Abstract
Background
The strongest known risk factor for endometrial cancer (EC) is obesity. To determine whether single nucleotide polymorphisms (SNPs) associated with increased body mass index (BMI) or waist-hip ratio (WHR) are associated with EC risk, independent of measured BMI, we investigated relationships between 77 BMI and 47 WHR SNPs and EC in 6,609 cases and 37,926 country-matched controls.
Methods
Logistic regression analysis and fixed-effects meta-analysis were used to test for associations between EC risk and (i) individual BMI or WHR SNPs, (ii) a combined weighted genetic risk score (wGRS) for BMI or WHR. Causality of BMI for EC was assessed using Mendelian randomization, with BMIwGRS as instrumental variable.
Results
The BMIwGRS was significantly associated with EC risk (P=3.4×10−17). Scaling the effect of the BMIwGRS on EC risk by its effect on BMI, the EC odds ratio (OR) per 5kg/m2 of genetically predicted BMI was 2.06 (95% confidence interval(CI)=1.89–2.21), larger than the observed effect of BMI on EC risk (OR=1.55, 95%CI 1.44–1.68, per 5kg/m2). The association attenuated but remained significant after adjusting for BMI (OR=1.22, 95%CI=1.10–1.39,P=5.3×10−4). There was evidence of directional pleiotropy (P=1.5×10−4). BMI SNP rs2075650 was associated with EC at study-wide significance (P<4.0×10−4), independent of BMI. EC was not significantly associated with individual WHR SNPs or the WHRwGRS.
Conclusions
BMI, but not WHR, is causally associated with EC risk, with evidence that some BMI-associated SNPs alter EC risk via mechanisms other than measurable BMI.
Impact
The causal association between BMI SNPs and EC has possible implications for EC risk modeling.
Keywords: endometrial cancer, genetic predisposition, risk score analysis, pleiotropy
Introduction
Endometrial cancer (EC: cancer of the lining of the uterine corpus) is the fourth most diagnosed cancer in European and North American women (1). Endometrial tumors are typically classified into two etiological types (2): hormonally driven Type 1, usually low grade endometrioid histology with ‘good’ prognosis (~80% of cases), and Type 2, non-endometrioid, largely serous or clear cell histologies with poorer prognosis. Overall, the strongest known risk factor is obesity (3), with every 5kg/m2 increase in body mass index (BMI) increasing EC risk by up to 60% (4). Women with a BMI ≥30 kg/m2 have a ~3-fold overall increased EC risk compared to non-obese women (BMI <25), increasing to an 8-fold risk in women with BMI ≥40 (5). Obesity is most commonly associated with endometrioid EC, and may also modestly increase the risk of non-endometrioid tumors (3, 6). Body fat distribution, measured as waist-hip ratio (WHR) or waist circumference (WC), may influence EC risk but the evidence is weaker (4, 7). Additionally, whether the WHR/WC associations are independent of BMI remains to be clarified.
Association studies assessing cancer risk with variants proven to be associated with obesity may inform our understanding of the biological relationship between obesity and cancer risk, and also identify variants/genetic loci that play a direct role in the etiology of obesity-associated cancers. Genome-wide association studies (GWAS) have now identified 97 loci associated with BMI and another 49 loci independently associated with WHR adjusted for BMI (8–11). Of these, a SNP in the FTO gene, in high linkage disequilibrium with obesity SNP rs1558902, is associated with a significantly increased risk of breast cancer (12), while combinations of BMI-associated variants summarised by a genetic risk score (GRS) have been associated with prostate and colorectal cancers (13, 14). A recent study of 3,376 European-ancestry EC cases and 3,867 controls found an association between a 97-SNP BMI GRS and EC which disappeared after adjusting for BMI (15). However, a 26-SNP BMI GRS was found to be significantly associated with EC in Chinese cases and controls independently of measured BMI (16). The relationship between WHR-associated SNPs and EC is as yet unknown for any population.
We have investigated whether SNPs known to influence BMI (N=77) or WHR adjusted for BMI (N=47) in Europeans, are also associated with the risk of EC using a large sample of 6,609 EC cases and 37,926 controls. We present the results of our association analyses for each SNP individually, and combined as a weighted genetic risk score (wGRS) (17) for each adiposity measure. Further, we investigated possibly pleiotropy of BMI risk SNPs using a Mendelian Randomization approach with a test for heterogeneity among the causal estimates from the different SNPs.
Material and Methods
Datasets
We analyzed four datasets from separate studies contributing to the Endometrial Cancer Association Consortium (ECAC), as detailed previously (18, 19), and as summarized in Supplementary Table 1). The first three comprised GWAS datasets genotyped using Illumina genotyping arrays, from Australia (“ANECS/QIMR/HCS”: 606 cases, 3,083 controls), and the UK (“SEARCH/WTCCC”, 681 cases, 5,190 controls (18, 20)); “NSECG/CORGI”, 919 cases, 894 controls(19, 21)). The fourth dataset (“iCOGS”) was genotyped using the ‘iCOGs’ custom Illumina Infinium iSelect genotyping array comprising 211,155 SNPs chosen for follow-up and fine-mapping of hormonal cancer GWAS hits, and included 4,402 cases recruited from 11 separate studies from 7 countries, and 28,758 controls from the same countries.
BMI information was available for subsets of cases and controls from the ANECS, SEARCH and iCOGS datasets (Table 1, Supplementary Table 2). Analyses that did not include BMI as a covariate included 6,609 cases and 37,296 controls; analyses including BMI as a covariate included 4,088 cases and 15,986 controls. The association between BMI and EC risk was assessed by meta-analysis of the ANECS, SEARCH and iCOGS datasets. There was modest evidence for heterogeneity (Ptrend All cases I2=73.4, P=0.02), driven by a lower estimate for the SEARCH dataset, with little difference between a fixed effects and random effects model (presented in Table 1).
Table 1.
Association between body mass index (BMI) and endometrial cancer risk overall, and for endometrioid and non-endometrioid histologies1
| Controls | All cases | Endometrioid cases | Non-endometrioid cases | ||||
|---|---|---|---|---|---|---|---|
| BMI2 Category |
N (%) | N (%) | OR (95% CIs) | N (%) | OR (95% CIs) | N (%) | OR (95% CIs) |
| <25kg/m2 | 7146 (45%) | 1159 (28%) | Reference | 964 (28%) | Reference | 195 (32%) | Reference |
| 25–29.9 | 5628 (35%) | 1252 (31%) | 1.06 (0.65–1.70) | 1053 (30%) | 1.05 (0.64–1.72) | 199 (33%) | 1.27 (1.07–1.48) |
| 30–34.9 | 2213 (14%) | 795 (19%) | 1.60 (0.95–2.71) | 684 (20%) | 1.62 (0.94–2.80) | 111 (18%) | 1.88 (1.64–2.12) |
| 35–39.9 | 693 (4%) | 472 (12%) | 3.522(2.35–4.43) | 413 (12%) | 3.26 (2.31–4.61) | 59 (10%) | 3.16 (2.85–3.48) |
| ≥40 | 294 (2%) | 409 (10%) | 6.10 (3.67–10.17) | 366 (10%) | 6.26 (3.55–11.06) | 43 (7%) | 5.92 (5.56–6.27) |
| Ptrend3 | 1.8×10−26 | 1.7×10−17 | 1.42×10−27 | ||||
| Per 5kg/m2 increase in EC risk |
1.55 (1.44–1.68) | 1.56 (1.42–1.72) | 1.50 (1.43–1.57) | ||||
Random effects model.
BMI range: Overall 15.24–75.00 (mean 27.18, SD 5.72); Cases 15.24–75.00 (mean 29.86, SD 7.45)’ Controls 15.94–67.90 (mean 26.52, SD 4.99).
Tests for heterogeneity: All cases I2=73.4%, Q=7.53, P=0.02; Endometrioid cases I2=81.1%, Q=10.57, P=0.005. Non-endometrioid cases are from the iCOGs dataset only and were not meta-analyzed.
WHR information was available only for a subset of WTCCC controls (the 1958 Birth Cohort, N=1259); the association between WHR wGRS and WHR was confirmed in this subset of individuals. Analyses assessing the association between WHR wGRS and EC risk included all cases and controls.
BMI and WHR SNP genotype imputation
Our analyses included 77 SNPs recently validated as associated with BMI at a genome-wide level of significance (P<5.0×10−8) in a large-scale meta-analysis including 339,224 individuals of European ancestry from 125 separate studies conducted by the Genetic Investigation of Anthropomorphic Traits (GIANT) consortium (8, 9). Only SNPs significant in the primary analysis were included (i.e. we did not include SNPs significant only in secondary or conditional analyses, or in the analysis including other ancestries). Using the same criteria, we included 47 SNPs associated with WHR after adjustment for BMI (WHRadjBMI) in a GIANT meta-analysis including 210,088 individuals from 101 studies (10, 11); 34 of these WHR SNPs had also reached genome-wide significance in analyses including only women (11). The BMI and WHR SNPs were non-overlapping. SNPs that were not directly genotyped on either the Illumina or iCOGS platforms were imputed to the 1000 Genomes dataset v3 (April 2012 release) using IMPUTE v2 (22) as described in (19). All target SNPs had imputation information scores >0.85 across datasets and minor allele frequencies >0.05.
Association of EC with individual BMI or WHR SNPs
The four datasets were analysed separately using unconditional logistic regression with a per-allele (1 degree of freedom) model using SNPTEST v2 (23), adjusting for principal components of the genomic kinship matrix as described previously (18, 19). The GWAS datasets were each analysed as a single stratum, the iCOGS dataset was adjusted for eight strata (six defined by country, while the large UK dataset was divided into ‘SEARCH’ and ‘NSECG’). Given no indication for heterogeneity between studies, betas and their standard errors for each dataset were combined using standard fixed-effects meta-analyses across studies in METAL (24). All statistical tests were 2-sided. P-values <4.0×10−4 (where P=0.05/124) were considered significant.
Association of EC with genetic risk scores for BMI and WHR
We next tested for associations between EC and the wGRS for BMI and WHR. For each individual in the study, the number of trait-increasing alleles at each SNP (between 0 and 2) was weighted by the reported effect size in the GIANT consortium meta-analysis (per-allele regression coefficient) on the relevant phenotype and then summed across SNPs (Supplementary Text) (8–11). As most WHR-associated SNPs showed a significant difference in effect between the sexes, we calculated the WHRwGRS using the effect size as reported for women (11). The weighted contributions from all SNPs were summed to give a BMIwGRS and two different WHRwGRS for each individual (a 34-SNP WHRwGRS including only SNPs reaching genome-wide significance in women, and a 47 SNP WHRwGRS including all WHR-associated SNPs for which we had data).
Associations between the BMIwGRS and BMI and the WHRwGRS and WHR were determined by linear regression, and associations between the BMIwGRS, WHRwGRS and case-control status by logistic regression. These analyses were performed separately for each study, and results combined using random effects meta-analysis. Associations between each wGRS and EC were performed per GRS unit (continuous) and after stratifying into quartiles based on the distribution in controls. All wGRS analyses were performed using the R software package (http://www.r-project.org/) with two-sided P-values <0.05 considered significant.
Finally, we used Mendelian Randomization (MR), with BMIwGRS as the instrumental variable, to assess the causality of BMI for EC. We genetically predicted the effect of a 5kg/m2 increase in BMI on EC risk by scaling the natural logarithm of the OR of EC per unit increase in the BMIwGRS on BMI. Using the MR approach, if BMI is causal for EC then the observed BMI OR for EC should be consistent with that predicted using the scaled BMIwGRS. A larger observed than predicted OR would suggest that at least part of the observed BMI-EC association is attributable to bias or confounding inflating the observed estimates. Conversely, a larger predicted than observed OR might indicate pleiotropy or bias or confounding that has reduced the observed estimate towards the null. To formally test the MR assumption of no pleiotropy, we used the MR adaptation of Egger’s test – a method originally developed for assessing small-study bias in meta-analysis (25). In this setting each point on the funnel plot represents the causal estimate derived from one BMI SNP, and we are testing whether the causal estimates from weaker SNPs (those less strongly associated with BMI) are skewed towards either high or low values, compared with stronger variants. We used Cochran’s Q-test as a further test for heterogeneity in the causal estimates of the individual SNPs (where the analysis is over the 77 SNPs rather than over multiple studies, as would be more usual in a meta-analysis context), and used the result of this test to guide whether the best estimate of the causal effect of BMI on EC is the combined estimate from the fixed-effects or from the random-effects inverse-variance weighted meta-analysis of the per-SNP causal estimates.
Results
There was evidence of association between EC and one BMI-associated SNP at P<4.0×10−4; SNP rs2075650 located within TOMM40 on chromosome 19 (per allele OR=1.13, 95% CI 1.05–1.21, P=2.4×10−4) (Supplementary Table 3). The signal was similar in the subset of samples with BMI information (OR=1.18, 95% CI 1.10–1.26; P=2.0×10−4), and remained significant after including BMI as a covariate (OR=1.16, 95% CI 1.07–1.24, P=3.7×10−4). For the individual SNPs, there was a very modest positive correlation between the published effect on BMI and the estimated effect on EC risk (Pearson R=0.26, P=0.02), which was attenuated when only samples with BMI information were included (Pearson R=0.19, P=0.09), and disappeared completely after conditioning on BMI (Pearson R=0.004, P=0.96) (Supplementary Figure 1).
There was also evidence for association with one WHR SNP, rs10842707 at the ITPR2-SSPN locus on chromosome 12 (OR=1.09, 95% CI 1.05–1.13, P=3.7×10−4) (Supplementary Table 4), although this signal fell below our study-wide significance threshold after adjusting for BMI (OR=1.08, 95% CI 1.01–1.14, P=1.1×10−2; unadjusted OR for the subset with BMI information was 1.07, 95% CI 1.01–1.13, P=2.4×10−2). There was no obvious correlation between published effect sizes for WHR SNPs and EC risk (Pearson R=−0.19, P=0.09; Supplementary Figure 2).
As expected, self-reported BMI was highly significantly associated with EC risk overall, with an OR=1.55 (95% CI 1.44–1.68, per 5kg/m2, P=1.8×10−26) for every 5kg/m2 increase in BMI (Table 1): ORs were somewhat greater for endometrioid (OR=1.56, 95% CI 1.42–1.72) HERE than non-endometrioid/mixed (OR=1.50, 95% CI 1.43–1.57) histologies. The association between BMI and the BMIwGRS was significant in both cases and controls (Supplementary Figure 3); overall each weighted allele (i.e. each unit increase in the BMIwGRS) was associated with a 4.83kg/m2 increase in BMI, 95% CI 4.33–5.32, P=1.2×10−81, indicating the suitability of this score as an instrumental variable for BMI in our dataset (F statistic on a pooled analysis adjusting for study 587.7).
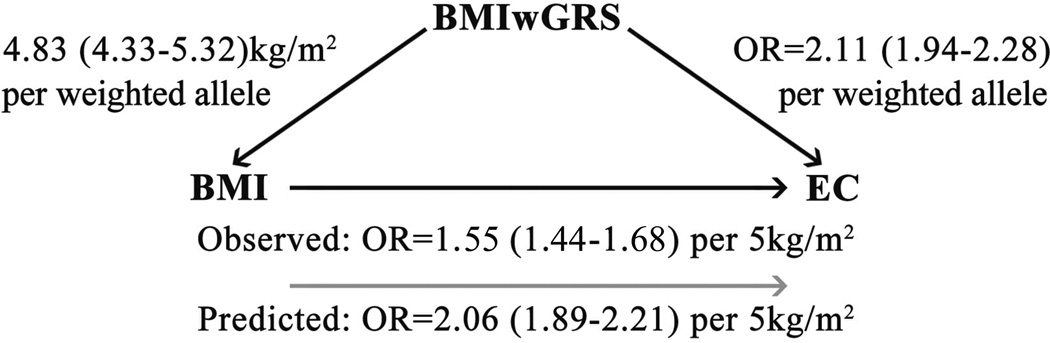
The BMIwGRS was significantly associated with EC risk in the entire dataset, with a per weighted allele OR=2.11 (95% CI 1.94–2.28, P=3.4×10−17: Table 2). Scaling according to the magnitude of the effect of the score on BMI (β=4.83kg/m2), we find the EC OR per 5kg/m2 of genetically predicted BMI to be 2.06 (95% CI 1.89–2.21) (Figure 1). This effect is apparently driven by an association with endometrioid disease (scaled OR=2.21, 95% CI 2.03–2.38, P=6.6×10−12). The overall association was similar for the subset with BMI information (scaled OR=2.18, 95% CI 1.96–2.41, P=4.2×10−12), and attenuated but remained significant after including BMI as a covariate in the model (scaled OR=1.22, 95% CI 1.12–1.34, P=5.3×10−4).
Table 2.
Association of the 77 SNP body mass index weighted genetic risk score (BMIwGRS) with endometrial cancer risk
| Total dataset | Dataset with BMI information | BMI-adjusted | ||||
|---|---|---|---|---|---|---|
| BMIwGRS1 Quartiles |
Odds Ratio (95% CI) | P-value | Odds Ratio (95% CI) | P-value | Odds Ratio (95% CI) | P-value |
| All cases | (6609 cases; 37926 controls) | (4062 cases; 15974 controls) | ||||
| Q1 | Reference | Reference | Reference | |||
| Q2 | 1.06 (0.98–1.15) | 1.1×10−2 | 1.03 (0.93–1.14) | 5.0×10−1 | 0.98 (0.88–1.09) | 8.0×10−1 |
| Q3 | 1.20 (1.12–1.28) | 4.0×10−6 | 1.23 (1.13–1.33) | 6.7×10−5 | 1.10 (1.00–1.21) | 6.2×10−2 |
| Q4 | 1.34 (1.27–1.42) | 2.3×10−14 | 1.38 (1.28–1.48) | 3.0×10−10 | 1.16 (1.06–1.26) | 3.7×10−3 |
| Per wGRS quartile increase in EC risk |
1.02 (0.01–1.03) | 9.2×10−8 | 1.06 (1.04–1.07) | 1.1×10−10 | 1.03 (1.02–1.05) | 1.3×10−4 |
| wGRS as a continuous variable |
2.11 (1.94–2.28) | 3.4×10−17 | 2.24 (2.01–2.48) | 4.2×10−12 | 1.23 (1.12–1.36) | 5.3×10−4 |
| Endometrioid | (5612 cases; 37926 controls) | (3484 cases; 15974 controls) | ||||
| Q1 | Reference | Reference | Reference | |||
| Q2 | 1.09 (1.00–1.17) | 5.6×10−2 | 1.05 (0.94–1.17) | 3.5×10−1 | 1.00 (0.88–1.11) | 9.8×10−1 |
| Q3 | 1.22 (1.13–1.30) | 4.1×10−6 | 1.28 (1.17–1.39) | 8.0×10−6 | 1.14 (1.03–1.26) | 1.8×10−2 |
| Q4 | 1.38 (1.30–1.46) | 1.7×10−14 | 1.43 (1.32–1.54) | 5.3×10−11 | 1.20 (1.09–1.31) | 1.2×10−3 |
| Per wGRS quartile increase in EC risk |
1.02 (0.01–1.03) | 2.1×10−7 | 1.06 (1.04–1.07) | 1.0×10−10 | 1.03 (1.02–1.05) | 7.8×10−5 |
| wGRS as a continuous variable |
2.27 (2.08–2.45) | 6.6×10−12 | 2.51 (2.30–2.72) | 3.3×10−17 | 1.26 (1.13–1.38) | 2.2×10−4 |
BMIwGRS range: Overall 1.19–2.57 (mean 1.85, SD 0.17); Cases 1.21–2.53 (mean 1.87, SD 0.17); Controls 1.19–2.57 (mean 1.85, SD 0.17).
Figure 1. Observed and predicted risks of increasing BMI on endometrial cancer.
The predicted effect of a 5kg/m2 increase in BMI on EC risk was estimated by scaling the effect of the per weighted allele increase in the BMIwGRS on BMI (4.83 kg/m2) by the effect of the per weighted allele increase in the BMIwGRS on EC (OR 2.11) in our dataset (exp[(4.83/5)*ln(2.11)]). The predicted effect (grey arrow) of a per 5kg/m2 increase in BMI on endometrial cancer risk (OR 2.06) is larger than that observed in our study (OR 1.55).
According to the Ptest, there was no significant evidence of directional pleiotropy (P=0.53), despite some possible asymmetry in the funnel plot (Supplementary Figure 4). However, Cochran’s Q-test did show some significant heterogeneity in the causal estimates from the individual SNPs (P=1.5×10−4), hence the causal effect of BMI on EC would be more appropriately estimated from the inverse-variance weighted random effects meta-analysis of the 77 BMI SNPs (P=1.8×10−9). Unfortunately, this effect estimate cannot be interpreted since the SNP-BMI regression coefficients presented by the GIANT consortium are for an inverse-normalised transformation of BMI, from which effects on the kg/m2 scale cannot be derived. Nevertheless, we note that the causal lnOR estimate from the random-effects analysis of individual SNPs is ~10% higher than that from the equivalent fixed-effects analysis (Supplementary Figure 4), thus we infer that the true causal effect of BMI on EC is slightly larger than our best estimate under the assumption of no directional pleiotropy i.e. OR >2.06 per 5kg/m2 as predicted in our dataset. This is somewhat larger than the observed OR=1.55 (95% CI 1.44–1.68) per 5kg/m2 of reported BMI in this dataset, and also larger than previously published estimates of the effect of reported BMI on EC in epidemiological studies (e.g. OR 1.54, 95% CI 1.47–1.61, per 5kg/m2 (4); OR=1.57, 95% CI 1.54–1.61, per 5kg/m2 for “Type1” largely endometrioid EC (3)).
Both WHRwGRS were significantly associated with WHR in the WTCCC control group (34-SNP WHRwGRS β=0.05, 95% CI 0.02–0.08, P=2.2×10−3; 47-SNP WHRwGRS β=0.05, 95% CI 0.03–0.08, P=1.8×10−4). As expected, neither WHRwGRS was associated with BMI (P=>0.80). The results for the 34-SNP WHRwGRS were very similar to those from secondary analyses using all 47 WHR SNPs, neither of which were significantly associated with EC risk (OR=1.02, 95% CI 0.99–1.04, P=0.09 and OR=0.97, 95% CI 0.63–1.31, P=0.86, respectively) (Table 3), or with risk stratified by histology (data not shown).
Table 3.
Association of the 34- and 47-SNP waist-hip ratio weighted genetic risk score (WHRwGRS) with endometrial cancer risk
| 34-SNP wGRS | 47-SNP wGRS | |||
|---|---|---|---|---|
| WHRwGRS1 Quartiles |
Odds Ratio (95% CI) | P-value | Odds Ratio (95% CI) | P-value |
| Q1 | Reference | Reference | ||
| Q2 | 1.00 (0.96–1.03) | 9.5×10−1 | 0.99 (0.96–1.02) | 4.5×10–1 |
| Q3 | 0.98 (0.96–0.01) | 7.0×10–2 | 098 (0.94–0.01) | 2.6×10–1 |
| Q4 | 0.99 (0.97–0.01) | 3.2×10–1 | 0.98 (0.95–0.01) | 1.3×10–1 |
| Per wGRS quartile increase in EC risk |
0.99 (0.99–1.00) | 1.7×10–1 | 0.99 (0.99–1.00) | 2.4×10–1 |
| wGRS as a continuous variable |
1.02 (0.99–1.04) | 9.0×10–2 | 0.97 (0.63–1.31) | 8.6×10–1 |
Range 34 SNP WHRwGRS: Overall 0.53–1.52 (mean 0.98, SD 0.12); Cases 0.57–1.36 (mean 0.98, SD 0.12); Controls 0.53–1.52 (mean 0.98, SD 0.12). Range 47 SNP WHRwGRS: Overall 0.67–1.76 (mean 1.21, SD 0.14): Cases 0.67–1.69 (mean 1.22, SD 0.14); Controls 0.68–1.76 (mean 1.20, SD 0.14).
Discussion
In this study we assessed whether SNPs associated with increased BMI or WHR are also associated with increased EC risk, either individually or in combination, and whether these genetic associations are independent of BMI. While BMI is clearly recognized as a major risk factor for EC, the role of WHR, independent of BMI, is less clear. Most studies including WHR have reported evidence for an association with EC (4, 26–32) but only four presented analyses adjusting for BMI, suggesting the WHR-EC risk association was attenuated in Caucasians (29–31), but not in Asians (32).
Combined as a wGRS, the 77 BMI-associated SNPs were highly significantly associated with BMI, even though the BMIwGRS explained only ~1% of the variance in BMI in our sample (less than the estimated 2.7% of the variance in BMI explained by 97 BMI-associated SNPs across ancestries in the discovery dataset (9)). The BMIwGRS was also significantly associated with EC, explaining ~0.1% of the variance in risk and confirming the causal nature of the association between BMI and EC. Indeed, the association between genetically-predicted BMI (based on the 77 SNP BMIwGRS) and EC risk was somewhat larger than that between observed BMI (i.e. that calculated from self-reported height and weight) and EC risk, and we identified significant heterogeneity in the per-SNP causal estimates, both of which suggest some modest degree of directional pleiotropy. Furthermore, the overall association signal attenuated but did not disappear when adjusting for BMI (OR=1.22 vs 2.06 per 5kg/m2 genetically predicted BMI), which also suggests that these SNPs mainly, but not entirely, operate to increase EC risk via BMI. In particular, we note that one BMI SNP, rs2075650, was found to be associated with EC risk independent of BMI in our dataset.
Our result could also (or instead) suggest that the aspect of body composition most relevant for EC risk is only partially captured by BMI; although BMI is widely used as a convenient proxy measure for adiposity, it is by no means a perfect measure (33). One would expect that the SNPs identified to date in GWAS of BMI, at least on aggregate, are more strongly associated with adiposity than with its proxy, BMI. Hence the combined effect of the 77 BMI SNPs might be a better predictor of risk due to adiposity than BMI self-reported at a single time point (which could be subject to regression dilution). The effect of BMI on EC risk has been reported to be attenuated among ever users of hormone replacement therapy (HRT), as compared to never users (34). Although we were unable to stratify our analyses according to HRT use, we are confident that the difference between the effects of observed and genetically-predicted BMI on EC risk seen in our study is not attributable to an interaction between BMI and HRT use, since both analyses were based on the same set of women, and so will necessarily have included the same proportions of current, previous and never HRT users. However, the discrepancy between the observed and predicted effects of BMI on EC could theoretically point to negative confounding between measured BMI and EC, via HRT use and some other factor (e.g. socioeconomic status) associated with both higher BMI and less frequent HRT use.
The evidence for modest pleiotropy for BMI SNPs and EC risk has been reported previously in a study of Chinese women. A study of 26 SNPs then reported as (nominally) associated with different measures of obesity in GWAS datasets (35) identified a GRS-EC association in Chinese women (16), which attenuated but remained significant after adjusting for BMI. Direct comparison to our findings is difficult, due to differences in SNP selection and overlap, and also because the relationship between BMI and percentage body fat differs among ethnic groups (36). However, the results from our European-ancestry study contrast with those from another recent analysis of 3,376 European-ancestry EC cases and 3,867 controls from the E2C2 consortium (15); neither cases nor controls from the E2C2 analysis overlap with those presented here. While the E2C2 analysis also identified a significant association between a 97-SNP GRS and EC risk (P=0.002), this association ablated after adjustment for BMI (P=0.78). The differences in findings between the two European studies may possibly reflect the BMI profiles of the two studies; while the mean BMI of the controls did not differ between the two studies (P=0.11), the mean BMI of cases in the E2C2 study was greater than that for cases in our study (P=0.017, mean difference of 0.43kg/m2). However, the differences are more likely to reflect the increased power of our larger study to detect modest effects. This is particularly pertinent to the single SNP findings. Although BMI SNP rs2075650 was found to be significantly associated with EC risk independent of BMI in our dataset (per allele OR of 1.13, 95% CI=1.05–1.21), this same SNP was not significantly associated with EC risk in the E2C2 analysis (15) (ORBMI-adjusted= 1.00, 95% CI=0.90–1.10), although there was some overlap between the 95% CIs. We note also that we find no evidence in support of the E2C2 tentative finding of a protective effect on EC of the subset of five BMI-risk alleles at loci known to be involved in Monogenic Obesity Syndromes, with OR point estimates above unity observed for the four loci we investigated in our study (rs6567160, MC4R, OR 1.02 (0.98–1.06), P=0.3; rs11030104, BDNF, OR 1.05 (1.01–1.09), P=0.05; rs10182181, POMC/ADCY3, OR 1.02 (0.98–1.06), P=0.4; Supplementary Table 3).
We also, for the first time, used a genetic approach to assess the influence of body fat distribution on EC risk, an epidemiological association which is less clear than that of adiposity as measured by BMI. Combined as a wGRS, 34 SNPs reported as significantly associated with WHR in women (11) were not significantly associated with EC in our sample. We focused on the 34-SNP WHRwGRS due to the marked sexual dimorphism amongst WHR-associated loci (11), however, the results did not differ when 47 SNPs were included in the WHRwGRS. Together, the 49 SNPs now reported as associated with WHR explain ~2.4% of the variance in WHR in women (~1.4% in both sexes combined) (11). As this is similar to the proportion of variation in BMI explained by currently known BMI-associated SNPs, it seems most likely that the lack of association between the WHRwGRS and EC is due to a true lack of association between WHR and EC, rather than the smaller number of SNPs included in the WHRwGRS, particularly as the WHR-EC association seen in epidemiological studies seems to be accounted for by BMI (4). Rather than WHR, waist circumference (WC) may be a more relevant measure of central adiposity, with evidence that the association between WC and EC is independent of BMI (4). However, analysis of the genetic association between WC and EC awaits the discovery of additional WC-associated SNPs, as only six have been reported to date (four in Caucasians), none of which reached genome-wide significance in women only (11).
In summary, our combined results from weighted genetic risk scores and Mendelian Randomization analysis provide a further line of evidence that increasing BMI has a direct effect on EC risk, and thus that interventions aimed at weight loss should reduce that risk (5). We also found that SNP alleles associated with increased BMI have an aggregate effect on EC risk that is over and above that predicted by their effects on BMI. This suggests a possible degree of pleiotropy in SNP functions, indicating that these SNPs, and potentially other BMI-associated SNPs yet to be discovered, would be more useful components in an EC risk prediction model than BMI itself. In contrast, our genetic findings indicate that WHR is not independently associated with EC risk. These findings support the value of genetic approaches to verify causal relationships between epidemiological risk factors and cancer risk.
Supplementary Material
Acknowledgments
The authors thank the many individuals who participated in this study and the numerous institutions and their staff who supported recruitment, detailed in full in the Supplementary Text. We also thank Felix Day and Stephen Burgess for helpful discussions.
Financial Support: The iCOGS endometrial cancer analysis was supported by NHMRC project grant [ID#1031333] to ABS, DFE and AMD. ABS and PW are supported by the NHMRC Fellowship scheme. IT is supported by Cancer Research UK and the Oxford Comprehensive Biomedical Research Centre. THTC is supported by the Rhodes Trust and the Nuffield Department of Medicine. Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement no 223175 [HEALTH-F2-2009-223175] [COGS], Cancer Research UK [C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565], the National Institutes of Health [CA128978] and Post-Cancer GWAS initiative [1U19 CA148537, 1U19 CA148065 and 1U19 CA148112 - the GAME-ON initiative], the Department of Defence [W81XWH-10-1-0341], the Canadian Institutes of Health Research [CIHR] for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund.
ANECS recruitment was supported by project grants from the NHMRC [ID#339435], The Cancer Council Queensland [ID#4196615] and Cancer Council Tasmania [ID#403031 and ID#457636]. SEARCH recruitment was funded by a programme grant from Cancer Research UK [C490/A10124]. Stage 1 and stage 2 case genotyping was supported by the NHMRC [ID#552402, ID#1031333]. This study makes use of data generated by the Wellcome Trust Case-Control Consortium (WTCCC). A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02 - funding for this project was provided by the Wellcome Trust under award 085475. NSECG was supported by the EU FP7 CHIBCHA grant, Wellcome Trust Centre for Human Genetics Core Grant 090532/Z/09Z, and CORGI was funded by Cancer Research UK. Recruitment of the QIMR Berghofer controls was supported by the NHMRC. The University of Newcastle, the Gladys M Brawn Senior Research Fellowship scheme, The Vincent Fairfax Family Foundation, the Hunter Medical Research Institute and the Hunter Area Pathology Service all contributed towards the costs of establishing the Hunter Community Study.
The Bavarian Endometrial Cancer Study (BECS) was partly funded by the ELAN fund of the University of Erlangen. The Leuven Endometrium Study (LES) was supported by the Verelst Foundation for endometrial cancer. The Mayo Endometrial Cancer Study (MECS) and Mayo controls (MAY) were supported by grants from the National Cancer Institute of United States Public Health Service [R01 CA122443, P30 CA15083, P50 CA136393, and GAME-ON the NCI Cancer Post-GWAS Initiative U19 CA148112], the Fred C and Katherine B Andersen Foundation, the Mayo Foundation, and the Ovarian Cancer Research Fund with support of the Smith family, in memory of Kathryn Sladek Smith. MoMaTEC received financial support from a Helse Vest Grant, the University of Bergen, Melzer Foundation, The Norwegian Cancer Society (Harald Andersens legat), The Research Council of Norway and Haukeland University Hospital. The Newcastle Endometrial Cancer Study (NECS) acknowledges contributions from the University of Newcastle, The NBN Children’s Cancer Research Group, Ms Jennie Thomas and the Hunter Medical Research Institute. RENDOCAS was supported through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet [numbers: 20110222, 20110483, 20110141 and DF 07015], The Swedish Labor Market Insurance [number 100069] and The Swedish Cancer Society [number 11 0439]. The Cancer Hormone Replacement Epidemiology in Sweden Study (CAHRES, formerly called The Singapore and Swedish Breast/Endometrial Cancer Study; SASBAC) was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institutes of Health and the Susan G. Komen Breast Cancer Foundation.
The Breast Cancer Association Consortium (BCAC) is funded by Cancer Research UK [C1287/A10118, C1287/A12014]. The Ovarian Cancer Association Consortium (OCAC) is supported by a grant from the Ovarian Cancer Research Fund thanks to donations by the family and friends of Kathryn Sladek Smith [PPD/RPCI.07], and the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge. Additional funding for individual control groups is detailed in the Supplementary Information.
P.A. Fasching reports receiving commercial research grants from Amgen and Novartis, and is a consultant/advisory board member for Novartis, Pfizer, Roche and Genomic Health.
Footnotes
Disclosure Statement: No potential conflicts of interest were disclosed by the other authors.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;36:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Tavassoli FA. Pathology and Genetics. Tumors of the breast and female genital organs. In: Tavassoli FA, Devilee P, editors. WHO/IARC Classification of Tumours. No 4 Lyon: IARC Press; 2003. [Google Scholar]
- 3.Setiawan VW, Yang HP, Pike MC, McCann SE, Yu H, Xiang YB, et al. Type I and II endometrial cancers: have they different risk factors? J Clin Oncol. 2013;31:2607–2618. doi: 10.1200/JCO.2012.48.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aune D, Navarro Rosenblatt DA, Chan DS, Vingeliene S, Abar L, Vieira AR, et al. Anthropometric factors and endometrial cancer risk: A systematic review and dose-response meta-analysis of prospective studies. Ann Oncol. 2015;6:1635–1648. doi: 10.1093/annonc/mdv142. [DOI] [PubMed] [Google Scholar]
- 5.Nagle CM, Marquart L, Bain CJ, O'Brien S, Lahmann PH, Quinn M, et al. Impact of weight change and weight cycling on risk of different subtypes of endometrial cancer. Eur J Can. 2013;49:2717–2726. doi: 10.1016/j.ejca.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 6.Yang HP, Wentzensen N, Trabert B, Gierach GL, Felix AS, Gunter MJ, et al. Endometrial cancer risk factors by 2 main histologic subtypes: the NIH-AARP Diet and Health Study. Am J Epidemiol. 2013;177:142–151. doi: 10.1093/aje/kws200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ju W, Kim HJ, Hankinson SE, De Vivo I, Cho E. Prospective study of body fat distribution and the risk of endometrial cancer. Cancer Epidemiol. 2015;39:567–570. doi: 10.1016/j.canep.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machiela MJ, Lindstrom S, Allen NE, Haiman CA, Albanes D, Barricarte A, et al. Association of type 2 diabetes susceptibility variants with advanced prostate cancer risk in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol. 2012;176:1121–1129. doi: 10.1093/aje/kws191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thrift AP, Gong J, Peters U, Chang-Claude J, Rudolph A, Slattery ML, et al. Mendelian randomization study of body mass index and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2015;24:1024–1031. doi: 10.1158/1055-9965.EPI-14-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prescott J, Setiawan VW, Wentzensen N, Schumacher F, Yu H, Delahanty R, et al. Body mass index genetic risk score and endometrial cancer risk. PloS One. 2015;10:e0143256. doi: 10.1371/journal.pone.0143256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delahanty RJ, Beeghly-Fadiel A, Xiang YB, Long J, Cai Q, Wen W, et al. Association of obesity-related genetic variants with endometrial cancer risk: a report from the Shanghai Endometrial Cancer Genetics Study. Am J Epidemiol. 2011;174:1115–1126. doi: 10.1093/aje/kwr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21:223–242. doi: 10.1177/0962280210394459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spurdle AB, Thompson DJ, Ahmed S, Ferguson K, Healey CS, O'Mara T, et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat Genet. 2011;43:451–454. doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Painter JN, O'Mara TA, Batra J, Cheng T, Lose FA, Dennis J, et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum Mol Genet. 2015;24:1478–1492. doi: 10.1093/hmg/ddu552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira T, Marchini J. Modeling interactions with known risk loci-a Bayesian model averaging approach. Ann Hum Genet. 2011;75:1–9. doi: 10.1111/j.1469-1809.2010.00618.x. [DOI] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dal Maso L, Tavani A, Zucchetto A, Montella M, Ferraroni M, Negri E, et al. Anthropometric measures at different ages and endometrial cancer risk. Br J Cancer. 2011;104:1207–1213. doi: 10.1038/bjc.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amankwah EK, Friedenreich CM, Magliocco AM, Brant R, Courneya KS, Speidel T, et al. Anthropometric measures and the risk of endometrial cancer, overall and by tumor microsatellite status and histological subtype. Am J Epidemiol. 2013;177:1378–1387. doi: 10.1093/aje/kws434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaki A, Gaber A, Ghanem E, Moemen M, Shehata G. Abdominal obesity and endometrial cancer in egyptian females with postmenopausal bleeding. Nutr Cancer. 2011;63:1272–1278. doi: 10.1080/01635581.2011.615973. [DOI] [PubMed] [Google Scholar]
- 29.Friedenreich C, Cust A, Lahmann PH, Steindorf K, Boutron-Ruault MC, Clavel-Chapelon F, et al. Anthropometric factors and risk of endometrial cancer: the European prospective investigation into cancer and nutrition. Cancer Causes Control. 2007;18:399–413. doi: 10.1007/s10552-006-0113-8. [DOI] [PubMed] [Google Scholar]
- 30.Conroy MB, Sattelmair JR, Cook NR, Manson JE, Buring JE, Lee IM. Physical activity, adiposity, and risk of endometrial cancer. Cancer Causes Control. 2009;20:1107–1115. doi: 10.1007/s10552-009-9313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves KW, Carter GC, Rodabough RJ, Lane D, McNeeley SG, Stefanick ML, et al. Obesity in relation to endometrial cancer risk and disease characteristics in the Women's Health Initiative. Gynecol Oncol. 2011;121:376–382. doi: 10.1016/j.ygyno.2011.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu WH, Matthews CE, Xiang YB, Zheng W, Ruan ZX, Cheng JR, et al. Effect of adiposity and fat distribution on endometrial cancer risk in Shanghai women. Am J Epidemiol. 2005;161:939–947. doi: 10.1093/aje/kwi127. [DOI] [PubMed] [Google Scholar]
- 33.Shah NR, Braverman ER. Measuring adiposity in patients: the utility of body mass index (BMI), percent body fat, leptin. PloS One. 2012;7:e33308. doi: 10.1371/journal.pone.0033308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crosbie EJ, Zwahlen M, Kitchener HC, Egger M, Renehan AG. Body mass index, hormone replacement therapy, and endometrial cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19:3119–3130. doi: 10.1158/1055-9965.EPI-10-0832. [DOI] [PubMed] [Google Scholar]
- 35.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.WHO Expert Consultation. Appropriate body-mass index for Asian populations and its implications for policy and intervention strategies. Lancet. 2004;363:157–163. doi: 10.1016/S0140-6736(03)15268-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.