand this work further. While our work here provides interesting findings on the relative yields of prompt strand break, it is important to emphasize that these are strictly qualitative, and given the experimental design employed here, cannot determine a quantified yield of strand break incidence at each site. Future studies will involve experimental designs that allow better quantification of strand break yields though careful titration of labeled DNA within the gel used to image strand break incidence. This will allow us to better compare our findings on prompt strand breaks determined through denaturing PAGE analysis to published strand break yields determined though free base release studies determined with HPLC (Swarts et al. 2007).
Conclusion
In this work we have established a novel method for evaluation of site-specific strand breaks in duplex DNA ODNs, generated by the direct effect of IR. Our method provides statistically significant information on the role of neighboring bases on strand break frequency at guanine bases. Although this study cannot give a yield of strand break occurrence in terms of a quantified chemical yield (nmol / J) as multiple titers would be needed with observations at increasing time intervals, these data do point to the possibility that guanine regions flanked by pyrimidine bases are protected from single-strand break formation, with an apparent increase in protection from thymine bases compared to cytosine bases. This behavior persists in both isolated G and triplet G regions. Given mechanisms of prompt strand break previously discussed, we propose that the mechanisms most likely contributing to the sequence dependent behavior are 1) base-sugar excited state radical cation transfer and 2) sugar-base hole transfer. Both mechanisms exhibit behavior that support our specific base context dependent findings and could be playing a significant role in our experimental system. In addition to our findings on sequence context, we have determined with this work that guanine triplets with analogous base context are less susceptible to prompt strand break formation from this type of damage when compared to isolated guanine bases. Additional studies using our methods will be integral to the determination of the effect of sequence context on single- and double-strand break formation, as well as the influence and nature of radical transport in these systems. Further studies will be required to determine the total susceptibility of G-rich sequences in G-quadruplex formation, to prompt strand break formation when compared to coding regions of the genome. Furthermore, quantitative data will be determined by testing a range of concentrations of dsDNA across the same time point range as this will give us information of the amount of strand breaks occur in nmols of DNA per joule of energy received. These future experiments will need to utilize an even proportion of damage events due to both direct and indirect effect of ionizing radiation damage. Finally, future studies should incorporate G-quadruplex structures in order to better approximate the relative damage contributions in vivo.
Acknowledgments
Compliance with Ethical Standards:
This study was funded by JJH (NIH Grant R01 GM052426)
The investigation was supported by PHS grant R01 CA32546, awarded by National Cancer Institute, DHHS. Miller currently supported by NIH Grant R01 GM057814. Hayes currently supported by NIH Grant R01 GM52426.
Footnotes
Conflict of Interest: Author ASM declares that he has no conflict of interest. Author PJB declares that he has no conflict of interest. Author JJH declares that he has no conflict of interest.
Ethical approval: This article does not contain any studies with human participants performed by any of the authors.
Declaration of Interest
The authors report no declarations of interest.
References
- Abdoul-Carime H, Gohlke S, Illenberger E. Site-specific dissociation of DNA bases by slow electrons at early stages of irradiation. Phys Rev Lett. 2004;16:168103. doi: 10.1103/PhysRevLett.92.168103. [DOI] [PubMed] [Google Scholar]
- Adhikary A, Becker D, Palmer BJ, Heizer AN, Sevilla MD. Direct formation of the C5′-radical in the sugar-phosphate backbone of DNA by high energy radiation. J Phys Chem B. 2012;116:5900–5906. doi: 10.1021/jp3023919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary A, Kumar A, Palmer BJ, Todd AD, Sevilla MD. Formation of S-Cl phosphorothioate adduct radicals in dsDNA-S-oligomers: Hole transfer to guanine vs. disulfide anion radical formation. J Am Chem Soc. 2013;135:12827–12838. doi: 10.1021/ja406121x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumainayagam CR, Lee HL, Nelson RB, Haines DR, Gunawardane RP. Low energy electron-induced reactions in condensed matter. Surf Sci Rep. 2010;65:1–44. doi: 10.1016/j.surfrep.2009.09.001. [DOI] [Google Scholar]
- Ayouaz A, Raynaud C, Heride C, Revaud D, Sabatier L. Telomeres: hallmarks of radiosensitivity. Biochimie. 2008;90:60–72. doi: 10.1016/j.biochi.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci U S A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Adhikary A, Sevilla MD. Physicochemical mechanisms of radiation-induced DNA damage. In: Hatano Y, Katsumura Y, Mozumder A, editors. Charged particle and photon interactions with matter: recent advances, applications, and interfaces. 1. New York: CRC Press; 2010. pp. 503–541. [Google Scholar]
- Becker D, Sevilla MD. The chemical consequences of radiation damage to DNA. Adv Radiat Biol. 1993;17:121–180. http://dx.doi.org/10.1016/B978-0-12-035417-7.50006-4. [Google Scholar]
- Bernhard WA. Radical reaction pathways initiated by direct energy deposition in DNA by ionizing radiation. In: Greenberg MM, editor. Radical and radical ion reactivity in nucleic acid chemistry. 1. New Jersey: John Wiley and Sons; 2009. pp. 41–68. [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA g-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candeias LP, Steenken S. Ionization of purine nucleosides and nucleotides and their components by 193-nm laser photolysis in aqueous solution: model studies for oxidative damage of DNA. J Am Chem Soc. 1992;114:699–704. doi: 10.1021/ja00028a043. [DOI] [Google Scholar]
- Caron LC, Sanche L. Theoretical studies of electron interactions with DNA and its subunits: from tetrahydrofuran to plasmid DNA. In: Čársky P, Čurik R, editors. Low-energy electron scattering from molecules, biomolecules and surfaces. 1. New York: CRC Press; 2012. pp. 161–230. [Google Scholar]
- Chung MH, Kiyosawa H, Ohtsuka E, Nishimura S, Kasai H. DNA strand cleavage at 8-hydroxyguanine residues by hot piperidine treatment. Biochem Biophys Res Commun. 1992;188:1–7. doi: 10.1016/0006-291X(92)92341-T. [DOI] [PubMed] [Google Scholar]
- Close DM. From the primary radiation induced radicals in DNA constituents to strand breaks: low temperature EPR/ENDOR studies. In: Shukla MK, Leszcyznski J, editors. Radiation-induced molecular phenomena in nucleic acids: a comprehensive theoretical and experimental analysis. 1. New York: Springer; 2008. pp. 493–529. [Google Scholar]
- Cullings HM, Fujita S, Funamoto S, Grant EJ, Kerr GD, Preston DL. Dose Estimation for Atomic Bomb Survivor Studies: Its Evolution and Present Status. Radiat Res. 2006;166:219–254. doi: 10.1667/RR3546.1. doi: http://dx.doi.org/10.1667/RR3546.1. [DOI] [PubMed] [Google Scholar]
- Cullis PM, Malone ME, Merson-Davies LA. Guanine radical cations are Precursors of 7,8-dihydro-8-oxo-2′-deoxyguanosine but are not precursors of immediate strand breaks in DNA. J Am Chem Soc. 1996;118:2775–2781. doi: 10.1021/ja9536025. [DOI] [Google Scholar]
- Ding LH, Shingyoji M, Chen F, Hwang JJ, Burma S, Lee C, Cheng JF, Chen DJ. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res. 2005;164:17–26. doi: 10.1667/rr3354. doi: http://dx.doi.org/10.1667/RR3354. [DOI] [PubMed] [Google Scholar]
- Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD. Hydroxyl radical footprinting. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- Doddridge ZA, Cullis PM, Jones GDD, Malone ME. 7,8-dihydro-8-oxo-2′-deoxyguanosine residues in DNA are radiation damage “hot” spots in the direct γ radiation damage pathway. J Am Chem Soc. 1998;120:10998–10999. doi: 10.1021/ja9816234. [DOI] [Google Scholar]
- Douple EB, Mabuchi K, Cullings HM, Preston DL, Kodama K, Shimizu Y, Fujiwara S, Shore RE. Long-term radiation-related health effects in a unique human population: lessons learned from the atomic bomb survivors of Hiroshima and Nagasaki. Disaster Med Public Health Prep. 2011;5:S122–S133. doi: 10.1001/dmp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, Herbig U, Longhese MP, D’adda diFagagna F. Telomeric DNA damage is irreparable and causes persistent DNA damage-response activation. Nat Cell Biol. 2012;14:355–365. doi: 10.1038/ncb2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates K. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22:1747–60. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genesca A, Martin M, Latre L, Soler D, Pampalona J, Tusell L. Telomere dysfunction: a new player in radiation sensitivity. BioEssays. 2006;28:1172–1180. doi: 10.1002/bies.20501. [DOI] [PubMed] [Google Scholar]
- Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadré J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, Riou JF. The g-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- Gu J, Leszczynski J, Schaefer HF., III Interactions of electrons with bare and hydrated biomolecules: from nucleic acid bases to DNA segments. Chem Rev. 2012;112:5603–5640. doi: 10.1021/cr3000219. [DOI] [PubMed] [Google Scholar]
- Hänsel R, Löhr F, Trantirek L, Dötsch V. High-resolution insight into g-overhang architecture. J Am Chem Soc. 2013;135:2816–2824. doi: 10.1021/ja312403b. [DOI] [PubMed] [Google Scholar]
- Henle ES, Han Z, Tang N, Rai P, Luo Y, Linn S. Sequence-specific DNA cleavage by Fe2+-mediated Fenton reactions has possible biological implications. J Biol Chem. 1999;274:962–971. doi: 10.1074/jbc.274.2.962. [DOI] [PubMed] [Google Scholar]
- Honda S, Hjelmeland L, Handa J. Oxidative stress-induced single-strand breaks in chromosomal telomeres of human retinal pigment epithelial cells in vitro. Invest Ophthalmol Vis Sci. 2001;42:2139–2144. [PubMed] [Google Scholar]
- Ilyenko I, Lyaskivska O, Bazyka D. Analysis of relative telomere length and apoptosis in humans exposed to ionising radiation. Exp Oncol. 2011;33:235–238. [PubMed] [Google Scholar]
- Kawanishi S, Oikawa S. Mechanism of telomere shortening by oxidative stress. Ann N Y Acad Sci. 2004;1019:278–284. doi: 10.1196/annals.1297.047. [DOI] [PubMed] [Google Scholar]
- Kawanishi S, Oikawa S, Murata M, Tsukitome H, Saito I. Site-specific oxidation at gg and ggg sequences in double-stranded DNA by benzoyl peroxide as a tumor promoter. Biochemistry. 1999;38:16733–16739. doi: 10.1021/bi990890z. [DOI] [PubMed] [Google Scholar]
- Krisch R, Flick M, Trumbore C. Radiation chemical mechanisms of single- and double-strand break formation in irradiated SV40 DNA. Radiat Res. 1991;126:251–259. doi: 10.2307/3577826. [DOI] [PubMed] [Google Scholar]
- Kumar N, Sahoo B, Varun K, Maiti S, Maiti S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nuc Acids Res. 2008;36:4433–4442. doi: 10.1093/nar/gkn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Sevilla M. Low-energy electron (LEE)-induced DNA damage: theoretical approaches to modeling experiment. In: Shukla M, Leszczynski J, editors. Handbook of computational chemistry volume III: applications–biomolecules. Berlin: Springer-Verlag; 2012. pp. 1215–1256. [Google Scholar]
- Lam E, Beraldi D, Tannahill D, Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun. 2013;4:1796. doi: 10.1038/ncomms2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Lu J, Liu Y. Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. EMBO J. 2010;29:398–409. doi: 10.1038/emboj.2009.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melvin T, Cunniffe S, O’Neill P, Parker A, Roldan-Arjona T. Guanine is the target for direct ionisation damage in DNA, as detected using excision enzymes. Nucleic Acids Res. 1998;26:4935–4942. doi: 10.1093/nar/26.21.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Kino K, Oyoshi T, Suzuki M, Kobayashi T, Miyazawa H. Analysis of guanine oxidation products in double-stranded DNA and proposed guanine oxidation pathways in single-stranded, double-stranded or quadruplex DNA. Biomolecules. 2014;4:140–59. doi: 10.3390/biom4010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naaman R, Sanche L. Low-energy electron transmission through thin-film molecular and biomolecular solids. Chem Rev. 2007;107:1553–1579. doi: 10.1021/cr040200j. [DOI] [PubMed] [Google Scholar]
- Oikawa S, Tada-Oikawa S, Kawanishi S. Site-specific DNA damage at the ggg sequence by UVA involves acceleration of telomere shortening. Biochemistry. 2001;40:4763–4768. doi: 10.1021/bi002721g. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates g-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol. 2008;15:598–604. doi: 10.1038/nsmb. [DOI] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of g-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Ray S, Daube S, Cohen H, Naaman R. Electron capturing by DNA. Isr J Chem. 2007;47:149–159. doi: 10.1560/IJC.47.2.149. [DOI] [Google Scholar]
- Purkayastha S, Milligan J, Bernhard W. The role of hydration in the distribution of free radical trapping in directly ionized DNA. Radiat Res. 2006;166:1–8. doi: 10.1667/RR3585.1. doi: http://dx.doi.org/10.1667/RR3585.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagstuen E, Hole E. Radiation produced radicals. In: Brustolon M, Giamello E, editors. Electron paramagnetic resonance: a practitioner’s toolkit. 1. New Jersey: John Wiley and Sons; 2009. pp. 325–381. [Google Scholar]
- Saito I, Nakamura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. Mapping of the hot spots for DNA damage by one-electron oxidation: efficacy of gg doublets and ggg triplets as a trap in long-range hole migration. J Am Chem Soc. 1998;120:12686–12687. doi: 10.1021/ja981888i. [DOI] [Google Scholar]
- Salin H, Ricoul M, Morat L, Sabatier L. Increased genomic alteration complexity and telomere shortening in B-CLL cells resistant to radiation-induced apoptosis. Cytogenet Genome Res. 2008;122:343–349. doi: 10.1159/000167821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Sharma K, Milligan J, Bernhard W. Multiplicity of DNA single-strand breaks produced in pUC18 exposed to the direct effects of ionizing radiation. Radiat Res. 2008;170:156–162. doi: 10.1667/RR1277.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Swarts S, Bernhard W. Mechanisms of direct radiation damage to DNA: the effect of base sequence on base end products. J Phys Chem B. 2011;115:4843–4855. doi: 10.1021/jp200902h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Tyagi R, Purkayastha S, Bernhard W. One-electron oxidation of DNA by ionizing radiation: competition between base-to-base hole transfer and hole-trapping. J Phys Chem B. 2010;114:7672–7680. doi: 10.1021/jp101717u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla L, Adhikary A, Pazdro R, Becker D, Sevilla M. Formation of 8-oxo-7,8-dihydroguanine-radicals in γ-irridated DNA by multiple one-electron oxidations. Nucleic Acids Res. 2004;32:6565–6574. doi: 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotheim-Maurizot M, Davidkova M. Radiation damage to DNA in DNA-protein complexes. Mutat Res. 2011;711:41–48. doi: 10.1016/j.mrfmmm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Steenken S, Jovanovic S. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J Am Chem Soc. 1997;119:617–618. doi: 10.1021/ja962255b. [DOI] [Google Scholar]
- Sutherland B, Bennett P, Saparbaev M, Sutherland J, Laval J. Clustered DNA damages as dosemeters for ionising radiation exposure and biological responses. Radiat Prot Dosim. 2001;97:33–38. doi: 10.1093/oxfordjournals.rpd.a006635. [DOI] [PubMed] [Google Scholar]
- Swarts S, Gilbert D, Sharma K, Razskazovskiy Y, Purkayastha S, Naumenko K, Bernhard W. Mechanisms of direct radiation damage in DNA, based on a study of the yields of base damage, deoxyribose damage, and trapped radicals in d (GCACGCGTGC)(2) Radiat Res. 2007;168:367–381. doi: 10.1667/RR1058.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts S, Sevilla M, Becker D, Tokar C, Wheeler K. Radiation-induced DNA damage as a function of hydration. I. release of unaltered bases. Radiat Res. 1992;129:333–344. doi: 10.2307/3578034. [DOI] [PubMed] [Google Scholar]
- von Sonntag C. The chemical basis of radiation biology. 1. New York: Taylor and Francis; 1987. [Google Scholar]
- von Sonntag C. DNA and double-stranded oligonucleotides. In: Shreck S, editor. Free Radical Induced DNA and Its Repair. 1. Berlin: Springer-Verlag; 2006. pp. 382–390. [Google Scholar]
- von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/S0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- von Zglinicki T, Martin-Ruiz C. Telomeres as biomarkers for ageing and age-related diseases. Curr Mol Med. 2005;5:197–203. doi: 10.2174/1566524053586545. [DOI] [PubMed] [Google Scholar]
- von Zglinicki T, Petrie J, Kirkwood T. Telomere-driven replicative senescence is a stress response. Nat Biotechnol. 2003;21:229–230. doi: 10.1038/nbt0303-229b. [DOI] [PubMed] [Google Scholar]
- von Zglinicki T, Saretzki G, Docke W, Lotze C. Mild hyperoxia shortens telomeres and inhibits proliferation of fibroblasts: a model for senescence. Exp Cell Res. 1995;220:186–193. doi: 10.1006/excr.1995.1305. [DOI] [PubMed] [Google Scholar]
- Wong K, Chang S, Weiler S, Ganesan S, Chaudhuri J, Zhu C, Artandi S, Rudolph K, Gottlieb G, Chin L, Alt F, Depinho R. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh R., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y, Kitagawa Y, Takano Y, Yamaguchi K, Nakamura T, Saito I. Experimental and theoretical studies on the selectivity of ggg triplets toward one-electron oxidation in B-form DNA. J Am Chem Soc. 1999;121:8712–8719. doi: 10.1021/ja991032t. [DOI] [Google Scholar]
- Zheng Y, Cloutier P, Hunting D, Sanche L, Wagner J. Chemical basis of DNA sugar-phophate cleavage by low-energy electrons. J Am Chem Soc. 2005;127:16592–16598. doi: 10.1021/ja054129q. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wagner J, Sanche L. DNA damage induced by low-energy electrons: electron transfer and diffraction. Phys Rev Lett. 2006;96:208101. doi: 10.1103/PhysRevLett.96.208101. [DOI] [PubMed] [Google Scholar]