Abstract
Purpose
This study was performed to further our understanding of the biological and genetic basis of follicular lymphoma (FL) and to identify potential novel therapy targets.
Experimental Design
We analyzed previously generated whole exome sequencing data of 23 FL cases and one transformed FL case and expanded findings to a combined total of 125 FL/ 3 t-FL. We modeled the 3D-location of RRAGC-associated hotspot mutations. We performed functional studies on novel RRAGC mutants in stable retrovirally transduced HEK293T cells, stable lentivirally transduced lymphoma cell lines and in Saccharomyces cerevisiae.
Results
We report recurrent mutations, including multiple amino acid hotspots, in the small G-protein RRAGC, which is part of a protein complex that signals intracellular amino acid concentrations to MTOR, in 9.4% of FL cases. Mutations in RRAGC distinctly clustered on one protein surface area surrounding the GTP/GDP binding sites. Mutated RRAGC proteins demonstrated increased binding to RPTOR (raptor) and substantially decreased interactions with the product of the tumor suppressor gene FLCN (folliculin). In stable retrovirally transfected 293T cells, cultured in the presence or absence of leucine, multiple RRAGC mutations demonstrated elevated MTOR activation as evidenced by increased RPS6KB/S6-kinase phosphorylation. Similar activation phenotypes were uncovered in yeast engineered to express mutations in the RRAGC homolog Gtr2 and in multiple lymphoma cell lines expressing HA-tagged RRAGC mutant proteins.
Conclusions
Our discovery of activating mutations in RRAGC in ~10% of FL provides the mechanistic rationale to study mutational MTOR activation and MTOR inhibition as a potential novel actionable therapeutic target in FL.
Keywords: Follicular lymphoma, exome sequencing, RRAGC mutations
Introduction
Follicular lymphoma (FL) constitutes the most common indolent B-cell lymphoma, with an incidence and prevalence of ~14,000 and ~100,000 cases, respectively in the US(1). FL remains incurable with conventional therapies and most patients receive multiple treatment regimens during the course of their illness. The development of targeted FL-directed therapies is in early stages (2–7) and lags behind progress made in other lymphoproliferative diseases such as CLL or MCL.
FL has a varied clinical course that is influenced by FL tumor cell-intrinsic and cell-extrinsic deregulations(2, 5, 6, 8–12). Cell-intrinsic factors, including genomic aberrations and epigenetic deregulations, prominently influence the FL phenotype. At the genomic level, FL is characterized by recurrent acquired structural abnormalities, including translocation (14;18) deregulating BCL2 expression, acquired genomic copy number aberrations (aCNA) and frequent acquired uniparental disomy (aUPD)/copy neutral LOH (cnLOH)(13–17). Furthermore, an expanding group of recurrently mutated genes has recently been shown to underlie the pathogenesis of FL (KMT2D/MLL2, CREBBP, EZH2, EP300, ARID1A, HIST1H1 B-E, STAT6 and others) and some of these afford novel opportunities for targeted therapy developments(18–26).
To further our understanding of the biological and genetic basis of FL and to identify targets for novel therapeutic approaches, we have further analyzed FL WES data and have identified recurrent mutations in the MTOR regulator RRAGC, a small G-protein. RRAGC forms heterodimers with either RRAGA or RRAGB and together these dimers are part of a multi-protein complex that under conditions of amino acid sufficiency facilitates recruitment of MTOR to lysosomal membranes resulting in MTOR activation through RHEB(27–31). Through sequencing of RRAGC in a combined 125 FL / 3 t-FL cases, we identified novel RRAGC mutations, including multiple mutation hotspots, in ~10% of FL. Detailed functional studies in multiple complementary experimental systems demonstrated that FL-associated RRAGC mutations are activating MTOR as evidenced by elevated phosphorylated (p)-RPS6KB/S6-kinase (S6K) levels. In aggregate these data provide novel insights into the properties of recurrent FL-associated RRAGC mutations and have implications for novel therapy developments targeting MTOR in genetically defined FL subsets.
Methods
Patient characteristics and study source material
Twenty-three FL cases and one transformed FL provided the source material for massive parallel sequencing (WES) as described and characteristics of these cases have been published(18, 22). Results from the Sanger-based re-sequencing of a combined total of 125 FL and 3 t-FL are reported here. Sixty-six FL / 3 t-FL patients (cohort I) were enrolled between 2007 and 2014 into two separate lymphoma repositories at the University of Michigan Comprehensive Cancer Center (IRBMED #HUM00007985 and IRBMED #HUM00017055). The study IRBMED #HUM00007985 allowed procurement of material from patients with lymphoma through fine needle aspiration of externally palpable lymph nodes. The study IRBMED #HUM00017055 allowed procurement of patient material during times of clinically indicated lymphoma surgeries. Genomic research on all specimens was approved through IRBMED #HUM00005467; Genomic analysis of B-cell Non-Hodgkin’s lymphoma. We furthermore analyzed by Sanger sequencing DNA from 59 sorted FL cases (cohort II) collected between 1990 and 2005 as previously described that were derived from de-identified leftover clinical material(14).
Exon resequencing of RRAGC in FL
Primers to amplify and sequence all coding exons and adjacent intronic sequences of RRAGC were designed using the primer 3 program (http://primer3.ut.ee/) and sequence information generated using direct sequencing as described. Mutations were confirmed to be somatically acquired using unamplified lymphoma cell-derived DNA and paired CD3 cell-derived DNA as templates isolated from highly pure flow-sorted cells.
RRAGC cDNA mutagenesis, retroviral and lentiviral vector generation, cell transfection and cell transduction to generate stable cell lines
Reagents and mutagenesis
A pCMV-SPORT plasmid containing the RRAGC cDNA (cat#: MHS6278-202757712; accession: BC016668) was purchased from ThermoScientific, and used as a template to generate mutant RRAGC cDNAs using the QuikChange® Lightning Site-Directed Mutagenesis Kit (Stratagene/Agilent, La Jolla, CA). Full-length wild type and mutant HA-tagged RRAGC were constructed using PCR and cloned into the PacI/NotI sites of the lentiviral vector FG9 (a gift from Dr. Colin Duckett, University of Michigan(32)). HA-tagged raptor (#8513) and RRAGB WT (#19301) was purchased from Addgene (Cambridge, MA)(27). Anti-HA antibody was purchased from Roche, Basel Switzerland (clone 3F10, #11867423001). Anti-raptor (05–1470) was from Millipore. Anti-beta-Actin, (#A544) was from Sigma Aldrich. Antibodies against RRAGC (#3360), anti-folliculin (#3697), anti-RRAGB (#8150), anti-RRAGA (#4357), anti-S6Kinase (#9202) and anti-pS6-Kinase-Thr 389 (#9205) were purchased from Cell Signaling Technology (Danvers, MA).
Transient transfection studies in HEK293T cells
HEK293T cells were transfected in 10 cm dishes with 1 or 2 µg of plasmids encoding either wild type (WT) or mutant forms of RRAGC or various other constructs as indicated using polyethylenimine (Polyscience Inc., #23966).
Generation of stable transduced HEK293T cell lines
RRAGC cDNAs excised from pCR Blunt topo vectors using EcoR1 were ligated into MSCV-IRES-PURO (MIP)(33) previously dephosphorylated with shrimp alkaline phosphatase (#78390; Affymetrix Santa Clara, CA). Retroviral production and infection of cells followed by puromycin selection were performed as described previously (33). The expression of transgenes and comparison with endogenous RRAGC expression was verified by immunoblotting.
Generation of stable lymphoma cell lines expressing HA-tagged wild type RRAGC and mutant RRAGC
HEK293T packaging cells were transfected with 5 µg FG9 (either the vector alone or vector expressing wild type or various mutant forms of RRAGC), together with 1 µg of the plasmids REV, RRE and VSVG. Viral supernatant fractions were collected 48–72 h after transfection by low speed centrifugation to remove cells and debris. The cell lines OCI-LY1, OCI-LY7 and SUDHL4 were infected by spin-inoculation at 30°C at 2600 rpm using 8µg/ml of Polybrene for 2 h before seeding into fresh medium. Enrichment for GFP-expressing cells was performed using FACS. Stably transfected cells were tested for expression levels of RRAGC proteins using RRAGC- and HA-directed antibodies and immunoblotting.
Culture of stable cell lines, leucine deprivation, immunoblotting and immuno-precipitations
OCI-LY1 and OCI-LY7 lines were obtained from the University of Michigan in 2005 with permission of the cell line originators at the Ontario Cancer Institute and stored cryopreserved. The SUDHL4 cell line was obtained from the University of Michigan in 2013 (Dr. Elenitoba-Johnson) and stored cryopreserved. Cell line authentication was performed through sequence analysis of six to eight gene mutations per line based on the COSMIC cell line project (catalogue of somatic mutations in cancer; http://cancer.sanger.ac.uk/cell_lines). Prior to leucine starvation, HEK293T cells and SUDHL4 cells were cultured in RPMI1640 medium supplemented with 10% FCS. LY1 and LY7 cells were cultured in DMEM supplemented with 20% FCS and 50µM beta-mercaptoethanol. For leucine starvation (60’), we employed leucine-free RPMI medium (#R1780; SIGMA Aldrich) supplemented with 10% dialyzed FCS using dialysis tubing (#D7884; Sigma-Alrich).
For immunoprecipitations cells were pelleted, washed and lysed on ice for 20 min in lysis buffer containing 0.3% CHAPS detergent (#DSC41010; Dot Scientific), 100mM NACL, 25mM Tris pH 8.0 (#T6066; Sigma Aldrich), 20mM NAF, 2mM EGTA, 2mM EDTA (#ED2SS; Sigma Aldrich), supplemented with protease inhibitors (#P8340) and phosphatase inhibitors (#P0044), and Sodium orthovanadate (#450243; Sigma-Aldrich, St. Louis, MO), and PMSF (#36978; Thermo Scientific). The detergent-soluble fraction of the cell lysates was cleared by centrifugation at 14,000 rpm for 10 min. For anti-HA immunoprecipitations, anti-HA-loaded beads (#A2095; Sigma Aldrich) were blocked with 5% BSA/TBST for 0.5 h, then washed three times with lysis buffer. Then, 20 µl anti-HA-conjugated beads were added to pre-cleared cell lysates and incubated with rotation for 5 h at 4°C. The beads were washed 4 times with lysis buffer containing 150 mM NaCl and protein liberated through boiling in 1×SDS-PAGE loading buffer. Protein was fractionated through SDS-PAGE and prepared for immunoblotting using standard procedures. For immunoblottings, the detergent used was 1% NP-40 instead of CHAPS (see immunoprecipitation conditions above) and NACL was used at 150 mM.
Computational methods
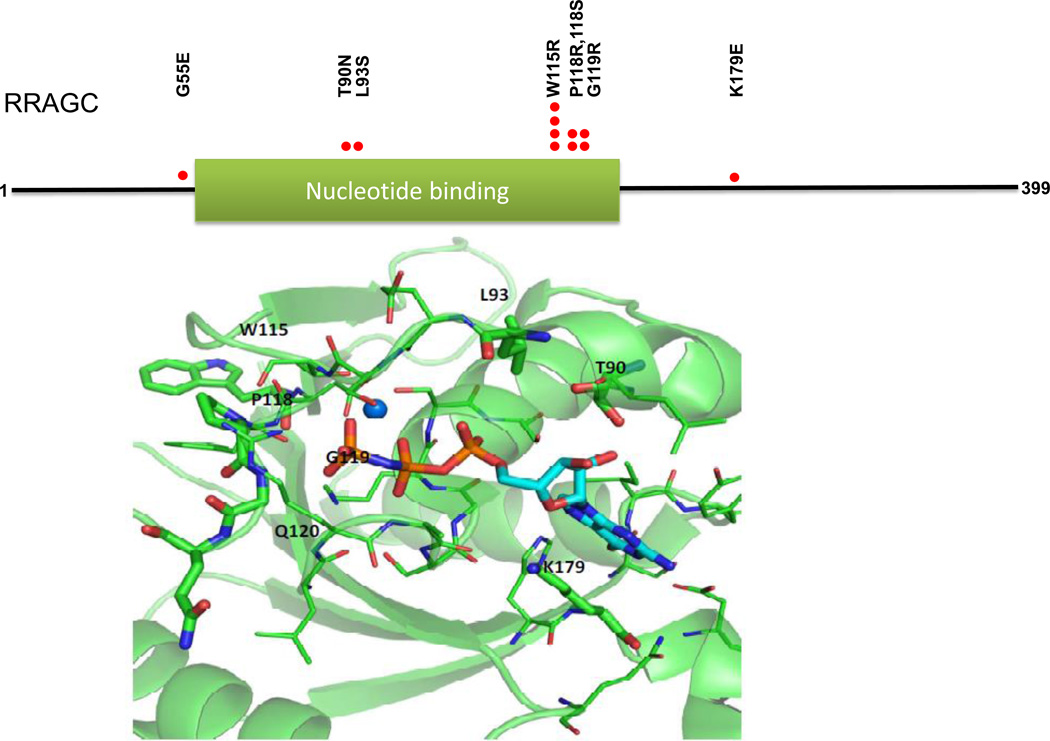
We located the identified RRAGC point mutations in the crystal complex (PDB: 3LLU) of the RRAGC nucleotide binding domain bound with the nucleotide analog phosphoamino-phosphonic acid guanylate ester. Residues undergoing mutation were labeled and are shown in stick representation, while other residues within 4 angstroms of GNP are shown in line representation. The magnesium ion is shown as a sphere. The Figure 1 was generated with PyMOL.
Figure 1.
FL-associated RRAGC mutations cluster on the protein surface surrounding the GTP/GDP binding site. Upper: Location of identified RRAGC missense mutations in a linear schema of RRAGC. Lower: Location of identified RRAGC missense mutations in the crystal complex (PDB:3LLU) of the RRAGC nucleotide binding domain (G-domain) bound with the nucleotide analog phosphoaminophosphonic acid guanylate ester (GNP), shown in stick representation. Amino acid residues undergoing mutation are labeled and shown in stick representation, while other residues within 4 angstroms of GNP are shown in line representation. A magnesium ion is shown as a sphere. The figure was generated with PyMOL.
The crystal structure of RRAGC was used for creating initial models of the protein in complex with GTP and GDP by modifying the GNP molecule in the original crystal structure. Complexes with point mutations of protein residues were also modeled using Molecular Operating Environment (MOE; 2013.08; Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2016). All complexes included a single monomer of RRAGC with the Mg ion from the crystal structure, and were initially subjected to energy minimization in AMBER (34) with the TIP3P solvent model, followed by MD simulations. After initial minimization of the solvent, the system was further relaxed with constraints on the backbone before final minimization. MD simulations involved a gradual increase in temperature to 300K over 30ps, while holding the solute constrained, followed by another 30ps of simulation with the constraint only on the backbone. Further equilibration was performed for 40ps before the production run for 10ns. Shake was applied to all bonds involving hydrogen permitting a time step of 0.002ps. The GTP and GDP parameters were obtained from published AMBER parameter database. Conformations were saved every 1ps for analysis. Energy decomposition was performed using MM/GBSA from the AMBER package utilizing conformers saved at 10ps intervals.
Yeast methods
Yeast strains used in this study are listed in Supplementary Table 1. Gene modifications were performed using standard methods (35, 36), and the primers used to introduce mutations into the GTR2 gene are listed in Supplementary Table 2. Yeast cells were cultured in YPD medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] glucose) to mid-log phase and then collected for the +N samples. Yeast cells grown in YPD were shifted into SD-N medium (0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% [w/v] glucose) for 2 h and then collected for the -N samples. The Pho8∆60 assay was performed to measure autophagy activity as previously described (37).
Results
Identification of recurrent somatic mutations in RRAGC in follicular lymphoma
We have previously reported initial results of paired-end massively parallel sequencing of DNA isolated from flow-sorted immunoglobulin light chain-restricted lymphomatous B-cells and paired CD3+ T-cells isolated from twenty-three cases of FL and one transformed FL (t-FL) (18, 22). Within this discovery group of 23 FL /1 t-FL cases, we identified two RRAGC mutations in cases L42 and L50 (Table 1). Re-sequencing of all RRAGC coding exons using Sanger sequencing in highly pure FL B cell DNA in a combined total of 125 FL / 3 t-FL cases identified a total of 9.4% (12/128) of cases with non-synonymous RRAGC mutations (Table 1). All RRAGC mutations were heterozygous.
Table 1.
Details of gene mutations in FL cases carrying RRAGC mutations.
| FL Case# |
Disease status |
RRAGC cDNA |
RRAGC PROTEIN |
STAT6 | CREBBP | KMT2D/MLL2 | TNFRSF14 | EZH2 | MEF2B | HIST1H1 B-E | ARID1A | OCT2 | TP53 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FL065 | unknown | c.164 G>A/G | p.55 G>E/G | WT | M | M | WT | WT | WT | WT | WT | WT | M |
| FL006 | unknown | c.269 C>A/C | p.90 T>N/T | WT | M | M | WT | WT | WT | M | M | WT | WT |
| ML007 | untreated | c.278 T>C/T | p.93 L>S/L | - | - | - | - | - | - | - | - | - | - |
| L026 | untreated | c.343 T>C/T | p.115 W>R/W | M | M | M | WT | WT | WT | WT | WT | WT | WT |
| L042 | untreated | c.343 T>C/T | p.115 W>R/W | WT | M | WT | WT | WT | M | WT | WT | WT | WT |
| L066 | untreated | c.343 T>C/T | p.115 W>R/W | WT | M | M | WT | WT | WT | WT | WT | WT | WT |
| ML107 | relapsed | c.343 T>C/T | p.115 W>R/W | - | - | - | - | - | - | - | - | - | - |
| L050 | untreated | c.353 C>C/G | p.118 P>P/R | WT | M | M | WT | M | WT | WT | WT | WT | WT |
| L055 | relapsed | c.352 C>C/T | p.118 P>P/S | WT | M | M | WT | WT | M | M | WT | M | WT |
| FL007 | unknown | c.355 G>A/G | p.119 G>R/G | WT | WT | M | WT | WT | WT | M | WT | WT | WT |
| FL015 | unknown | c.355 G>A/G | p.119 G>R/G | WT | WT | WT | WT | WT | WT | WT | M | WT | M |
| ML104 | relapsed | c.535 A>A/G | p.179 K>K/E | - | - | - | - | - | - | - | - | - | - |
Within the spectrum of RRAGC mutations, we identify multiple closely spaced novel RRAGC mutation hotspots in amino acid residues 115, 118 and 119 (Table 1 and Figure 1). In addition, a second group of mutations targeted RRAGC residues 90 and 93. The RRAGC mutation p.W115>R/W was the most common recurrent RRAGC mutation identified. FL-associated RRAGC mutations targeting amino acid residues 115–118 were also recently reported by Okosun et al (38). In their paper, the authors also identified relatively frequent mutations targeting residues 74 – 99 with a hotspot mutation at residue 90 (p.T90N), which we also detected albeit at lower frequencies.
A comparative analysis of the presence of other genes known to be recurrently mutated in FL (KMT2D/MLL2, CREBBP, STAT6, linker histones B-E, POU2F2, EZH2, TNFRSF14, TP53, MEF2B and ARID1A) in the RRAGC mutated cases did not disclose overt associations when taking into account the overall high mutation frequency for these genes in FL, with the possible exception of a lack of mutations in TNFRSF14.
Of the 12 RRAGC mutations reported here, 5 were detected in untreated FL patients and 3 in relapsed patients, while the clinical and treatment status of 4 patients with mutations detected in samples from FL cohort II is unknown (see Table 1 and methods). One RRAGC mutation occurred in a t-FL.
FL-associated RRAGC mutations are located on the surface of the RRAGC protein surrounding the GTP/GDP binding site
We generated 3-dimensional data based on the RRAGC crystal structure while bound to phosphoaminophosphonic acid guanylate ester, a non-reactive GTP analog, and mapped the location of the FL-associated RRAGC mutations onto the model; all RRAGC mutations were located on the surface of the G-domain of RRAGC, a region implicated in protein-protein interaction in addition to nucleotide binding, suggesting a potential mechanism of action (see below)(39). The RRAGC hotspot mutations in amino acid residues 115, 118 and 119 were located close to the third phosphate group of the nucleotide, while the mutations in amino acid residues 90, 93 and 179 were located alongside the nucleotide (Figure 1).
Analysis of the crystal structure of RRAGC in complex with GNP indicated that except for W115 all other mutant sites lie within 4 angstroms of the nucleotide analog (residue G55 lies outside the determined structure). As such, the mutant residues could potentially contribute to GTP/GDP binding either via their side chains or through backbone interactions. To investigate this further, we initially built models of the RRAGC-GTP/GDP complexes, and performed single in silico point mutations for each residue of interest, resulting in complexes of GTP and GDP with either wild type or mutated RRAGC. MD simulations of each complex were performed and the complexes were assessed for stability of the bound GTP/GDP nucleotide. We found minimal or no perturbation in the GTP/GDP binding orientation during the time course of the simulations (Supplementary Figures S1A &S1B, Table S3 and Supplementary Results).
We also performed a quantitative analysis of the change in interaction energy of the mutated residues with GTP/GDP (Supplementary Table S4), using energy decomposition analysis for each of the complexes. This further confirmed that the contributions of the individual residues, upon mutation, towards nucleotide binding are largely unaffected.
Overall, the changes in contributions to interaction energy for the mutants are similar for both GTP and GDP, suggesting that most of these would have similar effects, if any on the binding of either nucleotide.
Intrinsic activation of FL-associated RRAGC mutants as detected through HEK293T cell assays
HEK293T cells have been extensively used in the identification and functional characterization of components of the ragulator-RRAGA/B/C/D-MTOR pathway(27). In preliminary transient transfection assays performed in HEK293T or HELA cells, we noted that the co-transfection of RRAGC with RRAGB stabilized both proteins as well as stabilized RPTOR protein levels. This co-stabilization, however, was mostly independent of RRAGC mutant status.
To derive initial insights into the properties of FL-associated RRAGC mutant proteins, we next employed stably retroviral transduced HEK293T cells expressing HA-RRAGC WT and HA-RRAGC mutants (90N, 115R, 118R 118S and 119R; see Table 1). RRAGC expression levels were measured by immunoblotting using RRAGC and HA antibodies, which were aided by the slower migration of the HA-tagged RRAGC WT and mutant proteins in SDS-PAGE resulting in a doublet band (Figure 2). The expression levels of HA-RRAGC protein were approximately similar to the endogenous RRAGC levels facilitating subsequent data interpretation.
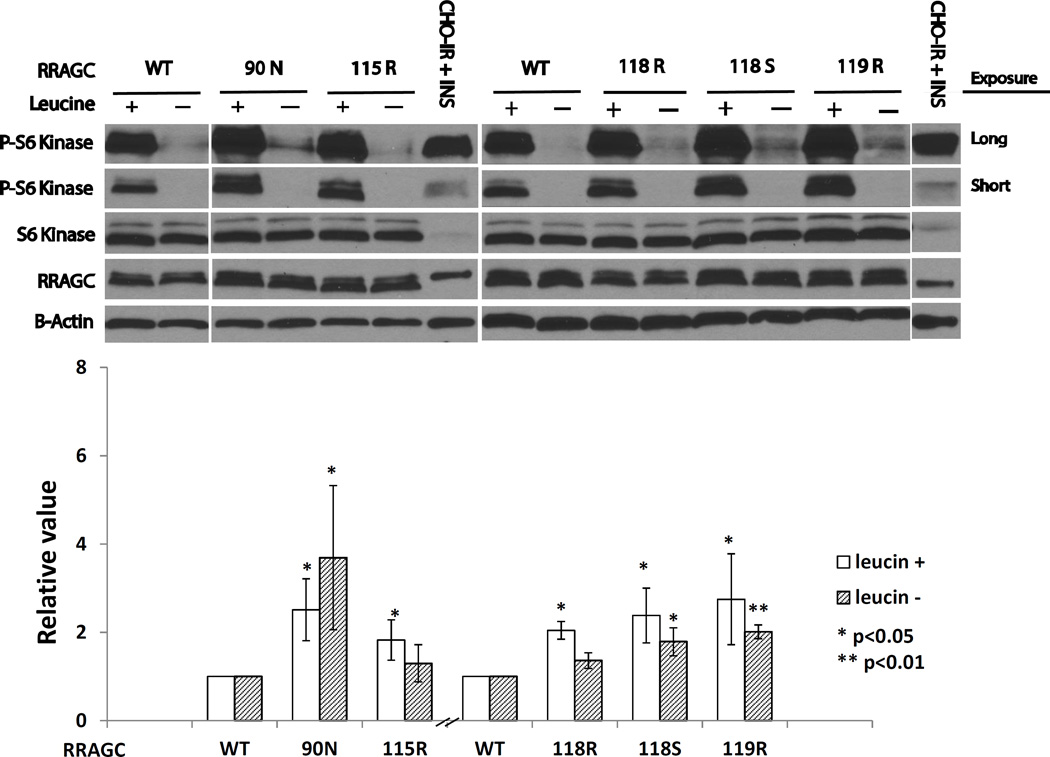
Figure 2.
FL-associated RRAGC mutations are intrinsically mTOR activating. Upper: HEK293T cells stably retrovirally infected expressing HA-tagged RRAGC wild type or various RRAGC mutants were grown in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS; +) or alternatively for the last 1 h of culture in RPMI1640 medium that was free of the amino acid leucine and supplemented with dialyzed 10% FBS (−). Cells were harvested and prepared for immunoblotting with various antibodies as indicated. Short and long exposures for RPS6KB/S6K (S6K) and p-Thr389-RPS6KB/S6K (p-S6K) are shown. CHO-IR + INS: Insulin receptor transfected CHO cells stimulated with insulin (immunoblot controls). Measurements of HA-RRAGC expression levels were aided by the slower migration of the HA-tagged RRAGC WT and mutant proteins in SDS-PAGE resulting in a doublet band. Lower: Displayed are combined quantitation results (p-S6K/total S6K) from 3 independent experiments using ImageJ densitometry results indexed to the measurements for RRAGC wt.
To identify effects of mutant RRAGC on MTOR activity as measured through RPS6KB/S6K phosphorylation at residue Thr389, we grew stably transduced HEK293T cells in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) or alternatively for the last 1 h of culture in RPMI1640 medium that was free of the amino acid leucine and supplemented with dialyzed 10% FBS. Cells were harvested and prepared for immunoblotting with various antibodies, in particular to monitor the activation status of RPS6KB/S6K. A representative experiment (N=3) is shown in Figure 2, upper panel. As a control we monitored CHO-IR cells treated with insulin, which activates MTOR signaling. As expected, we could detect p-Thr389-RPS6KB/S6K in extracts from these cells. Most pronounced across multiple experiments and cell lines tested (see below) was an activation phenotype for the RRAGC mutant p.90N as measured through p-Thr389-RPS6KB/S6K levels. MTOR is normally activated in the presence of leucine, resulting in phosphorylation of RPS6KB/S6K, whereas MTOR inhibition following leucine starvation results in reduced or near absent activity as seen for WT RRAGC. The RRAGC p.90N mutant displayed elevated MTOR activity (an increase in p-RPS6KB/S6K) in the presence of leucine (Figure 2, upper and lower panels). We also detected residual MTOR activity as measured through p-Thr389-RPS6KB/S6K levels even under leucine-free conditions; the latter is aberrant as evidenced by the relatively low level of activity in the RRAGC WT cells, and likely reflects residual MTOR activity. Modestly elevated p-Thr389-RPS6KB/S6K levels were also detected in all other RRAGC mutants overall indicative of a mild intrinsic activation phenotype of FL-associated RRAGC mutants towards MTOR.
Results of experiments performed in stable lentiviral transduced large B-cell lymphoma cell lines expressing HA-RRAGC-WT and various HA-RRAGC mutants
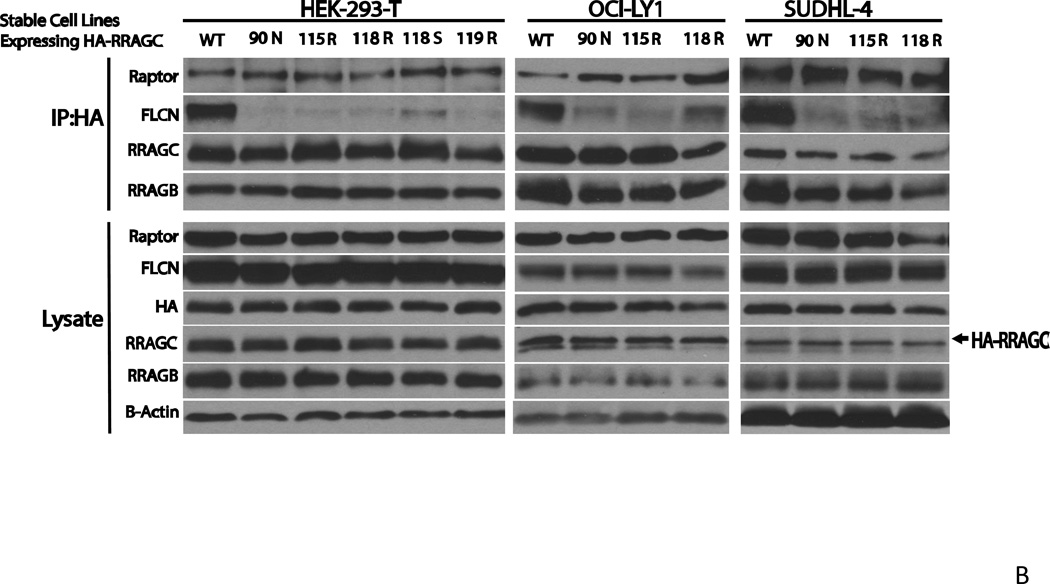
To analyze the properties of FL-associated RRAGC mutants (90N, 115R and 118R) in lymphoid cell lines that express endogenous wild type RRAGC, we generated stable lymphoma cell lines through lentiviral infection and cell sorting. Given the absence of FL cell lines, we resorted to commonly studied B cell lymphoma cell lines (OCI-LY1, LY7 and SUDHL4) that are designated as GCC-DLBCL lines. The resulting stable lines expressed appreciable quantities of exogenous RRAGC, which migrated slightly slower than endogenous RRAGC resulting in a doublet band by SDS-PAGE (Figure 3).
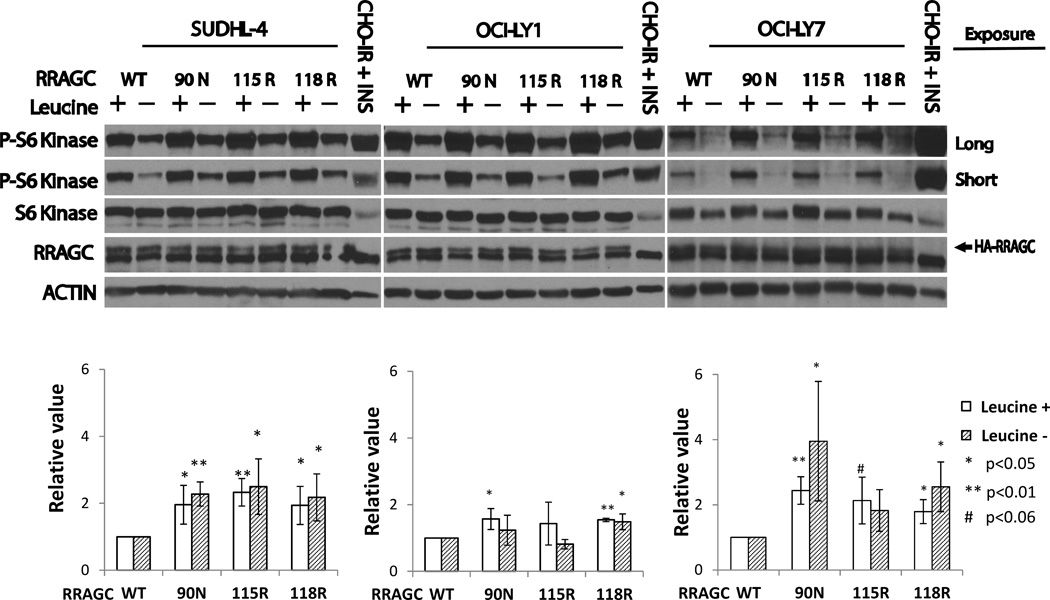
Figure 3.
Measurements of p-Thr389-RPS6KB/S6K levels in RRAGC WT or mutant stably lentivirally transduced lymphoma cell lines. Upper: Lymphoma cell lines stably lentivirally infected with constructs expressing HA-tagged RRAGC wild type or various RRAGC mutants as indicated were grown in RPMI or DMEM medium supplemented with 10% FBS (+) or alternatively for the last 1 h of culture in medium that was free of the amino acid leucine and supplemented with dialyzed 10% FBS (−). Cells were harvested and prepared for immunoblotting with various antibodies as indicated. Short and long exposures for p-Thr389-RPS6KB/S6K are shown. CHO-IR + INS: Insulin receptor transfected CHO cells stimulated with insulin (immunoblot controls). Measurements of HA-RRAGC expression levels were aided by the slower migration of the HA-tagged RRAGC WT and mutant proteins in SDS-PAGE resulting in a doublet band. Lower: Displayed are combined quantitation results (p-S6K/total S6K) from 3 independent experiments per cell line using ImageJ densitometry results indexed to the measurements for RRAGC wt.
Next, we measured p-Thr389-RPS6KB/S6K levels in lymphoma cell lines grown in fully supplemented medium or for the final 60 min in leucine-free medium supplemented with dialyzed FCS (N=3 independent experiments). In all three cell lines, grown in the presence or absence of the amino acid leucine, we detected an apparent increase in MTOR activation; MTOR activation was most pronounced in SUDHL4 and LY7 and less pronounced in LY1 (Figure 3, upper and lower panels).
We also analyzed the cell growth of these HA-RRAGC WT and mutant transduced large B-cell lymphoma cell lines when grown either in fully serum supplemented medium or in medium containing reduced serum concentrations or alternatively reduced leucine concentrations. Overall, we noticed no major effect of the RRAGC mutation status on cell numbers or viability.
Intrinsic activation of FL-associated RRAGC mutants as detected through assays in Saccharomyces cerevisiae
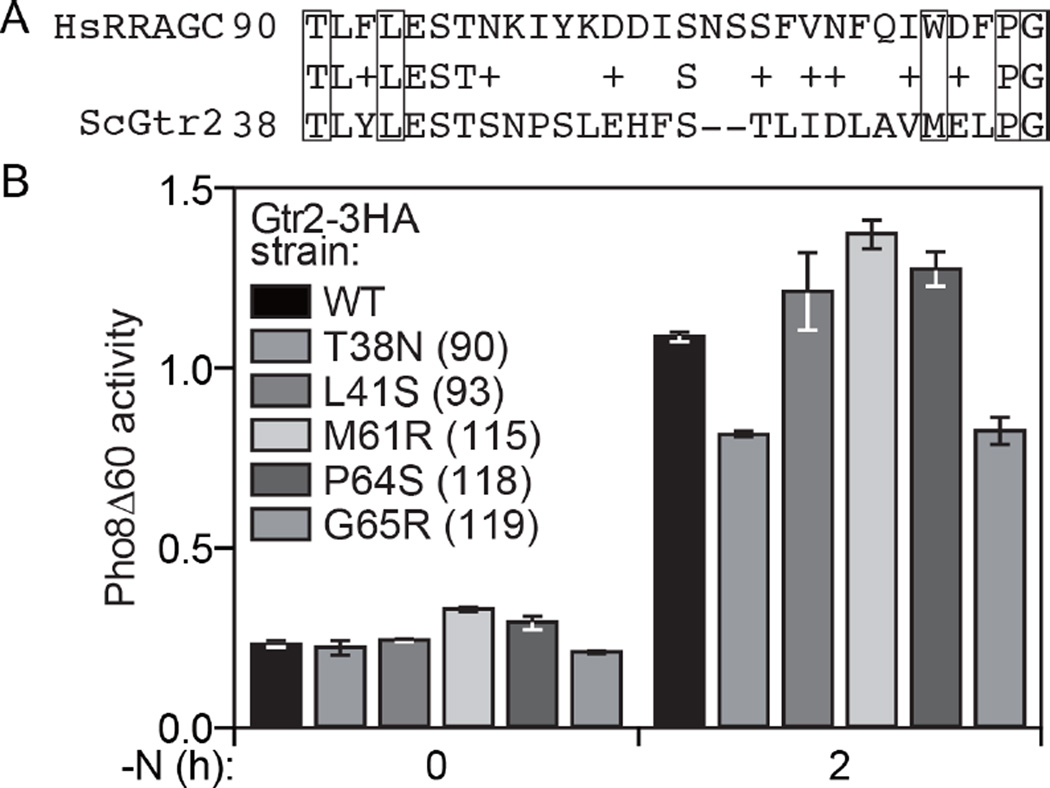
Next, we decided to extend these studies using a model organism and examined the effect of the corresponding mutations in the RRAGC homolog Gtr2 in the yeast Saccharomyces cerevisiae. The amino acid residues at positions 38/90 (yeast/human), 41/93, 64/118 and 65/119 are conserved, whereas yeast have a methionine residue at position 61 compared to tryptophan at residue 115 for human (Figure 4A). To monitor the effect of the mutations on TOR activity we took advantage of the quantitative Pho8∆60 enzymatic assay. In brief, Pho8∆60 is a modified form of a vacuolar phosphatase that remains in the cytosol due to the removal of its signal sequence. Under conditions of reduced TOR activity there is an upregulation of macroautophagy, which results in the vacuolar delivery and activation of the Pho8∆60 zymogen (40). Thus, in WT cells there was only a low level of basal Pho8∆60 activity in growing conditions (0 h of nitrogen starvation [-N]; Figure 4B). Inhibition of TOR resulting from removal of nitrogen from the medium resulted in a substantial increase in activity. A mutation of T38N or G65R caused a substantial reduction in Pho8∆60 activity, corresponding to elevated TOR activity. This result agrees in particular with the elevated TOR activity seen with the p.90N mutation of RRAGC (Figure 2). In contrast, the other three Gtr2 mutations displayed essentially WT, or even slightly elevated levels of Pho8∆60 activity. This apparent difference may reflect differences in the structure of Gtr2 compared to RRAGC, or differences in the binding partners between these two proteins. Nonetheless, these data support the hypothesis that mutations at Gtr2/RRAGC positions 38/90 and 65/119 in particular affect the activity of TOR/MTOR, resulting in hyperactivation.
Figure 4.
Analysis of TOR activation resulting from Gtr2 mutations in yeast. (A) Alignment of human and yeast RRAGC and Gtr2 proteins, respectively. Mutated residues are boxed. (B) Yeast cells with the PHO8 locus replaced with pho8∆60 were chromosomally-tagged with Gtr2-3HA WT and the indicated mutants. Protein extracts were prepared from cells in growing conditions (0 h) or after 2 h of nitrogen starvation, and assayed for Pho8∆60-dependent phosphatase activity.
Co-immunoprecipitation studies of RRAGC-WT or various HA-RRAGC mutants and RPTOR, RRAGB or FLCN detects increased RPTOR and decreased FLCN binding by mutant RRAGC
To begin addressing the mechanism of MTOR activation by RRAGC mutations we focused on interactions with RPTOR and FLCN, the former a known binding protein for RRAG heterodimers and the later a known tumor suppressor implicated in MTOR signaling and the causative gene for Birt-Hogg-Dube syndrome(41). We performed two sets of immunoprecipitation experiments: i) In HEK293T cells transiently co-transfected with plasmids encoding HA-tagged RPTOR, RRAGB wt and RRAGC WT or RRAGC mutants to test the in vivo complex formation between these proteins; and ii) in stable transduced HEK293T cells or lymphoma cell lines expressing HA-tagged RRAGC WT or HA-RRAGC mutants. Note that in the former experiment the immunoprecipitation (IP) bait was HA-RPTOR, while in the latter set of experiments the IP bait was HA-RRAGC.
In preliminary transient co-transfection experiments in HEK293T cells, we noted that FLCN co-immunoprecipitated with HA-RPTOR even under conditions lacking exogenous RRAGC (Supplementary Figure 2) suggesting direct FLCN-RPTOR binding.
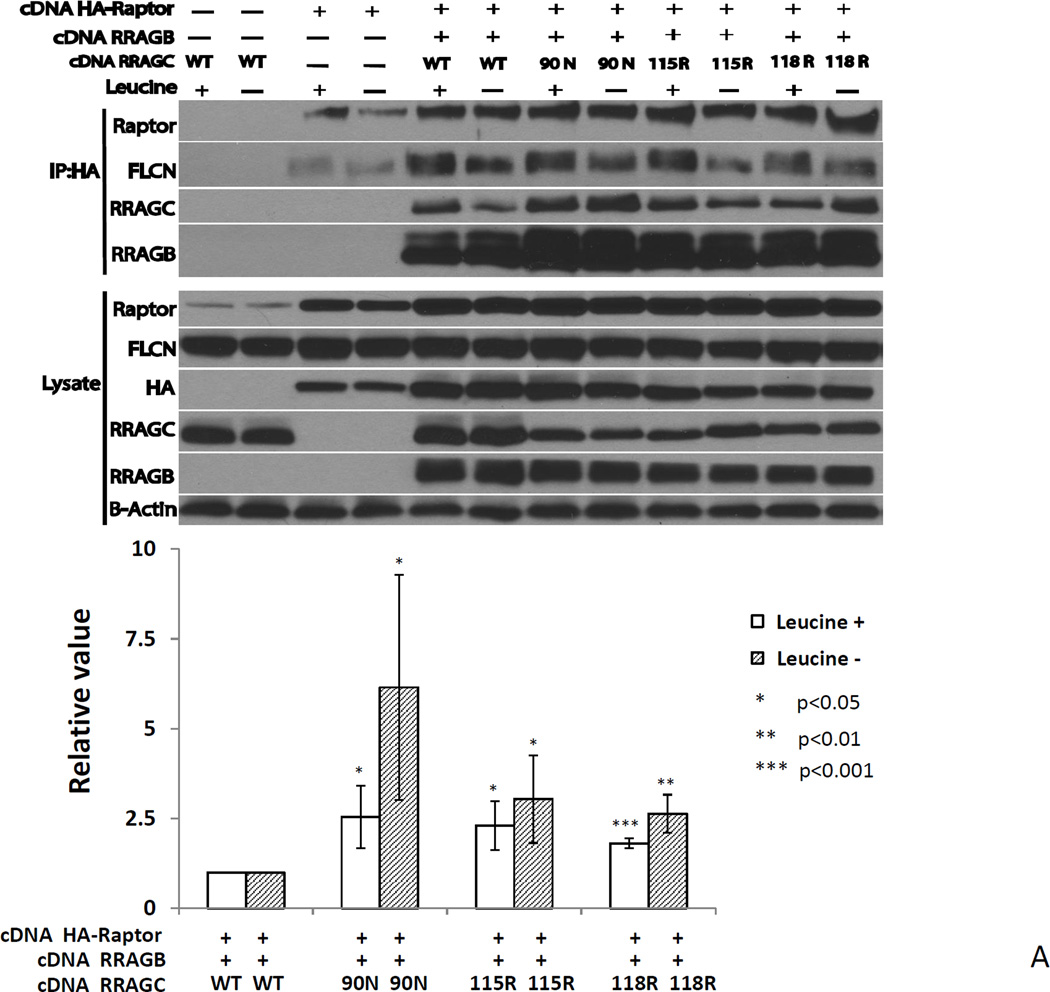
Next, we transiently co-transfected HEK293T cells with plasmids encoding HA-tagged RPTOR, RRAGB WT and RRAGC WT or RRAGC mutants and performed IPs using anti-HA covalently bound to beads. After washing and protein fractionation by SDS-PAGE, we performed immunoblotting on proteins liberated from HA beads as well as on aliquots of cell lysate not subjected to IPs (Figure 5A). Two principle findings emerged: 1) HA-RPTOR co-immunoprecipitated RRAGB and RRAGC WT as expected (Figure 5A, lanes 5 and 6). In comparison, greater amounts of the RRAGC mutants were co-immunoprecipitated, and this was particularly the case with the p.90N mutant (Figure 5A, compare lanes 7 and 8 with lanes 5 and 6; for quantification see lower panel). There was a similar increase in the amount of co-immunoprecipitated RRAGB, which likely reflects the interaction of this protein with RRAGC and stabilization of the complex. Prior leucine starvation did not influence the heightened RRAGC-RPTOR interaction. Okosun et. al. had similarly demonstrated an increased mutant RRAGC-RPTOR binding suggesting a mechanisms for the observed increased mTOR activation(38). 2) HA-RPTOR co-immunoprecipitated FLCN in the absence of co-transfected RRAGB/C (Figure 5A, lanes 3 and 4), but the FLCN amount was increased in the presence of the RRAGB/C heterodimer. Of interest, under leucine-starved conditions, a slower migrating FLCN band was much lower in intensity and differential FLCN phosphorylation, as previously described, may underlie this observation(42).
Figure 5.
A RRAGC mutants demonstrate increased HA-RPTOR binding in co-immunoprecipitation studies in transiently transfected HEK293T cells. Upper: HEK293T cell were transiently co-transfected as indicated with expression plasmids coding for HA-RPTOR, RRAGB, RRAGC WT or the indicated RRAGC mutants. CHAPS detergent lysates were prepared and subjected to anti-HA-bead conjugate-mediated immunoprecipitations. Bound protein was eluted and fractioned by SDS-PAGE and prepared for immunoblotting with the indicated antibodies. Leucine starvation (Leu −) was performed in parallel transfections. Lysate: aliquots of lysates prior to immunoprecipitation. Lower: Displayed are combined quantitation results (RRAGC IP band intensities divided by RRAGC detergent cell lysates band intensities normalized to raptor IP band densities) from 3 independent experiments using ImageJ densitometry results indexed to the measurements for RRAGC wt.
B Decreased co-immunoprecipitation of FLCN with HA-RRAGC mutants in stably transduced HEK293T cells or lymphoma cell lines. Stable HA-RRAGC WT or mutant transduced HEK293T cells or the LY1 and SUDHL4 lymphoma cell lines were used. CHAPS detergent lysates were prepared and subjected to anti-HA-bead conjugate-mediated immunoprecipitations. Bound protein was eluded and fractioned by SDS-PAGE and prepared for immunoblotting with the indicated antibodies. Lysate: aliquots of lysates prior to immunoprecipitation. The position of transduced HA-RRAGC is marked by an arrow.
Next, we performed IPs out of CHAPS detergent lysates from stably transduced HEK293T cells or the LY1 and SUDHL4 lymphoma cell line expressing HA-tagged RRAGC WT or HA-RRAGC mutants, with the goal of detecting protein interactors expressed endogenously. After washing and protein fractionation by SDS-PAGE, we performed immunoblotting on proteins liberated from HA beads as well as on aliquots of cell lysate not subjected to IPs. Under these conditions, we again observed increased binding of RPTOR but, more strikingly, a very substantially impaired ability for mutant RRAGC proteins to bind FLCN, overall suggesting an alternative mechanism for increased MTOR activity in RRAGC mutant cells (Figure 5B). The much impaired ability of RRAGC mutant proteins to interact with FLCN provides impetus for future in-depth study of this impaired interaction in the pathophysiology of RRAGC mutants in FL.
Discussion
In this manuscript, we present the discovery and initial functional characterization of heterozygous mutations in RRAGC, an mTOR regulator, in ~10% of follicular lymphoma. FL-associated RRAGC mutations predominantly targeted three novel mutation hotspots in amino acid residues 115, 118 and 119 and also target amino acid residues 90, 93 and 179, all of which are located on a protein surface surrounding the RRAGC GTP/GDP binding site.
Given extensive prior work on the biology of RRAGC, in particular the involvement of RRAGC as part of a multi protein complex in amino acid sensing pathways that regulate the MTOR activation state, we focused our initial attention on the effect of FL-associated RRAGC mutations on MTOR activity(27–29). In various experimental systems, we demonstrate that FL-associated RRAGC mutations confer a mild MTOR activation phenotype as measured through the phosphorylation state of the widely used substrate RPS6KB/S6K; we also detected a similar result for the corresponding Gtr2 mutations by monitoring TOR activity in yeast. The nucleotide state of RRAGC, when part of a heterodimer with GTP-loaded RRAGA, has been implicated as a regulatory mechanism of MTOR activation in some but not all published studies(43, 44), but we do not know if this is the explanation for the phenotype we observed and our extensive structural modeling studies do not support strong effects of RRAGC mutations on GTP/GDP binding.
It is also possible that the RRAGC mutations result in altered interactions with MTOR binding partners. In subsequent experiments, aiming at examining whether such a mechanism operates for mutant RRAGC-mediated MTOR activation, we observed increased binding of RRAGC mutants to RPTOR, which likely contributes to the observed MTOR activation phenotype. In addition, we make the unexpected novel observation of substantially decreased binding of all RRAGC mutants to the product of the tumor suppressor gene FLCN, the causative gene for the genetic syndrome Birt-Hogg-Dube, suggesting that indeed this impaired interaction is also involved in MTOR regulation and underlies the pathobiology of FL-associated RRAGC mutations. This hypothesis fits well with the known pro-growth pathologies observed in patients with Birt-Hogg-Dube syndrome (41) or mice with engineered deletion of the BHD gene(45).
While this work was in progress, Okuson and colleagues reported on a 17% incidence of RRAGC mutations in FL (but very few or no such mutations in other lymphoid neoplasias), with a comparatively higher incidence of mutations targeting residues surrounding amino acid 90 in RRAGC(38). The authors also observed MTOR activation properties of the RRAGC mutations and increased RPTOR binding. Together, these two studies identify a previously unrecognized connection between the amino acid signaling pathway to MTOR and the pathobiology of FL B cell lymphoma.
The functional data presented in this study support the conclusion that FL-associated RRAGC mutations are activating and like other initial reports open avenues for novel research aiming at an in-depth understanding of the effects of these mutants on FL B cells and possibilities for therapeutic applications. Here, analysis of primary FL B cells carrying RRAGC mutations may be of value as may be the introduction of mutant RRAGC alleles into the mouse germline or into mice sensitized for FL development(46). Of additional interest would be studies aiming at understanding if indeed the RRAGC mutations result in an addiction to MTOR in so afflicted FL B cells and if such potential addiction could result in therapeutic opportunities in FL subsets. Given the absence of untransformed FL cell lines, such studies await availability of sufficient quantities of RRAGC-mutated primary FL cases to perform such experiments and the development of relevant animal models.
There are, however, additional considerations worth mentioning with regard to mutant RRAGC function: First, it was surprising that only RRAGC is mutated in FL while mutations in RRAGA, RRAGB or RRAGD were absent in the examined cohort. This is noteworthy, as in multiple studies RRAGA is the dominant regulator of RPTOR interactions and MTOR activation in cells. What is unique to RRAGC that selects for mutations in FL? Our studies offer one hypothesis centered on the impaired interaction of RRAGC mutants with FLCN and imply that indeed FLCN would have a negative regulatory role in this context and follow-up studies are needed to explore this further. Second, the measured MTOR activation for some of the RRAGC mutations was relatively mild potentially implying that other RRAGC mutant-regulated pathways could be discovered. Are RRAGC mutations compensating in a unique manner for other FL changes, including frequent gene mutations that are present in FL B cells? Finally, given the tight control of MTOR activity in cells and the complexity of regulatory pathways, context-dependent findings may emerge in the future.
In summary, the data provided here in aggregate provide insights in the mutational activation of the MTOR pathway in FL and provide fertile grounds for future research, including but not limited to the exploration of the MTOR pathway as a therapeutic target for small molecules, including MTOR inhibitors in genetically defined subsets of FL (47–49).
Supplementary Material
TRANSLATIONAL RELEVANCE.
Follicular Lymphoma (FL) constitutes the second most common NHL in the United States, with over 100,000 patients living with the disease. Almost all patients with FL need therapy within years from diagnosis, and most patients receive multiple chemo- or immunotherapies over their lifetimes. Despite recent progress, the development of targeted therapies in FL is in its early stages. Here, we report on recurrent activating mutations in the MTOR regulator RRAGC, a small G-protein and a component of the cellular amino acid sensing and sufficiency signaling pathway in ~10% of FL. FL-associated RRAGC mutations activated MTOR as evidenced by elevated phosphorylated RPS6KB/S6-kinase phosphorylation in multiple experimental systems. These data provide the mechanistic rationale to study mutational MTOR activation in FL and MTOR inhibition as a potential novel actionable therapeutic target in FL subsets.
Acknowledgments
We are grateful for services provided by the genomics and bioinformatics core of the University of Michigan Comprehensive Cancer Center. We are also grateful to the contribution of the hematological malignancy group of the University of Michigan. We thank Dr. Ken Inoki (Life Sciences Institute, University of Michigan) for the generous donation of plasmids.
FUNDING SOURCES
This work was supported by R01CA190384 (to SNM), the Weatherhall foundation (to MK), R01GM053396 (to DJK) and a Scholar in Clinical Research Award by the Leukemia and Lymphoma Society (SNM).
Footnotes
Presented as part of a podium presentation at the American Society of Hematology Meeting, Orlando FL, 2015.
INDIVIDUAL CONTRIBUTIONS
Mark Kaminski, Alfred Chang and Sami N. Malek enrolled patients and analyzed clinical data.
Zhang Xiao Ying, Luke F. Peterson, Mehmet Yildiz, Kamlai Saiya Cork and Sami Malek performed the laboratory research.
Denzil Bernard and Shaomeng Wang assisted with the structural modeling.
Meiyan Jin and Daniel Klionsky performed and/or interpreted all yeast experiments.
Sami Malek conceived the study and wrote the paper, which was edited by Daniel Klionsky.
CONFLICT OF INTEREST
There is no conflict of interest to disclose.
REFERENCES
- 1.Morton LM, Wang SS, Devesa SS, Hartge P, Weisenburger DD, Linet MS. Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 3.Cheung KJ, Johnson NA, Affleck JG, Severson T, Steidl C, Ben-Neriah S, et al. Acquired TNFRSF14 mutations in follicular lymphoma are associated with worse prognosis. Cancer Res. 2010;70:9166–9174. doi: 10.1158/0008-5472.CAN-10-2460. [DOI] [PubMed] [Google Scholar]
- 4.Stevenson FK, Stevenson GT. Follicular lymphoma and the immune system: from pathogenesis to antibody therapy. Blood. 2012;119:3659–3667. doi: 10.1182/blood-2011-11-367730. [DOI] [PubMed] [Google Scholar]
- 5.Relander T, Johnson NA, Farinha P, Connors JM, Sehn LH, Gascoyne RD. Prognostic factors in follicular lymphoma. J Clin Oncol. 2010;28:2902–2913. doi: 10.1200/JCO.2009.26.1693. [DOI] [PubMed] [Google Scholar]
- 6.Kridel R, Sehn LH, Gascoyne RD. Pathogenesis of follicular lymphoma. J Clin Invest. 2012;122:3424–3431. doi: 10.1172/JCI63186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salles G, Ghesquieres H. Current and future management of follicular lymphoma. Int J Hematol. 2012;96:544–551. doi: 10.1007/s12185-012-1202-y. [DOI] [PubMed] [Google Scholar]
- 8.Roulland S, Faroudi M, Mamessier E, Sungalee S, Salles G, Nadel B. Early steps of follicular lymphoma pathogenesis. Adv Immunol. 2011;111:1–46. doi: 10.1016/B978-0-12-385991-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 9.Leich E, Ott G, Rosenwald A. Pathology, pathogenesis and molecular genetics of follicular NHL. Best Pract Res Clin Haematol. 2011;24:95–109. doi: 10.1016/j.beha.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Bende RJ, Smit LA, van Noesel CJ. Molecular pathways in follicular lymphoma. Leukemia. 2007;21:18–29. doi: 10.1038/sj.leu.2404426. [DOI] [PubMed] [Google Scholar]
- 11.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112:E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myklebust JH, Irish JM, Brody J, Czerwinski DK, Houot R, Kohrt HE, et al. High PD-1 expression and suppressed cytokine signaling distinguish T cells infiltrating follicular lymphoma tumors from peripheral T cells. Blood. 2013;121:1367–1376. doi: 10.1182/blood-2012-04-421826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgibbon J, Iqbal S, Davies A, O'Shea D, Carlotti E, Chaplin T, et al. Genome-wide detection of recurring sites of uniparental disomy in follicular and transformed follicular lymphoma. Leukemia. 2007;21:1514–1520. doi: 10.1038/sj.leu.2404696. [DOI] [PubMed] [Google Scholar]
- 14.Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13:4777–4785. doi: 10.1158/1078-0432.CCR-07-0456. [DOI] [PubMed] [Google Scholar]
- 15.Viardot A, Moller P, Hogel J, Werner K, Mechtersheimer G, Ho AD, et al. Clinicopathologic correlations of genomic gains and losses in follicular lymphoma. J Clin Oncol. 2002;20:4523–4530. doi: 10.1200/JCO.2002.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Bentz M, Werner CA, Dohner H, Joos S, Barth TF, Siebert R, et al. High incidence of chromosomal imbalances and gene amplifications in the classical follicular variant of follicle center lymphoma. Blood. 1996;88:1437–1444. [PubMed] [Google Scholar]
- 17.Bouska A, McKeithan TW, Deffenbacher KE, Lachel C, Wright GW, Iqbal J, et al. Genome-wide copy-number analyses reveal genomic abnormalities involved in transformation of follicular lymphoma. Blood. 2014;123:1681–1690. doi: 10.1182/blood-2013-05-500595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz M, Li H, Bernard D, Amin NA, Ouillette P, Jones S, et al. Activating STAT6 mutations in follicular lymphoma. Blood. 2015;125:668–679. doi: 10.1182/blood-2014-06-582650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6:130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oricchio E, Ciriello G, Jiang M, Boice MH, Schatz JH, Heguy A, et al. Frequent disruption of the RB pathway in indolent follicular lymphoma suggests a new combination therapy. J Exp Med. 2014;211:1379–1391. doi: 10.1084/jem.20132120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okosun J, Bodor C, Wang J, Araf S, Yang CY, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green MR, Gentles AJ, Nair RV, Irish JM, Kihira S, Liu CL, et al. Hierarchy in somatic mutations arising during genomic evolution and progression of follicular lymphoma. Blood. 2013;121:1604–1611. doi: 10.1182/blood-2012-09-457283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galban S, Hwang C, Rumble JM, Oetjen KA, Wright CW, Boudreault A, et al. Cytoprotective effects of IAPs revealed by a small molecule antagonist. Biochem J. 2009;417:765–771. doi: 10.1042/BJ20081677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson LF, Wang Y, Lo MC, Yan M, Kanbe E, Zhang DE. The multi-functional cellular adhesion molecule CD44 is regulated by the 8;21 chromosomal translocation. Leukemia. 2007;21:2010–2019. doi: 10.1038/sj.leu.2404849. [DOI] [PubMed] [Google Scholar]
- 34.Case TAD DA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. San Francisco: University of California; 2012. [Google Scholar]
- 35.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Toulmay A, Schneiter R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast. 2006;23:825–831. doi: 10.1002/yea.1397. [DOI] [PubMed] [Google Scholar]
- 37.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 38.Okosun J, Wolfson RL, Wang J, Araf S, Wilkins L, Castellano BM, et al. Recurrent mTORC1-activating RRAGC mutations in follicular lymphoma. Nat Genet. 2016;48:183–188. doi: 10.1038/ng.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, et al. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 2011;25:1668–1673. doi: 10.1101/gad.16968011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt LS, Linehan WM. Molecular genetics and clinical features of Birt-Hogg-Dube syndrome. Nat Rev Urol. 2015;12:558–569. doi: 10.1038/nrurol.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oshiro N, Rapley J, Avruch J. Amino acids activate mammalian target of rapamycin (mTOR) complex 1 without changing Rag GTPase guanyl nucleotide charging. J Biol Chem. 2014;289:2658–2674. doi: 10.1074/jbc.M113.528505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, et al. Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci U S A. 2009;106:18722–18727. doi: 10.1073/pnas.0908853106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–564. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buglio D, Georgakis GV, Hanabuchi S, Arima K, Khaskhely NM, Liu YJ, et al. Vorinostat inhibits STAT6-mediated TH2 cytokine and TARC production and induces cell death in Hodgkin lymphoma cell lines. Blood. 2008;112:1424–1433. doi: 10.1182/blood-2008-01-133769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hao Y, Chapuy B, Monti S, Sun HH, Rodig SJ, Shipp MA. Selective JAK2 Inhibition Specifically Decreases Hodgkin Lymphoma and Mediastinal Large B-cell Lymphoma Growth In Vitro and In Vivo. Clin Cancer Res. 2014;20:2674–2683. doi: 10.1158/1078-0432.CCR-13-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.