Abstract
Incidence and mortality rates for prostate cancer (PCa) are higher in African-American (AA) men than European American (EA) men, but the biological basis for this disparity is unclear. We carried out a detailed analysis of gene expression changes in PCa compared to their matched benign tissues in a cohort of AA men and compared them to existing data from EA men. In this manner, we identified MNX1 as a novel oncogene upregulated to a relatively greater degree in PCa from AA men. Androgen and AKT signaling play a central role in the pathogenesis of PCa and we found that both of these signaling pathways increased MNX1 expression. MNX1 in turn upregulated lipid synthesis by stimulating expression of SREBP1 and fatty acid synthetase. Our results define MNX1 as a novel targetable oncogene increased in AA PCa that is associated with aggressive disease.
Keywords: prostate cancer, MNX1, androgen receptor, AKT, African-American
INTRODUCTION
African-American (AA) men have both a higher incidence and significantly higher mortality rates from prostate cancer (PCa) than European American (EA) men (1). Several groups have found that AA patients have greater tumor volumes in radical prostatectomies in comparison to similarly staged EA patients (2,3). While some of the difference in mortality due to PCa can be attributed to socioeconomic factors, a number of studies have shown that there is a still a higher mortality rate from PCa in AA men even after adjustment for socioeconomic factors(4). Thus biological differences account for a significant portion of the disparity in incidence and mortality from PCa in AA men in comparison to EA men (4),.
There have been a limited number of studies comparing PCa tissues from AA and EA men. Several studies have evaluated differential expression of specific proteins among AA and EA cohorts including epidermal growth factor receptor, the androgen receptor (AR), PGEM1, MDM2 and caveolin-1(5–8). More recently, the TMPRSS2/ERG fusion gene has been shown to occur at lower rate in AA PCa (9–12). Several studies have compared gene expression in AA and EA PCa using expression microarrays. The largest of these studies examined gene expression profiles of 13,000 genes in 33 AA and 36 EA PCas and identified 162 gene transcripts that were differentially expressed between racial groups(13). Interestingly many of these transcripts were related to immune response, stress response, cytokine signaling and chemotaxis.
In this study we have carried out a detailed analysis of gene expression changes in PCas from a cohort of AA men compared to their matched benign tissues. By comparing our data to existing EA expression data we identified MNX1 as an androgen and AKT regulated oncogene that is upregulated to a greater degree in AA PCa compared to EA PCa. MNX1 regulates lipid synthesis, which has been linked to aggressive PCa. Thus our studies have identified a new oncogenic pathway that is important in AA and to a lesser extent in EA pathway which results in potentially targetable changes in tumor metabolism.
MATERIALS AND METHODS
Prostate and prostate cancer tissues
Tissue samples were obtained from the Human Tissue Acquisition and Pathology Core of the Dan L. Duncan Cancer Center and were collected from fresh radical prostatectomy specimens after obtaining informed consent under an Institutional Review Board approved protocol. Cancer samples contained a minimum of 70% cancer and benign tissues were free of cancer on pathological examination. RNAs were extracted using Qiagen DNA/RNA Mini kit according to manufacturer’s instruction. RNAs with RIN number ≥ 7 were chosen for gene expression arrays.
Gene expression microarrays
The quality of isolated RNAs was confirmed on an Agilent 2200 TapeStation system. Twenty-five ng of total RNA was amplified and labeled with Cy3 dye using Low Input Quick Amp Labeling Kit (Agilent Technologies). The labeled cRNA from each sample’s labeling reaction was hybridized to individual microarrays. For microarray hybridization, 825 ng of cyanine 3-labeled cRNA was fragmented and hybridized on the Agilent SurePrint G3 Human GE 8 × 60K V2 Microarrays at 65 °C for 17 hours using the Agilent Gene Expression Hybridization Kit. The hybridized microarrays were dissembled at room temperature in Gene Expression Wash Buffer 1, then washed in Gene Expression Wash Buffer 1 at room temperature for 1 minute. This was followed by a wash for 1 minute in Gene Expression Wash Buffer 2 at an elevated temperature. The processed microarrays were scanned Agilent High-Resolution SureScan microarray scanner and data was extracted using Agilent Feature Extraction Software (11.5.1.1). Expression patterns were visualized as color maps using Java TreeView(14). Array data have been deposited into the Gene Expression Omnibus GSE71016.
Cell culture
Human PCa cells LNCaP, DU145 and PC3 cells, were all maintained in RPMI-1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) LAPC4 cells were cultured in RPMI-1640 medium with 10% FBS supplemented with 10nM R1881 (Sigma). VCaP and 293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) with 10% FBS. The cells were maintained in BRFF-HPC1 medium (Biological Research Faculty and Facility, Inc.) supplemented with 20% FBS. All cell culture medium contained 1x Antibiotic-Antimycotic (Gibco). Normal human prostate epithelial cells (PrEC) were cultured using Clonetics Prostate Epithelial Cell Systems from Lonza Group Ltd. MDA PCa 2a and MDA PCa2b cells were a kind gift from Dr. Nora Navone, MD Anderson Cancer Center. All other cell lines were obtained from the American Type Culture Collection. Cell were obtained between 2001 and 2012, expanded, frozen and stored as stocks in liquid nitrogen. All cell lines are authenticated by STR analysis at MD Anderson Cancer Center Characterized Cell Line Core Facility.
Quantitative RT-PCR
To examine expression of mRNA we carried out quantitative real-time RT-PCR (Q-RT-PCR on an Applied Biosystems StepOne (Lifetechnologies). Total RNA was extracted from cell lines using the RNeasy kit (Qiagen). cDNA was synthesized using an iScript cDNA Synthesis kit (BioRad) with OligodT in a PTC-200 thermocycler (5 min at 25°C; 30 min at 42°C; 5 min at 85°C). MNX1, SREBF1, FASN, and β-actin TaqMan probes (ABI). PCR conditions were set using standard 2-step manufacturer’s protocol. Differences in mRNA levels were analyzed using the 2−ΔΔCT method normalized to β-actin expression. Each measurement point was repeated at least in duplicate.
In situ localization of MNX1 mRNA
Prostate cancer and benign prostate tissues were collected immediately after radical prostatectomy, formalin-fixed and paraffin embedded. Sections were prepared, deparrafinized and MNX1 mRNA was detected in situ using a RNAscope probe designed by Advanced Cell Diagnostics. Pretreatment was a described by the manufacturer with Pretreatment 2 being carried out for 17 min. Hybridization, amplification and signal detection using diaminobenzidine were carried out as described by the manufacturer. After signal detection, slides were stained with Mayer’s Hematoxylin and cover slipped. Incubations were carried out in a HybEZ oven (Advanced Cell Diagnostics) at 40° C.
Stable knockdown of MNX1
Stable MNX1 knockdown LNCaP cells were created by utilizing MNX1-shRNAs in lentiviral vector pGFP-C-shLenti (Origene). 4 unique human 29mer shRNAs for MNX1 constructs in lentiviral GFP vector were purchased from Origene (Cat # TL319509). Lentiviruses carrying these stable shRNAs were produced by 293FT cells using Lenti-vpak Packaging kit (Origene) following manufacture’s instruction. LNCaP cells were infected by these viruses and were selected with 0.5 μg/ml puromycin (Sigma).
Stable overexpression of MNX1 in DU145 cells
The pCMV6-MNX1 vector was obtained from Origene (Cat# RC212606) and after infection DU145 cells were selected with media containing 200ug/ml G418.
Androgen regulation studies
LNCaP cells were cultured in RPMI-1640 media with 10% FBS in 10-cm culture dishes. The medium was changed to RPMI-1640 supplemented with 10% charcoal–dextran stripped serum (CSS) 24h after plating and cells were incubated for an additional 24, 48 or 72 hrs. Synthetic androgen 10 nM R1881 was added to another 2 dishes of cells after incubation in CCS medium for 48h and cells were grown for an additional 24 hours. Enzalutamide (40 uM) was added for 24 hrs Pellets were collected and analyzed for MNX1 protein expression by Western blotting.
AKT inhibitor studies
LNCaP cells were cultured in RPMI-1640 media with 10% FBS in 10-cm culture dishes. Cells were treated in triplicate with 500nM AZD5363, 20 UM LY294002, 1uM rapamycin or vehicle. Cells were collected after treatment and analyzed for MNX1, P-AKT (S473), P-S6, and β-actin expression by Western blotting.
Cell Proliferation Assays
Cells were plated at 2.5 or 5×103 cells per well in 96 well plates. Proliferation was determined using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega) as described by the manufacturer. The absorbance was read at 490 nm with VERSAmax Tunable microplate reader.
Matrigel Invasion Assays
2.5× 104 cells were plated in the top chamber of Matrigel-coated membrane (24-well insert; pore size, 8 mm; BD Biosciences). The cells on the apical side of each insert were then scraped off after 24h or 36 h. The wells were washed with PBS, fixed with 100% methanol and stained with DAPI. After staining, membranes were removed from the insert and mounted on slides, and the invading cells were counted under the Nikon Eclipse TE2000-U microscope. Matrigel assays were performed in triplicate.
Western Blot
Total cellular protein lysate was prepared as described previously (15). Primary antibody against MNX1 was purchased from Origene (Cat #TA307213). Mouse antibody against β-actin was obtained from Sigma-Aldrich. Primary antibodies were used at 1:1000 dilution for Western blotting using procedures described previously (16)
Mouse Xenograft Studies
Experiments were carried out on male SCID mice. Tumor xenografts were established by subcutaneous injection over each flank in 50ul volume mixed with 50ul Matrigel (BD Bioscience). The tumor size was measured weekly using calipers. Tumors were harvested 8 weeks after inoculation and weighed. All procedures were approved by the Baylor College of Medicine Institutional Animal Use and Care Committee.
Sample preparation for LC-MS/MS
The lipid analyses were carried out using the protocol described previously (17) using 5 control and 5 MNX1 knockdown tumors. For each tumor, 100 mg was homogenized in 1:4 ice cold water: methanol mixture. This was followed by sequential addition of ice cold chloroform and water in 3:1 ratio and separation of the organic (methanol and chloroform) and aqueous solvents (water:methanol:chloroform:water; ratio 1:4:3:1). The aqueous extract was de-proteinized using a 3 KDa molecular filter (Amicon Ultracel -3K Membrane, Millipore Corporation) and the filtrate containing metabolites was dried under vacuum (Genevac EZ-2plus).
Measurement of lipids
Targeted profiling using single reaction monitoring (SRM) for the fatty acids, the RP chromatographic method employed a gradient containing water (solvent A) with 10 mM ammonium acetate (pH 8) and 100% methanol (solvent B). Further separation of metabolites was performed on a Luna Phyenyl Hexyl (3 um, 2×15 0 mm) maintained at 40°C. The binary pump flow rate was 0.2 ml/min with a gradient spanning 40% B to 50% B over a 8 minute, 50% B to 67% B over 5 min, hold 67%B for 9 min, 67% B to 100% B over 1 min, hold 100%B for 6 minutes time period, 100% B to 40% B over 1 min and hold 40%B for 7 minutes time period. The following lipids were present at detectable amounts: palmitic acid (saturated form), stearic acid, heptadecanoic acid, octanoic acid, dihomolinoleic acid, myristic acid, heptanoic acid aminoheptanoic acid, docosahexaenoic acid, adrenic acid, arachidonic acid, docosapentaenoic acid, eicosapentaenoic acid, lauric acid, oleic acid, palmitoelic acid and linoleic acid.
RESULTS
Identification of a novel oncogene in AA PCa
Expression array analysis was performed using RNAs from 48 high purity tumors from AA men and matched benign tissue using Agilent 60K expression arrays. This analysis revealed a total of 4341 probes altered in cancer Vs benign (at p<.01, t-test) in AA PCa corresponding to 1803 genes. Of these, 469 genes were upregulated >1.5 fold and 1146 were decreased 0.7 fold or more. Inspection of the data revealed multiple genes that are well known to be upregulated (AMACR(18), Hepsin (19), TFF3(20) and TDRD1(21)) and downregulated (GSTP1(22)) in PCa (Fig 1), confirming the overall quality of the data.
Figure 1. Expression array analysis of genes known to be altered in PCa and MNX1.
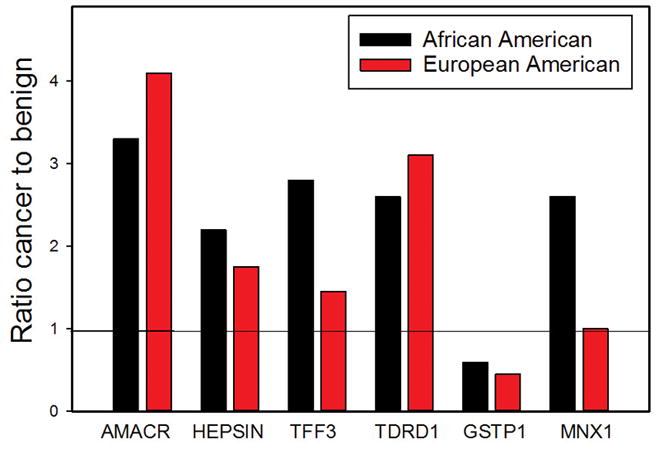
Fold change (cancer versus benign) of specific gene expression in PCa tissues from African-American men and for same genes in the Taylor dataset for EA men.
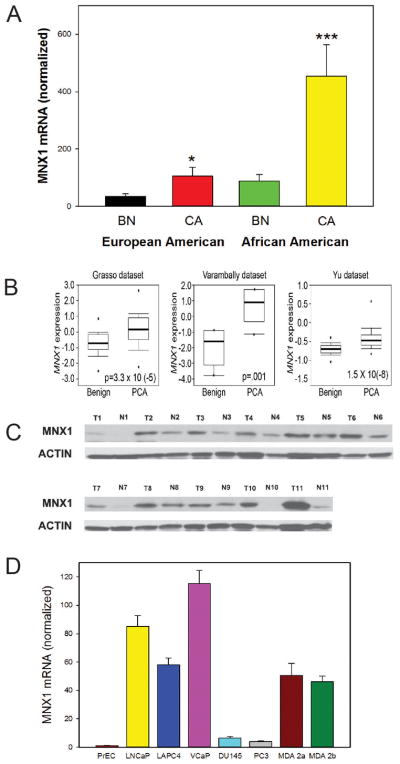
To identify genes that were specifically upregulated (>1.5-fold) or down regulated (at least 0.7-fold) in PCa from AA PCa we compared expression of all such genes (all significant at p<.01, t-test) with the same genes in EA patients (Taylor dataset (23)). While comparison of gene expression using two different platforms can be problematic, the majority of significant genes were significant in both groups, indicating there are not major biases using this approach. The most highly upregulated protein coding gene specifically upregulated in AA PCa was the homeobox gene MNX1/HLXB9. This gene plays an important role in pancreas development (24) and is a causative oncogene in infantile AML (25). However it has not been shown to be an oncogene in an epithelial cancer. As shown in Figure 1, MNX1 is increased 2.6 fold by array analysis, similar to AMACR and PSGR; (p<.001, t-test) in AA PCa but not significantly altered in EA PCa (1.03 fold, p=.16; Taylor dataset). Upregulation of MNX1 has been confirmed by Q-RT-PCR of AA RNAs (5.1 fold; p<.002, t-test, Figure 2A). Direct analysis of RNAs from EA PCa and matched benign tissues shows a smaller but still significantly increased expression of MNX1 mRNA in PCa. Consistent with this finding, 3 of 13 Oncomine PCa tissue datasets (predominantly EA) showed statistically significant increases in MNX1 mRNA (p≤.001), although the Taylor dataset was not among those with MNX1 upregulation (Figure 2B). No dataset showed downregulation. As a comparison, ERG was upregulated in 10 of 19 datasets at p≤.001. We then carried out Western blots using protein extracts from matched PCa and benign tissues from AA men (Fig 2C). While variable expression was seen in both cancer and benign tissues, the mean increase in MNX1 protein in cancer compared to matched benign tissues was 12.4-fold (range 0.83 to 81.1-fold) by densitometry. Increased MNX1 protein was also seen in extracts from EA PCa and matched benign tissues. RNA in situ hybridization showed expression of MNX1 in PCa cells and to a much lesser extent benign epithelial cells but no expression in benign or tumor associated stroma (Supplementary Fig. 1) Expression of MNX1 mRNA was almost undetectable in benign prostate epithelial cells (PrECs) but was markedly elevated in the androgen receptor (AR) expressing LNCaP, LAPC4, VCaP, MDA PCa 2a and MDA PCa 2b PCa cells but not in AR negative DU145 and PC3 (Fig 2D). Of note, MDA PCa 2a and MDA PCa 2b were derived from the PCa of an AA man. Thus MNX1 is expressed at increased levels in PCa, with more marked increases in cancers from AA men.
Figure 2. Expression of MNX1 in PCa.
A. MNX1 mRNA levels in EA and AA PCa and benign tissues by Q-RT-PCR. Asterisks indicate statistically significant differences between cancer and benign tissues. Mean +/−SEM; * p<.05; *** p<.001 by t-test B. Examples of Oncomine datasets with increased expression of MNX1 relative to benign tissues; p value shown in bar and whisker plot. C. Western blot on MNX1 from PCa and matched benign tissues from AA men. β-actin is a loading control. D. Expression of MNX1 mRNA in primary prostatic epithelial cells (PrEC) and PCa cell lines.
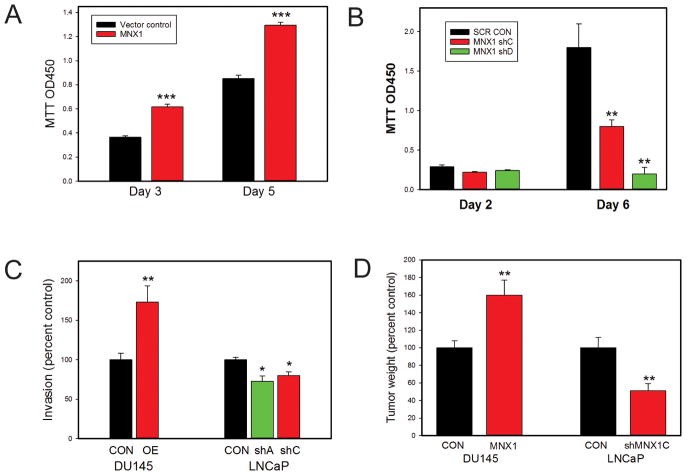
To assess the biological activity of MNX1 in PCa cells we established DU145 cells expressing MNX1 protein and assessed the impact on proliferation (Fig 3A). MNX1 expression resulted in significantly increased proliferation. Similarly, knockdown of MNX1 in LNCaP cells resulted in significant decreases in proliferation (Fig 3B). Increased MNX1 expression in DU145 cells increased invasion and knockdown in LNCaP decreased invasion through Matrigel (Fig 3C). See Supplementary Fig 2 for confirmation of protein overexpression and knockdown by Western blotting. Most critically, MNX1 expression resulted in larger xenograft tumors in SCID mice using DU145 cells with MNX1 overexpression (versus controls) while knockdown in LNCaP cells decreased tumor size (Fig 3D). Thus MNX1 is an oncogene which expressed at higher levels in both AA and EA PCa, but is significantly higher in AA PCa.
Figure 3. Biological effects of MNX1 on PCa cells.
A. Proliferation of DU145 cells overexpressing MNX1 versus vector controls. B. Proliferation of LNCaP cells with knockdown of MNX1 using two different shRNAs versus scrambled control. C. Invasion through Matrigel of DU145 cells overexpressing MNX1 versus vector controls and LNCaP cells with knockdown of MNX1 using two different shRNAs versus scrambled control. D. Final tumor weight of xenograft tumors using DU145 cells overexpressing MNX1 versus vector controls and LNCaP cells with knockdown of MNX1 using an shRNA versus scrambled control. For A–D, mean +/− SEM is shown; * p<.05; **<p<.01; *** p<.001 by t-test or Mann Whitney. See also Supplementary Figure 2.
MNX1 expression is regulated by androgens
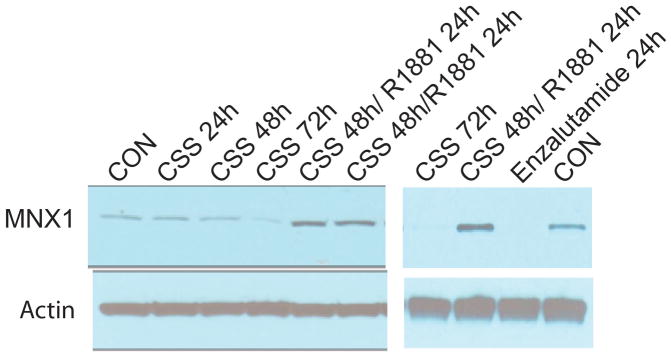
As noted above, the AR expressing cell lines show much higher expression of MNX1, suggesting that MNX1 expression may be regulated by androgens. Incubation of LNCaP cells in charcoal stripped serum (which depletes androgens) results in decreased MNX1 protein and stimulation with the androgen R1881 increases expression (Fig 4). Inhibition of AR signaling with enzalutamide also markedly decreased MNX1 level (Fig 4, right).
Figure 4. Control of expression of MNX1 by androgens.
LNCaP cells were incubated in charcoal stripped serum (CSS) for 24, 48 or 72 hrs. Two plates of cells were treated with 10nM R1881 after 48 hrs in CSS; cells were collected 24 hrs after treatment. Western blot with anti-MNX1 antibody and anti β-actin is shown. In a separate experiment cells were treated with ezalutamide for 24 hrs or vehicle only (CON). Cells were also placed in CSS for 72 hrs or placed in CSS for 48 hrs and then treated with R1881 for 24 hrs as an additional set on controls.
MNX1 expression is increased by AKT signaling
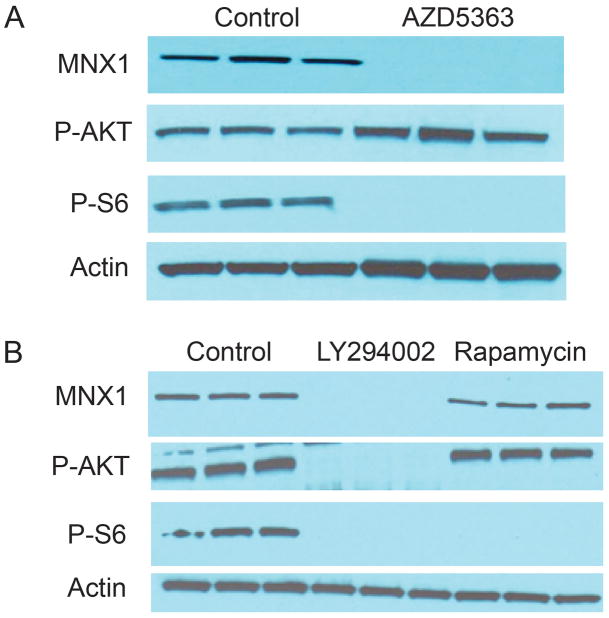
Molecular alterations resulting in increased activity of the AKT pathway are among the most common molecular alterations in aggressive PCa (23). We therefore sought to determine whether AKT signaling can regulate modulate expression of MNX1. LNCaP cells were treated with AZD5363, an AKT kinase inhibitor or vehicle only (Fig 5A). As a direct AKT kinase inhibitor, AZD5363 results in feedback upregulation of AKT phosphorylation (phospho-S473) but decreased activation of AKT downstream targets such as phospho-S6 (Fig 5A). MNX1 protein is completely lost in response to treatment with AZD4563. Similar effects were seen with the PI-3 kinase inhibitor LY294002 (Fig 5B), which also markedly decreased MNX1 expression. In contrast, rapamycin had no effect on MNX1 levels (Fig 5B). These results indicate that MNX1 regulation by AKT must be dependent on one of the other targets of AKT signaling other than mTOR.
Figure 5. Control of expression of MNX1 by AKT.
A. LNCaP cells were treated with 500nM AZD5363 for 24 hrs and cell lysates prepared. Western blot for MNX1, P-AKT (S473) and P-S6 were performed. Actin is a loading control. Note increased AKT phosphorylation is due to feedback induced by inhibition of AKT kinase activity.
B. LNCaP cells were treated with either 20 uM LY294002 or 1uM rapamycin or vehicle only for 24 hrs and cell lysates prepared. Western blot for MNX1, P-AKT (S473) and P-S6 were performed. Actin is a loading control.
MNX1 downstream target genes
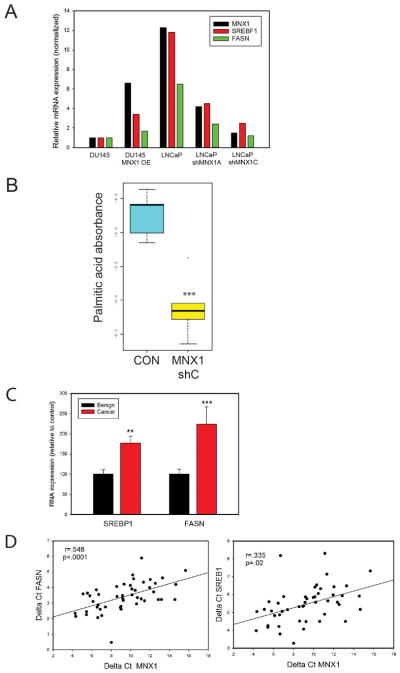
It has shown that fatty acid synthetase (FASN) mRNA is increased in AA PCa relative to EA PCa (26). In addition FASN is increased by androgens (27). We therefore examined whether expression of FASN and its regulatory factor sterol regulatory element binding transcription factor 1 (28) were modulated by MNX1. As shown in Fig 6A, DU145 cells express lower levels of MNX1, FASN and sterol regulatory element binding transcription factor 1 (SREBP1) mRNAs than LNCaP cells. DU145 cells with overexpression of MNX1 show increased expression of both FASN and SREBP1, while LNCaP cells with MNX1 knockdown show a 60–70% decrease in FASN and SREBP1. Of note, MNX1 was knocked down to a similar extent as FASN and SREBP1 in LNCaP cells, indicating that MNX1 plays a significant role in driving these two genes in PCa. To confirm the connection between MNX1 and lipid synthesis we analyzed LNCaP MNX1 knockdown tumors and controls (see Fig 3D) for levels of 17 lipids as described in Materials and Methods. While some differences in the content for various lipids was seen between controls and knockdowns, only palmitic acid, a major product of FASN, was statistically significantly altered (p<.001, t-test; Fig 6B). To confirm that MNX1 drives SREBP1 and FASN in human PCa we analyzed FASN in RNAs from benign and PCa tissues from AA men. As shown in Fig 6C, both SREBP1 and FASN are significantly increased in AA PCa compared to benign tissues (p<.01, Mann-Whitney). We then examined the correlation between MNX1 and FASN in the cancer samples. The correlation was extremely high (r=.548, p<.001, Pearson’s) in AA samples (Fig 6D). There was a similar significant, but somewhat lower correlation, of MNX1 with SREBP1 mRNA levels (r=.335, p=.02, Pearson’s). As a positive control we examined the correlation of SREBP1 and FASN, since FASN is a known direct target of SREBP1. As predicted the correlation was quite high (r=.63. p<.001). Overall this data strongly supports the idea that one important pathway by which MNX1 promotes oncogenesis is by increasing expression of SREBP1 and its target FASN in AA PCa.
Figure 6. MNX1 regulates lipid synthesis.
A. Expression of MNX1, SREBF1, and FASN in DU145 overexpressing MNX1 or LNCaP with MNX1 knockdown using two different shRNAs versus controls. Data is expressed as percent of vector control for DU145 for each gene. Means of duplicate or triplicate determinations are shown. B. Palmitic acid content of LNCaP xenograft tumors versus scrambled controls (see Fig 3D); *** p<.001 by t-test. C. SREBP1 and FASN mRNA expression in AA benign prostate and PCa tissues. Mean +/− SEM is shown. ** p<.01; *** p<.001; Mann Whitney. D. Correlation between SREBP1 or FASN with MNX1 mRNA in AA PCa by Pearson Product Moment. Correlation coefficient and p-value are shown.
DISCUSSION
In this report we show that MNX1 is an oncogene that is upregulated in AA PCa and to a lesser extent in EA PCa. MNX1 is a homeobox gene that is known to be an oncogene in the rare malignancy infantile AML (25). It has also been reported to be increased in poorly differentiated hepatocellular carcinoma cell lines (29). It has never been linked to PCa previously, perhaps since most prior molecular genetic studies of PCa have been dominated by EA PCa, where its expression is lower overall. Our data also indicates that at least part of the oncogenic activity of MNX1 is mediated via upregulation of SREBP1 and FASN. It has been shown that FASN is oncogenic in prostate epithelial cells (30) and is associated with aggressive PCa (30,31). SREBP1 is an important mediator of the induction of lipid synthesis and FASN by androgens both in normal tissues (32) and in PCa (33) and presumably the effect of MNX1 on FASN is mediated mainly by its upregulation of SREBP1. Importantly, lipid biosynthesis is potentially targetable (31) and our data supports the hypothesis that agents targeting lipid synthesis may be even more important in treatment of AA PCa. It should be noted that the extent of SREBP1 and FASN in LNCaP cells closely parallels the extent of MNX1 knockdown implying that MNX1 may be an important intermediate regulator of AR induction of lipid synthesis. It should be noted that MNX1 increases expression of FASN and SREBP1 in DU145 cells, which are AR negative. Thus this activity does not require AR. However, it is possible that other activities of the AR inducible MNX1 protein may require AR. Further studies across a broad array of PCA cell lines are needed to clarify this point. In addition to increasing lipid biosynthesis, SREBP1 can have other oncogenic affects such as increasing reactive oxygen species (28). MNX1 almost certainly affects other oncogenic pathways as well in PCa, which we are currently investigating.
Our studies show that MNX1 protein levels are controlled by AR signaling, which is well known to play a central role PCa pathogenesis and remains the most important therapeutic target in PCa (34). There is significant evidence that AA PCa may have higher AR signaling (7,10,12). Thus at least some of the increase in MNX1 in AA PCa may reflect higher activity of AR signaling in AA PCa. In addition, MNX1 is regulated by AKT signaling. Activation of PI3-kinase/AKT signaling is very common in localized PCa and is almost universal in advanced PCa (23). Little is known about whether AKT signaling is more active in AA PCa compared to EA PCa. It has been shown that a G10398A mitochondrial polymorphism is more common in AA men with PCa (35)and in breast cancer this polymorphism leads to higher AKT activation (36). However further studies are needed to determine if AKT signaling alterations play a role in the higher expression of MNX1 in AA PCa.
In summary we show MNX1 is upregulated in AA and, to a lesser extent, EA PCa and promotes transformation at least in part by increasing lipid synthesis. Our data supports the emerging concept that significant oncogenic pathways are similar in AA and EA PCa but are altered at different rates and extents in AA and EA PCa (11,12,37). Further studies are needed to fully define the biology of MNX1 in AA and EA in PCa and determine the clinical impact of MNX1 expression.
Supplementary Material
Acknowledgments
Financial support: This work was supported grants from the Department of Defense W81XWH-12-1-0046 (M. Ittmann) and W81XWH-12-546 1-0130 (A. Sreekumar); the National Cancer Institute U01 CA167234 (A. Sreekumar, N. Putluri) and the Dan L. Duncan Cancer Center (P30 CA125123) supporting Human Tissue Acquisition and Pathology and Genomic and RNA Profiling Shared Resources; CPRIT Core Facility Support Award RP120092 (N. Putluri, A. Sreekumar); funds from the Alkek Center for Molecular Discovery (A. Sreekumar), the Prostate Cancer Foundation (M. Ittmann) and by the use of the facilities of the Michael E. DeBakey VAMC.
The assistance of Billie Smith with RNA in situ localization is gratefully acknowledged. We would also like to acknowledge the assistance of Barry Davies with the AZD5363 studies. Information related to Agilent Technologies products described in this manuscript are for research use only and not for use in diagnostic procedures.
Footnotes
Conflicts of interest: There are no conflicts of interest.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez-Ortiz RF, Troncoso P, Babaian RJ, Lloreta J, Johnston DA, Pettaway CA. African-American men with nonpalpable prostate cancer exhibit greater tumor volume than matched white men. Cancer. 2006;107:75–82. doi: 10.1002/cncr.21954. [DOI] [PubMed] [Google Scholar]
- 3.Moul JW, Connelly RR, Mooneyhan RM, Zhang W, Sesterhenn IA, Mostofi FK, et al. Racial differences in tumor volume and prostate specific antigen among radical prostatectomy patients. J Urol. 1999;162:394–7. [PubMed] [Google Scholar]
- 4.Freedland SJ, Isaacs WB. Explaining racial differences in prostate cancer in the United States: sociology or biology? Prostate. 2005;62:243–52. doi: 10.1002/pros.20052. [DOI] [PubMed] [Google Scholar]
- 5.Yang G, Addai J, Ittmann M, Wheeler TM, Thompson TC. Elevated caveolin-1 levels in African-American versus white-American prostate cancer. Clin Cancer Res. 2000;6:3430–3. [PubMed] [Google Scholar]
- 6.Shuch B, Mikhail M, Satagopan J, Lee P, Yee H, Chang C, et al. Racial disparity of epidermal growth factor receptor expression in prostate cancer. J Clin Oncol. 2004;22:4725–9. doi: 10.1200/JCO.2004.06.134. [DOI] [PubMed] [Google Scholar]
- 7.Gaston KE, Kim D, Singh S, Ford OH, 3rd, Mohler JL. Racial differences in androgen receptor protein expression in men with clinically localized prostate cancer. J Urol. 2003;170:990–3. doi: 10.1097/01.ju.0000079761.56154.e5. [DOI] [PubMed] [Google Scholar]
- 8.Petrovics G, Zhang W, Makarem M, Street JP, Connelly R, Sun L, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004;23:605–11. doi: 10.1038/sj.onc.1207069. [DOI] [PubMed] [Google Scholar]
- 9.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. The Prostate. 2011;71:489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 10.Farrell J, Petrovics G, McLeod DG, Srivastava S. Genetic and molecular differences in prostate carcinogenesis between African American and Caucasian American men. Int J Mol Sci. 2013;14:15510–31. doi: 10.3390/ijms140815510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlins SA, Alshalalfa M, Davicioni E, Erho N, Yousefi K, Zhao S, et al. Characterization of 1577 Primary Prostate Cancers Reveals Novel Biological and Clinicopathologic Insights into Molecular Subtypes. European urology. 2015 doi: 10.1016/j.eururo.2015.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamoah K, Johnson MH, Choeurng V, Faisal FA, Yousefi K, Haddad Z, et al. Novel Biomarker Signature That May Predict Aggressive Disease in African American Men With Prostate Cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2015 doi: 10.1200/JCO.2014.59.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 14.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Feng S, Dakhova O, Creighton CJ, Cai Y, Wang J, et al. FGFR-4 Arg(3) enhances prostate cancer progression via extracellular signal-related kinase and serum response factor signaling. Clin Cancer Res. 2011;17:4355–66. doi: 10.1158/1078-0432.CCR-10-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Shao L, Creighton CJ, Zhang Y, Xin L, Ittmann M, et al. Function of phosphorylation of NF-kB p65 ser536 in prostate cancer oncogenesis. Oncotarget. 2015 doi: 10.18632/oncotarget.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta S, Putluri N, Long W, Zhang B, Wang J, Kaushik AK, et al. Coactivator SRC-2-dependent metabolic reprogramming mediates prostate cancer survival and metastasis. The Journal of clinical investigation. 2015;125:1174–88. doi: 10.1172/JCI76029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Weng J, Cai Y, Penland R, Liu M, Ittmann M. The prostate-specific G-protein coupled receptors PSGR and PSGR2 are prostate cancer biomarkers that are complementary to alpha-methylacyl-CoA racemase. Prostate. 2006;66:847–57. doi: 10.1002/pros.20389. [DOI] [PubMed] [Google Scholar]
- 19.Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, et al. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 2001;61:5692–6. [PubMed] [Google Scholar]
- 20.Garraway IP, Seligson D, Said J, Horvath S, Reiter RE. Trefoil factor 3 is overexpressed in human prostate cancer. The Prostate. 2004;61:209–14. doi: 10.1002/pros.20096. [DOI] [PubMed] [Google Scholar]
- 21.Boormans JL, Korsten H, Ziel-van der Made AJ, van Leenders GJ, de Vos CV, Jenster G, et al. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. International journal of cancer Journal international du cancer. 2013;133:335–45. doi: 10.1002/ijc.28025. [DOI] [PubMed] [Google Scholar]
- 22.Lin X, Tascilar M, Lee WH, Vles WJ, Lee BH, Veeraswamy R, et al. GSTP1 CpG island hypermethylation is responsible for the absence of GSTP1 expression in human prostate cancer cells. The American journal of pathology. 2001;159:1815–26. doi: 10.1016/S0002-9440(10)63028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nature genetics. 1999;23:71–5. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 25.von Bergh AR, van Drunen E, van Wering ER, van Zutven LJ, Hainmann I, Lonnerholm G, et al. High incidence of t(7;12)(q36;p13) in infant AML but not in infant ALL, with a dismal outcome and ectopic expression of HLXB9. Genes, chromosomes & cancer. 2006;45:731–9. doi: 10.1002/gcc.20335. [DOI] [PubMed] [Google Scholar]
- 26.Powell IJ, Dyson G, Land S, Ruterbusch J, Bock CH, Lenk S, et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2013;22:891–7. doi: 10.1158/1055-9965.EPI-12-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer research. 2008;68:4447–54. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang WC, Li X, Liu J, Lin J, Chung LW. Activation of androgen receptor, lipogenesis, and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Molecular cancer research: MCR. 2012;10:133–42. doi: 10.1158/1541-7786.MCR-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkens L, Jaggi R, Hammer C, Inderbitzin D, Giger O, von Neuhoff N. The homeobox gene HLXB9 is upregulated in a morphological subset of poorly differentiated hepatocellular carcinoma. Virchows Archiv: an international journal of pathology. 2011;458:697–708. doi: 10.1007/s00428-011-1070-5. [DOI] [PubMed] [Google Scholar]
- 30.Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G, et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. Journal of the National Cancer Institute. 2009;101:519–32. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flavin R, Peluso S, Nguyen PL, Loda M. Fatty acid synthase as a potential therapeutic target in cancer. Future oncology. 2010;6:551–62. doi: 10.2217/fon.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heemers H, Vanderhoydonc F, Roskams T, Shechter I, Heyns W, Verhoeven G, et al. Androgens stimulate coordinated lipogenic gene expression in normal target tissues in vivo. Molecular and cellular endocrinology. 2003;205:21–31. doi: 10.1016/s0303-7207(03)00205-3. [DOI] [PubMed] [Google Scholar]
- 33.Swinnen JV, Ulrix W, Heyns W, Verhoeven G. Coordinate regulation of lipogenic gene expression by androgens: evidence for a cascade mechanism involving sterol regulatory element binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12975–80. doi: 10.1073/pnas.94.24.12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer research. 2013;73:4599–605. doi: 10.1158/0008-5472.CAN-12-4414. [DOI] [PubMed] [Google Scholar]
- 35.Mims MP, Hayes TG, Zheng S, Leal SM, Frolov A, Ittmann MM, et al. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2006;66:1880. doi: 10.1158/0008-5472.CAN-05-3774. author reply 80–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007;249:249–55. doi: 10.1016/j.canlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Farrell J, Young D, Chen Y, Cullen J, Rosner IL, Kagan J, et al. Predominance of ERG-negative high-grade prostate cancers in African American men. Mol Clin Oncol. 2014;2:982–86. doi: 10.3892/mco.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.