Abstract
The Cancer Genome Atlas (TCGA) network study of 12 cancer types (PanCancer 12) revealed frequent mutation of TP53, and amplification and expression of related TP63 isoform ΔNp63 in squamous cancers. Further, aberrant expression of inflammatory genes and TP53/p63/p73 targets were detected in the PanCancer 12 project, reminiscent of gene programs co-modulated by cREL/ΔNp63/TAp73 transcription factors we uncovered in head and neck squamous cell carcinomas (HNSCC). However, how inflammatory gene signatures and cREL/p63/p73 targets are co-modulated genome-wide is unclear. Here, we examined how inflammatory factor TNF-α broadly modulates redistribution of cREL with ΔNp63α/TAp73 complexes and signatures genome-wide in the HNSCC model UM-SCC46 using chromatin immunoprecipitation sequencing (ChIP-seq). TNF-α enhanced genome-wide co-occupancy of cREL with ΔNp63α on TP53/p63 sites, while unexpectedly promoting redistribution of TAp73 from TP53 to Activator Protein-1 (AP-1) sites. cREL, ΔNp63α, and TAp73 binding and oligomerization on NF-κB, TP53 or AP-1 specific sequences were independently validated by ChIP-qPCR, oligonucleotide-binding assays, and analytical ultracentrifugation. Function of the binding activity was confirmed using TP53, AP-1, and NF-κB specific response elements, or p21, SERPINE1, and IL-6 promoter luciferase reporter activities. Concurrently, TNF-α regulated a broad gene network with co-binding activities for cREL, ΔNp63α, and TAp73 observed upon array profiling and RT-PCR. Overlapping target gene signatures were observed in squamous cancer subsets and in inflamed skin of transgenic mice overexpressing ΔNp63α. Furthermore, multiple target genes identified in this study were linked to TP63 and TP73 activity and increased gene expression in large squamous cancer samples from PanCancer 12 TCGA by CircleMap. PARADIGM inferred pathway analysis revealed the network connection of TP63 and NF-κB complexes through an AP-1 hub, further supporting our findings. Thus, inflammatory cytokine TNF-α mediates genome-wide redistribution of the cREL/p63/p73, and AP-1 interactome, to diminish TAp73 tumor suppressor function and reciprocally activate NF-κB and AP-1 gene programs implicated in malignancy.
Keywords: TNF-alpha, cRel, ΔNp63α, TAp73, ChIP-seq
Introduction
The Cancer Genome Atlas (TCGA) project aims to provide a comprehensive catalog of the key genomic changes in major cancer types, in order to foster effective diagnosis, treatment and prevention. The recently published TCGA HNSCC (head and neck squamous cell carcinoma) project, including 279 patient tissues, revealed that more than 84% of HPV negative HNSCC harbor genetic alterations in tumor suppressor TP53, concurrent with amplification and expression of related family member ΔNp63 in ~47% cases, and altered TNF-α-NF-κB/REL, inflammation, and death pathways in ~60% cases (1). In addition, the TCGA network PanCancer 12 project performed integrative analyses on 3,527 specimens of 12 cancer types, using five genome-wide high throughput platforms (2). The project revealed a unique convergence that classified squamous cell carcinoma (SCC) of head and neck, lung and a subset of bladder cancers into a common subtype, called “squamous-like” (2). This genomic classification suggests that those squamous cancer types shared more molecular similarities than other tumor types from the same tissue-of-origin. Consistent with findings in the HNSCC TCGA project, the hallmarks of these squamous cancers include high rates of TP53 mutation, amplification and over-expression of oncogenic isoform ΔNp63, and altered activation of TP53/TP63/TP73 and immune pathway genes linked to NF-κB/REL transcription factors. These newly identified SCC signatures raised several critical questions, including how and to what extent do TP63/TP73 and NF-κB/REL family transcription factors interact to regulate global gene programs in squamous cancers?
The TP53 family comprises TP53, TP63, and TP73 and their isoforms. TP53 is the most frequently mutated tumor suppressor gene in cancer, especially in squamous cancers, and is described as the “guardian of the genome.” Mutation or inactivation of TP53 promotes genomic instability by disrupting cell cycle arrest, DNA repair, senescence, apoptosis, and autophagy of irreversibly damaged cells (3). Interestingly, however, the other TP53 family members, TP63 and TP73, are infrequently mutated, and can potentially compensate for disrupted TP53 (4, 5), but their shared and distinct functions appear to be more complex than those of TP53 in tumorigenesis, as suggested by TCGA findings. TP63 and TP73 isoforms share a core structural architecture, sequence homology, and potential but usually weaker tumor suppressor function than TP53, while they also exhibit distinct roles in development, adhesion, tumor promotion, and inflammatory responses in normal and malignant epithelia (4, 6-8). The TA isoforms contain full-length N-terminal transactivating domains that function as TP53 homologs, whereas ΔN isoforms having a truncated N-terminus can serve as antagonists of TP53 and its TA counterparts, as well as promote cancer gene programs (9, 10). However, the genome-wide role and mechanisms whereby ΔNp63α/TAp73 promote tumorigenesis and linked to inflammation in cancer are undefined.
We recently revealed that inflammatory cytokine TNF-α stimulates binding of NF-κB subunit cREL with ΔNp63 to form a protein complex and displacement of TAp73 DNA binding, leading to the repression of growth arrest and apoptotic genes CDKN1A (p21), NOXA, and PUMA (6). Complexes between cREL, RELA and ΔNp63 were also implicated in reciprocal activation of several NF-κB regulated genes (8). Interestingly, the NF-κB family proto-oncogene cREL is amplified and overexpressed in a subset of HNSCC and other cancers (11), but its functions are less well characterized (12). These experimental data suggest the hypothesis that NF-κB and TP53 family members could coordinate wider global and reciprocal cross-talk between cell death, survival and inflammatory gene programs. Specifically, are these interactions between cREL, ΔNp63α and TAp73 part of a novel mechanism that reciprocally regulates a genome-wide oncogenic program? Do the reciprocal interactions between cREL and ΔNp63α with TAp73 indicate their capability to bind TP53, NF-κB or other DNA response elements (REs)? Finally, what is the fate and functional significance of TAp73 displaced by the cREL/ΔNp63α complexes?
To answer these questions, we performed genome-wide chromatin immunoprecipitation assays, followed by high-throughput sequencing (ChIP-seq), and microarray profiling in HNSCC cell lines, to explore the genome-wide regulatory role of cREL/ΔNp63α and TAp73 complexes. Furthermore, bioinformatic analysis of in vivo data from TCGA, immunohistochemistry and tissue array of HNSCC, and a ΔNp63α transgenic mouse model supports their contribution in the regulation of the cancer gene program. These findings present a new paradigm for how TNF-α and these TFs orchestrate gene programs implicated in cancer-related inflammation, survival, and migration, and help explain the mechanisms underpinning the dysregulated network of TP53/TP63 and inflammation observed in the Pan-TCGA project.
Results
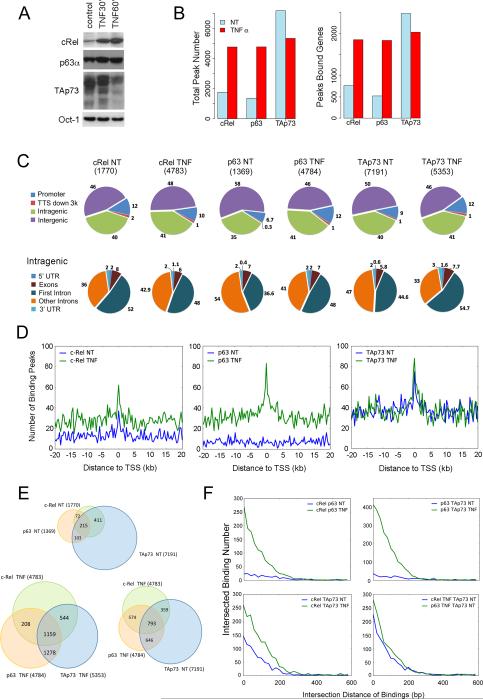
TNF-α promotes enrichment and co-localized binding of cREL, TP63α and TAp73 in the regulatory regions and around transcription start sites genome-wide
To investigate cREL, p63α and TAp73 binding activity genome-wide, ChIP-seq was performed using UM-SCC46 cells, previously shown to exhibit higher expression of mtTP53, TP63 and TP73 proteins, and TNF-α modulation of their interactions in target gene regulation representative of HNSCC with mtTP53 (6). We confirmed that TNF-α induced cREL and ΔNp63α, and partially decreased multiple TAp73 isoforms in nuclear extracts (Figure 1A), where TAp73 is predominantly detected in UM-SCC46 and other HNSCC lines under baseline conditions (Figure S1A). TNF-α increased total genomic binding peak numbers and peak associated genes by cREL and p63α, while partially decreasing TAp73 binding activity (Figure 1B, Table S1), and similar patterns were seen on individual chromosomes (Figure S1B, S2). We observed that the percentage of genome-wide bindings were disproportionately enriched near or within genes (promoter, intragenic, transcriptional termination site region) compared with much larger intergenic regions (Figure 1C, upper panels). Cumulatively, over half of the binding peaks were within the promoter (~7-12%) and intragenic regions (35-41%). These peri-genic region peak distributions were significantly enriched compared with the whole genome background distribution, tested using the exact binomial test (Figure S3). The binding activities within the intragenic regions were enriched in the first intron (Figure 1C, lower panels). After TNF-α treatment, cREL and p63α binding were substantially induced near the TSSs, while the basal TAp73 TSS binding peak did not appreciably change in the presence of TNF-α (Figure 1D). The intersection sets of three TF binding peaks within 1 kb distance were identified, showing that intersecting binding peaks under basal conditions (215) were significantly increased after TNF-α treatment (1159) (Figure 1E, upper and lower left; Fisher's exact test, p=2.2e-16). 793 TAp73 basal binding peaks intersected with TNF-induced cREL and p63α binding (Figure 1E, lower right). After combination of all binding activities, 1,217 candidate binding peaks were observed, which aligned within the regulatory loci of genes and generated 530 genes that displayed co-binding by the three TFs (Figure S4). Furthermore, the physical distance between the intersections of three TF binding peaks was examined, defined as within 1kb distance on the same chromosome (Figure 1F). TNF-α increased intersection binding peaks between cREL and p63α, or between p63α and TAp73, around the TSS within <200 bp (Figure 1F, upper two panels), when compared with other intersections (Figure 1F, lower panels). More detailed results were included in the supplemental information. Together, these data show that TNF-α modulated the co-localization of cREL, p63α, and TAp73 binding enriched around TSSs.
Figure 1. TNF-α promotes genome-wide cREL, p63α and TAp73 binding activities in the regulatory regions.
(A) UM-SCC46 cells were treated with TNF-α (20 ng/ml) for 1 h to induce altered cREL, p63, and p73 protein expression in the nucleus. Oct-1 was used as the loading control. (B) ChIP-seq was performed using antibodies against cREL, p63α, and TAp73 for the UM-SCC46 cells treated (TNF) and untreated (NT) with TNF-α. The pulled-down DNAs were sequenced by the high-throughput sequencer GAIIx from Illumina. Total peak numbers (left) and peak-bound genes (right) are presented in the bar graph. (C) Characterization of the modulation of TF binding in the regulatory regions. The distribution of binding peaks in different regions of the genome is presented in the pie charts, and peak numbers are labeled at the top. The upper panel shows the percentages of the peaks distributed among regulatory (promoter, TTS, intragenic) and intergenic regions. The lower panel shows the percentages of the peaks distributed only among the different intragenic regions. (D) The binding peak numbers of each TF were plotted within 20 kb upstream (−) and downstream of the TSSs across the whole genome. The blue line is the basal transcription factor binding without TNF-α treatment; while the green line is after TNF-α treatment. (E) The transcription factor binding sites within 1 kb distance were identified, and the number of overlapping sites are presented in the Venn diagrams. (F) Distance relationships between two peak sets under different conditions in the ChIP-seq experiments were analyzed. Intersected peaks were defined to be within 1kb distance on the same chromosome. Y axis shows the quantity of intersected peaks at different intersection distance in bp (x axis). Blue, untreated; green, TNF-α treated.
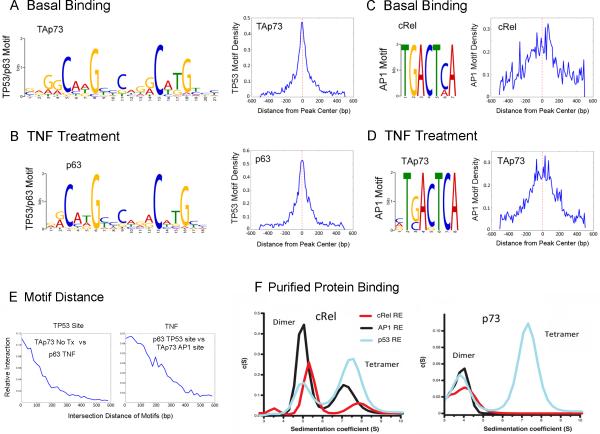
De novo motif search defines genome-wide binding on TP53 and AP-1 consensus elements modulated by TNF-α
Next, the top motifs most frequently bound by the three TFs under different treatment conditions were identified using Gibb's motif sampler and summarized (Figure 2A-D, Figure S5). The basal binding of TAp73 (Figure 2A, left), and TNF-α induced p63α binding (Figure 2B, left), were enriched for a TP53/p63 consensus sequence. Surprisingly, basal cREL (Figure 2C, left) and TNF-α induced TAp73 bindings (D, left) were enriched for AP-1 consensus motifs. A narrow distribution of ~200 bp for the basal TAp73 (Figure 2A, right) and TNF-α induced p63α binding to TP53 motifs (Figure 2B, right) were observed. However, broader distribution patterns (~400 bp) were observed for the basal and TNF-α induced cREL (Figure 2C, right and Figure S5), and TNF-α induced TAp73 binding on AP-1 sites (Figure 2D, right). On TP53/p63 motifs, there was abundant overlapping binding activity of basal TAp73 versus TNF-α induced p63α, ranging within ~200-300 bp (Figure 2E, left). TNF-α also induced concurrent binding of p63α on TP53 sites and TAp73 on AP-1 motifs, located in a broader range of ~400 bp (Figure 2E, right).
Figure 2. De novo motif search identified TP53 and AP-1 consensus sequences.
The motifs most frequently bound by p63, TAp73 and c-REL transcription factors were identified by MEME-ChIP (55) or Gibbs Motif Sampler (54). They are shown as the sequence logos with known consensus (A-D left). (A, left) The predominant TP53/p63 motifs consistent with the TP53.02 consensus sequences were identified in basal binding of TAp73. (B, left) The predominant motif of TP63 was identified by TP63α binding activities after TNF-α treatment. (C, left) AP-1 motif was identified in basal cREL binding. (D, left) The AP-1 motif was observed in TNF-α induced TAp73 binding. (A-D, right) Distance relationships between different motifs detected by ChIP-Seq were examined. Motifs are considered to be intersecting if they are located within 1 kb distance. The corresponding motifs were mapped back to peak sequences. Motif density (y axis) was plotted against the distance from the center of the binding peak (x axis), showing the distribution pattern of specific motifs in peaks (scales are labeled as ×102). (E) The relative interaction on the TP53 motif (y axis) was plotted against the distance of basal TAp73 binding vs. TP63α after TNF-α treatment. Under TNF treated condition, the relative interaction (x axis) of TP63 binding on TP53 motifs and TAp73 binding on AP-1 motif was plotted against the distance between the two binding motifs (x axis, right). (F) Sedimentation coefficient distribution of purified cRel Rel homology domain (RHD, left) and p73 DNA binding domain (DBD, right) binding to fluorescein-labeled AP-1 (black), cREL (red) and TP53 (blue) response elements (REs). Fluorescein absorbance was detected at 488 nm. The peaks corresponding to the protein dimers and tetramers are indicated. No tetrameric binding of p73 DBD was observed to the AP-1 and cREL REs. For clarity the peak at 2 S for the unbound DNA species is not displayed.
Binding of purified cREL homology domain and TP73 DNA-binding domain to cREL, TP53 and AP-1 response elements (REs)
Next, we purified recombinant cREL Rel homology domain (RHD) and TP73 DNA binding domain (DBD) proteins and tested their ability to bind oligonucleotides with minimal consensus sequences from cREL, TP53 and AP-1 response elements (REs) (Figure 2F). Analytical ultracentrifugation with fluorescein-labeled DNA allowed us to measure sedimentation velocities of the protein-DNA complexes and classify their oligomerization state. Remarkably, cREL exhibited binding activities for all three REs, with a higher affinity for its own RE (Figure 2F, left). TP73 DBD exhibited dimeric DNA-protein complexes for all three REs tested; however, the tetramer complex was only observed for the TP53 RE (Figure 2F, right). Thus, our binding experiments with purified proteins confirmed that both cREL RHD and p73 DBD are able to bind not only to their own consensus sequences, but also to the other DNA REs, supporting the binding to motifs identified in the ChIP-seq experiment.
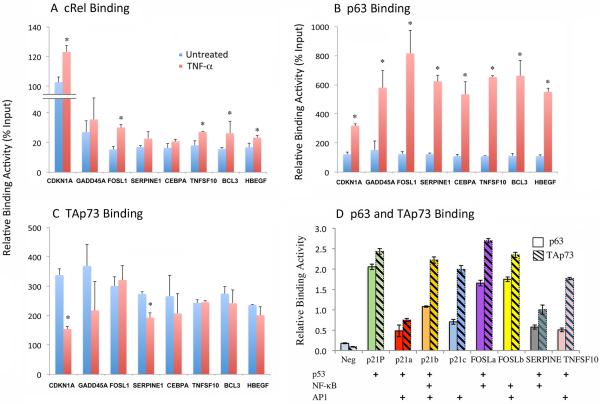
Validation of co-localized binding on individual gene promoters by ChIP-qPCR and oligo-based binding assays
Representative peak binding activities of four of the genes detected by ChIP-seq are presented in Figure S6. We further validated the specific binding activities of the three TFs on regulatory regions of eight representative genes detected by ChIP-seq and confirmed the binding by ChIP-PCR (Figure 3A-C, Table S2). Treatment with TNF-α significantly increased cREL and/or p63α binding (Figure 3A, B), while TAp73 binding was partially decreased or not significantly changed (Figure 3C). Next, we performed a different binding experiment using short ~50-70 bp synthetic oligonucleotides containing 10-20bp sequences of individual or a combination of predicted consensus motifs (Figure 3D, Table S3). We observed a relatively stronger basal TAp73 binding activity when compared with p63α binding, consistent with the ChIP-seq and ChIP-qPCR results. Interestingly, the oligonucleotide containing a predicted AP-1 motif (p21-c) without the other motifs also exhibited TAp73 binding. These binding activities detected by ChIP-seq were validated by two different experimental methods. Thus our current study provided evidence for the co-localization of cREL, ΔNp63 and TAp73 to multiple target promoters and response elements in response to TNF-α, consistent with our previous publications (6, 8).
Figure 3. Validation of co-localized binding activities of cREL, p63α and TAp73.
Based on overlapped binding peaks in the regulatory regions, we predicted core binding sequences containing potential p63/TP53 or NF-κB/cREL binding motifs using a bioinformatics approach. PCR primer sequences were designed to flank the regions containing the binding motifs (Table S2). In the genes of interest, the overlapping binding peaks were located in promoter/enhancer regions (SERPINE1, BCL3, CEBPA, HBEGF) or in first (CDKN1A, FOSL1, TNFSF10) or other (GADD45A) introns. Quantitative PCR was performed in independent ChIP experiments, and isotype antibody served as the negative control. (A-C) ChIP binding activity of cREL (A), p63α (B), and TAp73 (C). Blue, untreated; red, TNF-α treated. (D) The DNA sequences were extracted from ChIP-seq peaks and the core p53, p63, AP-1, NF-κB, cREL consensus motifs were predicted and depicted. Nuclear extracts were isolated from UM-SCC46 cells and the binding assays were performed using a 96-well colorimetric binding assay with 50-70mer oligos containing the 10-20bp motifs synthesized and labeled with biotin (Table S3). Open bars, anti-p63α antibody; dashed bars, anti-TAp73 antibody. Neg: negative control using p21 control oligo without lysate. p21-P: positive control using a p21 oligo containing the known TP53/TP63 site. The data are presented as the mean ± SD calculated from three replicates from one representative experiment.
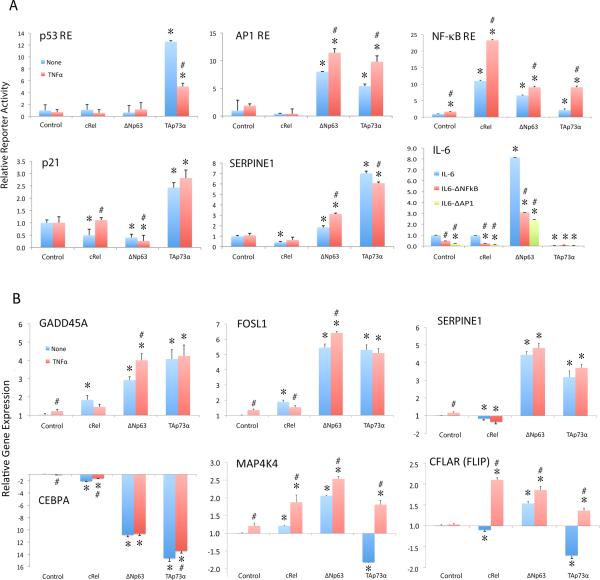
cREL, ΔNp63α, and TAp73 modulate transcriptional regulation and gene expression
Next, we performed functional validation using luciferase reporters containing p53, AP1 and NF-κB/REL specific consensus REs after expression of ΔNp63α, TAp73, cREL or empty control vectors (Figure 4A). Overexpressed TAp73 was a strong inducer of classical TP53 RE reporter activity, which was down-modulated by TNF-α (Figure 4A, upper left). Both ΔNp63α and TAp73 also induced significant AP-1 RE inducing activity, and TNF-α further enhanced it (upper middle). cREL induced strong NF-κB reporter activity that was increased by TNF-α, while ΔNp63α and TAp73 exhibited less pronounced effects (upper right). Then the individual promoters of CDKN1A (p21) containing TP53, p63 REs, SERPINE1 with AP-1 RE, and IL-6 with AP-1 and NF-κB REs were tested for their activities (Figure 4A, lower panels). TAp73 was the strongest inducer of CDKN1A (p21) and SERPINE1. ΔNp63α was the strongest inducer of IL-6, while mutation of either the NF-κB or the AP-1 RE significantly suppressed the activity. Together, these data further confirm the overlapping capability of these TFs to modulate p53, AP1 and NF-κB/REL transcriptional activities.
Figure 4. cREL, ΔNp63α, and TAp73 modulate transcriptional regulation and differential gene expression.
(A) UM-SCC46 cells were transfected with luciferase reporter plasmids containing specific TP53, AP-1, or NF-κB REs (upper panels), or the promoter sequences of CDKN1A (p21) (TP53, TP63 REs), SERPINE1 (AP-1 REs) or IL-6 (AP-1 and NF-κB REs; lower panels). Overexpression of cREL, ΔNp63α, or TAp73α was induced by TNF-α treatment (20 ng/ml) for 48 h. IL-6 binding site-specific point mutant promoter constructs included the deletion mutation of NF-κB or AP-1 binding sites without TNF-α (bottom right panel). The relative reporter activity was normalized to the corresponding β-gal activity and/or compared with the control vectors. Blue, untreated; red, TNF-α treated, except in IL-6 reporter assay (bottom right panel). In IL-6 reporter assay without TNF-α treatment, blue: reporter with full length IL-6 promoter; red: IL-6 promoter with the NF-κB binding motif deleted; green: IL-6 promoter with the AP-1 binding motif deleted. (B) The newly identified target genes were validated by q-RT-PCR 48 h after cREL, ΔNp63α, or TAp73α overexpression under TNF-α treatment (20 ng/ml). Blue, untreated; red, TNF-α treated. The data are presented as the mean ± SD of three replicates from one representative experiment. Statistical significance was calculated using a two-tailed Student's T-Test, p<0.05. * indicates the statistical significance when comparing the conditions with overexpressed plasmids versus control plasmid. # indicates the statistical significance when comparing untreated versus TNF-α treated condition.
We examined how TNF-α and three TFs modulated expression of 6 candidate genes identified through ChIP-seq and confirmed by microarray (Figure 5 below and Figure S6, S7). ΔNp63α and TAp73 strongly induced GADD45A, a known TP53 target (13), as well as AP-1 subunit FOSL1 and SERPINE1 (Figure 4B, upper panels). CEBPA was strongly repressed by ΔNp63α and TAp73 (Figure 4B, lower left panel). The modulation of MAP4K4 and CFLAR (FLIP) by the three TFs was distinct, in that cREL plus TNF-α, ΔNp63α alone, and ΔNp63α plus TNF-α significantly induced gene expression. Collectively, TNF-α-modulated cREL, ΔNp63α, and TAp73 can differentially regulate a functionally diverse gene repertoire.
Figure 5. TNF-α modulates expression of a global gene repertoire bound by cREL, p63α, and TAp73.
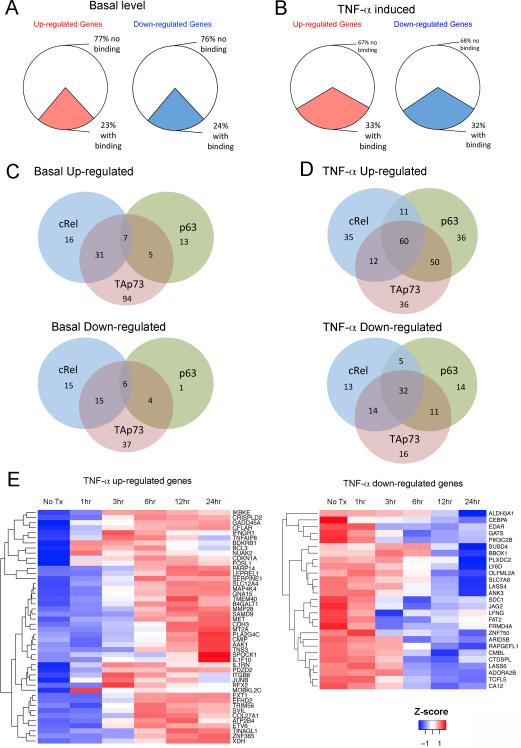
RNA was isolated from cells treated with TNF-α for 1, 3, 6, 12, and 24 h and the differential gene expression was examined by an Illumina bead-based array. (A) Differential expression of up- and down-regulated genes in UM-SCC46 cells under basal level conditions when compared with normal human oral keratinocyte cells, and (B) altered gene expression of UM-SCC46 cells upon TNF-α treatment. The colored sections represent the percentages of differentially expressed genes with binding activities for cREL, p63, or TAp73. (C) Gene numbers with individual and intersecting binding activities among the three TFs at the basal level, or (D) after TNF-α treatment. Top Venn diagrams represent up-regulated genes, and bottom Venn diagrams represent down-regulated genes. (E) Heat maps of hierarchical cluster analysis of 46 up-regulated (left) and 27 down-regulated (right) genes with overlapped TF binding activities induced by TNF-α. Red, increased expression; blue, decreased expression (compared with untreated controls). Color key, Z-score, reflects the relationship of the value of gene expression in a specific sample to the mean of the expression values of the same gene in all the samples. Euclidean distance with complete linkage was used to constitute the gene cluster.
TNF-α globally modulates expression of genes with co-localized binding of TFs
Next we tested if TNF-α globally modulates expression of genes exhibiting co-localized binding of cREL, p63α, and TAp73 (Figure 5). A total of 1,050 genes with ≥1.5 fold differential expression were identified during at least one time point after TNF-α treatment. Without TNF-α, a quarter of altered genes exhibited TF binding, while TNF-α significantly increased up- and down-regulated genes bound by the TFs by ~10%, together accounting for a third of differentially expressed genes (Figure 5A, B, colored sections). TNF-α treatment significantly altered gene numbers with binding activities (Chi-square, up-regulated genes, p=4.3E-07; down-regulated genes, p=5.5E-05). The numbers of differentially expressed genes with individual or overlapping binding activities for the three TFs are presented for basal activities (Figure 5C), or TNF-α modulated activities (Figure 5D). From these, we identified those genes displaying co-binding of all three transcription factors that were up- or down-regulated by TNF-α from Figure 5C, D, and removed the redundant genes. Altogether, TNF-α up-regulated 46 genes and down-regulated 27 genes with overlapping binding of cREL, p63α, and TAp73, which were selected for hierarchical gene clustering and displayed in heatmaps (Figure 5E). Supporting the potential for bidirectional gene modulation, the expression of a selection of target genes up- and down-modulated by TNF-α treatment was validated by qRT-PCR (Figure S7). Using a less stringent difference of 1.3 fold change in gene expression by TNF-α treatment, we detected 84 up-regulated genes and 61 down-regulated genes bound by all three transcription factors (Table S4a and 4b). Using either 1.5 or 1.3-fold criteria, the TNF-α modulated genes bound by the TFs include multiple molecules implicated in inflammation, oncogenesis, and NF-κB and AP-1 mediated signaling.
cREL, ΔNp63, and TAp73 expression and altered gene and protein signatures in HNSCC tissues and ΔNp63α transgenic mouse skin
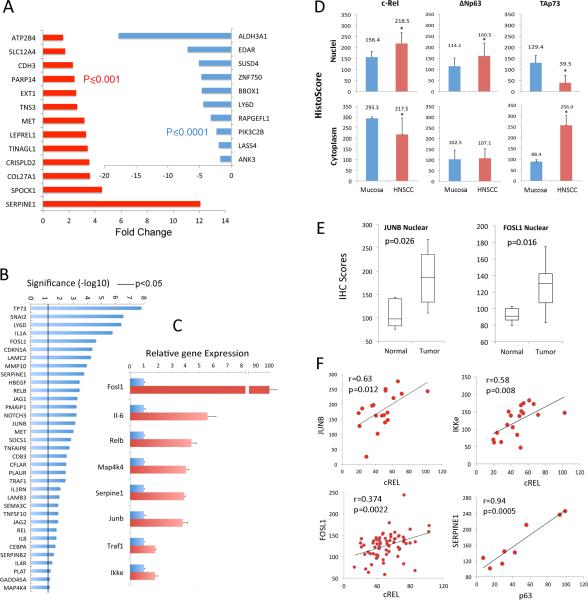
We examined the 46 TNFα-upregulated genes and 27 down-regulated genes co-bound by all 3 TFs from Figure 5E using the HNSCC mRNAseq expression dataset recently published for 279 primary tumor and 16 mucosa specimens from The Cancer Genome Atlas (1). Using these data, we found 23/73 genes (31.5%), including 13 of 46 activated genes, and 10 of 27 repressed genes, showed significant concordance when comparing primary HNSCC tumor to mucosa specimens (Figure 6A; fold change ≥1.5, p<0.05, student t-test with FDR correction <1% cut off). An additional 23 genes (31.5%) were also expressed in the same direction as in cell lines, but below the significance threshold (Table S6). Thus, overall 63% of genes showed consistent changes in direction of expression when comparing primary tumors to mucosa between cell line data and TCGA human tissue data. Consistent with this, an unsupervised cluster analysis and heatmap of the TCGA data for all 73 genes confirms that many of the TNFα inducible and down-regulated genes in cell lines co-cluster among 4 major tumor subsets (Figure S8, clusters a-d). Among the TNF-α inducible genes, several of the most significantly upregulated genes co-cluster predominantly in major clusters a and/or b (SERPINE1, MET, SPOCK1, CRISPLD2, TINAGL1, LEPREL1, TNS3, PARP14, CDH3), which have previously been reported in PubMed to be deregulated in cancer and/or metastasis. MET and SERPINE1 are implicated in cell migration, invasion and metastasis, and SERPINE1 is also a downstream target of MET signaling (14-17).
Figure 6. cREL, ΔNp63, and TAp73 nuclear and cytoplasmic localization and target gene expression in human HNSCC tissues and skin of ΔNp63α transgenic mice.
(A) 46 up-regulated and 27 down-regulated genes from Figure 5D were queried using the mRNA expression dataset from TCGA HNSCC project, which includes 279 tumor and 16 mucosa specimens. Significant up-regulation of 13/46 activated genes (red), and down-regulation of 10/27 repressed genes (blue) are detected in TCGA tumors compared to normal samples (fold change ≥1.5, student t-Test with FDR < 1% multivariate comparison correction cut off). (B) Three publicly available datasets of gene profiling microarrays were investigated, which included a total of 125 (67 metastatic and 58 non-metastatic) HNSCC tissues. Pearson correlation coefficients of gene expression between p63 and potential target genes were calculated, and genes exhibiting statistically significant correlation are presented. The line indicates p<0.05. (C) RNA was isolated from the skins of ΔNp63α transgenic mice (red). qRT-PCR was performed to quantify gene expression levels with a statistically significant increase compared with their age-matched non-transgenic littermates (blue, p<0.05). The data were calculated from triplicates of one representative experiment and presented as the mean ± SD. (D) Human HNSCC and normal mucosa frozen sections were stained for cREL, ΔNp63, and TAp73, and intensities within nuclei or cytoplasm in three 200X fields per slide were acquired and quantified using an Aperio Scanscope and Cell Quantification Software (Vista, CA, USA), and presented as mean histoscores ± SD for 8-13 samples. * p<0.05, HNSCC vs normal mucosa by t-tests. (E) Immunohistochemistry comparing transcription factors JUNB and FOSL1 nuclear staining in human HNSCC tissue array. Images were acquired using an Aperio Scanscope at 200X magnification, and staining intensity was quantified using Aperio Cell Quantification Software. Tumor protein expression of evaluable specimens for JUNB and FOSL1 (n=66) and mucosa (n=11) samples. Student t-test, p<0.05 (F) Associations in expression levels between the transcription factors nuclear cREL with targets nuclear JUNB and IKKε (stage III tumors), nuclear cREL with nuclear FOSL1 (in all tumor stages), and nuclear TP63 with nuclear SERPINE1/PAI1 (metastatic tumors). A non-directional test for the significance of the Pearson Product-Moment Correlation Coefficient with the computed histoscores for each protein was used.
As additional TP63 bound and modulated genes identified in our dataset are implicated in cancer, inflammation and metastasis, we also examined 48 TP63 targets, including 31 curated from the literature, and 17 from our prior (8) or present study of TNF-α modulated genes (Figure S7), as described in supplemental Methods. An expression-based method was used to cluster a group of genes with common profiling patterns and TP63 transcription factor motifs, to computationally infer co-expressed networks, using three independent publicly available microarray data sets containing 125 HNSCC samples that include 58 non-metastatic and 67 metastatic HNSCC tissues (18-20). Among these genes, a statistically significant correlation was established between expression of TP63 and 35 genes (71.4%, Pearson correlation coefficient p<0.05, Figure 6B). This analysis provided evidence for differential expression of multiple TP63-linked genes that are implicated cancer cell phenotype (e.g. JUNB/FOSL1, CDKN1A, JAG2/NOTCH2), invasion and metastasis (e.g., SERPINE1, MET, SNAI2, MMP10), and inflammation (e.g. IL1A, IL8, RELB). Several of these TP63 target genes also co-modulated by TNF-α co-cluster together in major subsets of TCGA HNSCC tumors (Figure S8, e.g., FOSL1, CDKN1A (p21), SERPINE1, MET, TNFAIP8).
We further compared ΔNp63 bound and up-regulated genes in a conditional K5-ΔNp63α transgenic mouse model, where increased tnf-α expression and a similar nuclear redistribution of cREL/ΔNp63 and TAp73 was observed (21, 22). Array profiling detected increased expression of a panel of up-regulated genes, which were validated by qRT-PCR. These genes included AP-1 subunits Fosl1 and Junb, NF-κB pathway related genes Traf1, Ikkε and Relb, and their downstream genes, Il-6, and Serpine1 (Figure 6C).
We next validated if proteins of TNF-α co-modulated TFs and four target genes identified in multiple platforms above, display corresponding alterations in human HNSCC tumor and tissue arrays. The nuclear-cytoplasmic distribution of cREL, ΔNp63, and TAp73 in human specimens was quantified by immunohistochemistry score. We observed increased cREL and ΔNp63, and decreased TAp73 nuclear distribution, with a reciprocal decrease in cytoplasmic cREL and increased TAp73 cytoplasmic staining in HNSCC (Figure 6D, S9), consistent with TNF-α modulated nuclear redistribution and DNA binding of these TFs in HNSCC in vitro in Figure 1A, B, and prior studies (6) (8). Next, we examined the protein expression of target genes in human HNSCC tissue array, which contains 61 primary tumors of stage I-IV at different anatomic locations, 8 lymph node metastatic tumors and 11 normal mucosa tissues (Supplemental Methods). Notably, key AP-1 family transcription factor targets JUNB and FOSL1, showed significantly elevated nuclear staining in HNSCC tumors relative to mucosa (Figure 6E), consistent with altered AP-1 promoter binding and expression of JUNB and FOSL1 observed above (Figures 3-5). Further, the nuclear staining of JUNB and FOSL1 was significantly correlated when compared with cREL (Figure 6F). Increase in staining for two additional targets, IKKε and SERPINE1, were also significantly correlated with cREL and p63 staining, respectively. Together, these data from human and mouse tissues support our hypothesis for the altered cellular distribution and functional co-modulation of cREL, ΔNp63α, TAp73, with AP-1 subunits JUNB/FOSL1, and a subset of other target genes detected in different in vitro and in vivo systems.
CircleMap and Pearson correlation analyses support TP53/p63/p73 modulated gene signatures in squamous cancers
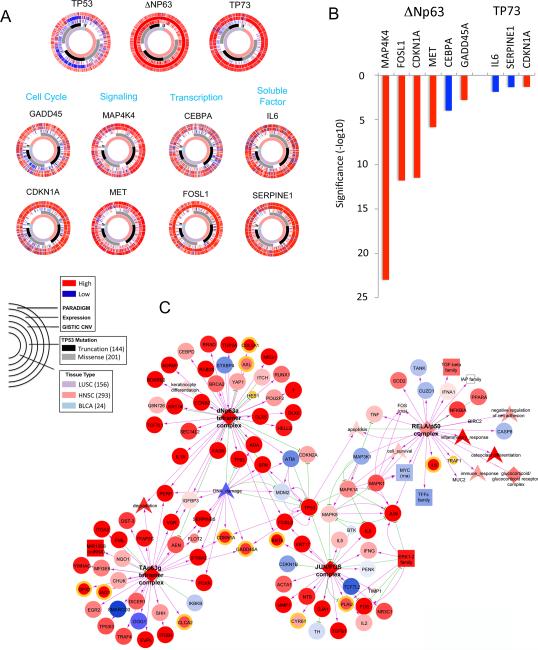
We next examined how our current findings may relate to large datasets and analyses from the recently published TCGA PanCancer 12 project, which identified inflammatory as well as a “TP53-like” compensatory gene expression activity pattern associated with increased ΔNp63 and TP73 activities, and frequent genomic inactivation of TP53 in the squamous-like tumors (2). These squamous cancers included 293 HNSCC, 156 lung SCC and 24 bladder SCC, which were analyzed for TP53/63/73 status and expression and function of 8 genes identified among their known and new targets in the present study. Figure 7A shows CircleMaps, which enable a multi-dimensional comparison of the genomic alterations, expression, and inferred pathway activities for TP53, ΔNp63, TP73, and these target genes, using the annotated PARADIGM bioinformatics analytic platform (23, 24). The TP53 CircleMap shows that the majority of SCC tissues contain TP53 mutations (second circle), and/or copy number loss (third circle) with corresponding decrease in TP53 expression (fourth circle). However, they retain weak inferred PARADIGM activity for TP53 (the outer circle), which cannot be explained by inactivated TP53, but could be compensated by overlapping function of other family members. Consistent with this, the TP63 CircleMap exhibited high copy number gain, expression, and PARADIGM gene signatures, supporting a role for copy number gain and expression in TP63 PARADIGM gene activity. Similarly, TP73 overexpression and PARADIGM activity are also congruent. We further analyzed eight targets identified in this study, including genes involved in cell cycle (CDKN1A (p21), GADD45), signal receptor and kinase (MET, MAP4K4), transcription factors (AP-1/FOSL1, CEBPA), and secreted factors (SERPINE1, IL6). The expression and PARADIGM activity of ΔNp63, TP73, and these target genes are higher across SCC tumors compared with other cancer types. By Pearson correlation analysis (Figure 7B), ΔNp63 exhibited significant positive correlation with 5 genes bound and inducible by TNF-α, including MAP4K4, FOSL1, CDKN1A (p21), and GADD45, whose activation ΔNp63 and TNF-α promotes, and a negative correlation with CEBPA, repressed by ΔNp63 and TNF-α (Figures 4B, 5E). TP73 exhibited a negative association with IL6, which TAp73 suppresses, and a positive correlation with CDKN1A (p21), which it activates (Figure 4A). SERPINE1 showed only a weak negative association with TP73, which was observed with TNF-α treatment by reporter but not RT-PCR (Figure 4A, B).
Figure 7. Squamous cancer signatures by CircleMaps and PARADIGM SuperPathway analyses.
(A) CircleMap of PARADIGM-Shift differences associated with SCC tissue origins and TP53 mutation status were created for TP53, TP63, TP73 and target genes identified in this study. Samples were ordered first by tissue origin of SCC (innermost ring), then by TP53 mutation status (second ring), GISTIC score (indicating CNV), mRNA expression level, and finally by PARADIGM activities (outer ring). The Red-blue color intensity reflects magnitude of CNV, expression and PARADIGM activities (red: high, blue: low). TP53 mutation is highlighted (black: truncation, gray: missense). Samples were restricted to the C2-Squamous-like cluster-of-cluster-assignments (COCA), including 156 lung SSC, 293 HNSCC, and 24 bladder SCC samples. Each plot illustrates multiple data types across many samples for a given gene. (B) The PARADIGM activity of ΔNp63 or TP73, and expression of eight target genes are higher across SCC tumors compared with other cancer types as shown in (A). Pearson correlation coefficients between ΔNp63 or TP73 PARADIGM activity and eight target genes presented in (A) were calculated, and the significance of p value was presented in y axis. The PARADIGM activity of ΔNp63 exhibited significant positive correlations with expression of five target genes bound and inducible by TNF-α, and a negative correlation with CEBPA expression. The PARADIGM activity of TP73 exhibited negative associations with with IL-6 and SERPINE1, and a positive association with CDKN1A (p21). (C) PARADIGM SuperPathway subnetwork defining C2-Squamous-like Pan-Cancer 12 integrative subtype. Zoom-in view of network neighborhood surrounding the ΔNp63α tetramer, TAp63γ tetramer, RelA/p50 and Jun/Fos complexes. Color of the nodes reflects activation (red) or repression (blue) within the squamous subtype when compared with the mean of other tumor types. Edge color denotes interaction type: inhibitory (green) and activating (yellow). Node shape reflects feature type: protein (circle), complex (diamond), family or miRNA or RNA (square), abstract concepts (arrowhead). The target genes with cRel, p63 and TAp73 binding identified in this study are highlighted with different colored outer rings as showing in supplemental Figure S10.
PARADIGM SuperPathway analyses connect p63, NF-κB, and AP-1 subnetworks in squamous cancers
Our current and prior experimental data indicate that ΔNp63 and REL NF-κB family members bind and modulate inflammation and survival genes (Figures 3-5) (6, 8, 25). Analyses of TCGA PanCancer 12 datasets also revealed a strong TP63 gene signature associated with altered immune and inflammatory gene signatures in squamous cancers (2). However, the network(s) linking TP63 and NF-κB family members and target genes has not been well elucidated. We thus searched within the interconnected network of differentially activated PARADIGM pathways, linked through regulatory hubs with >15 downstream targets, derived from a larger interconnected network displaying differentially activated proteins inferred between the C2-Squamous-like subtype and other tumors, as detailed in Supplemental Methods, Table S5, Figure S10 (26). Interestingly, we observed increased activation of the network neighborhood surrounding TP63, connected with NF-κB complex (RELA/p50) through AP-1 (JUN/FOS) complexes within the squamous cancers (Figure 7C). The activation of these networks and targets are consistent with our evidence that TNF-α orchestrates cREL/ΔNp63 binding and induction of AP-1 subunits, reprograming of cREL/p73 binding via AP-1 binding consensus sites, and their related gene signatures in vitro and in vivo (Figure 2-6). Several genes shown on the network identified in this study have been independently validated previously, such as TP63 regulated CDKN1A, GADD45A, IL1A, CHUK, YAP1, IGFBP3, KRT14; as well as NF-κB and AP-1 related genes IL-6 and IL-8, and MAPK1/ERK/JUN/FOSand pathways. Most of these related genes in squamous cancers are activated (red) when compared with other cancer types. Furthermore, a more extended network connecting ΔNp63α, TP53 and AP-1 transcription complexes (among other regulatory hubs), links a larger number of target genes identified in this study by existing experimental data (Figure S10A). Although not all target genes identified from our current study are linked by this expanded network (Figure S10A-B), they are significantly enriched among the 4213 differentially activated PARADIGM proteins inferred between squamous and other cancers (hypergeometric test p = 6E-9 for basal targets, and p = 1.22E-10 for TNF-α responsive targets) Table S5.
Discussion
Here we provide evidence for a novel and dynamic genome-wide TF binding paradigm, whereby the inflammatory cytokine TNF-α modulated binding activities of cREL, ΔNp63α, and TAp73 genome-wide, intersecting a diverse repertoire of genes implicated in the malignant phenotype. This modulation was linked to attenuated expression of specific TP53 tumor suppressor targets and promotion of REL/NF-κB and AP-1 oncogenic and inflammatory gene programs (Figure 8). Our study provides a link between global TP63/TP73, AP-1 and NF-κB mediated survival and inflammatory gene programs, which helps explain the finding of unique TP63/TP73 gene signatures compensating for mutant TP53 identified recently in squamous cancers from TCGA HNSCC and PanCancer projects (1, 2). Furthermore, our current data are consistent with previous mechanistic studies demonstrating how TNF-α induced changes alter the DNA binding of c-REL, ΔNp63 and TAp73 family members (6, 8). Our studies showed that these transcription factors regulate genes involved in several aspects of cell fate including cell proliferation, stemness, survival/apoptosis, and migration, as well as, epithelial cell growth, inflammation and immune responses in murine keratinocyte and ΔNp63 transgenic mouse models (27, 28).
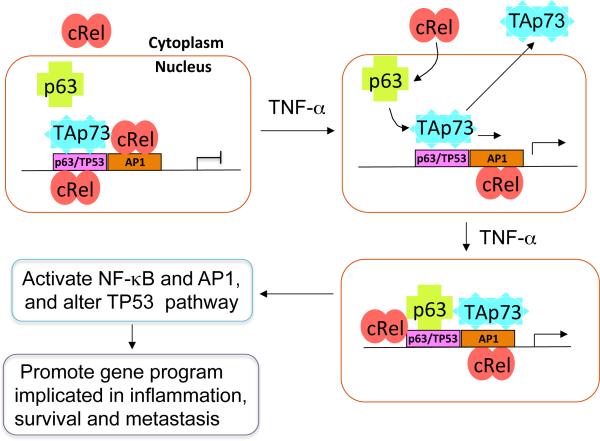
Figure 8. Mechanistic model illustrating effects of TNF-α modulation of cREL, ΔNp63α and TAp73 chromatin occupancy and reprogramming of TAp73 from TP53 to AP-1 sites to promote inflammatory and cancer gene programs.
Most head and neck and solid cancers have high frequencies of TP53 mutation, where TAp73 can serve as a potential tumor suppressor. In this model, without TNF-α, TAp73 predominantly occupies TP53 or p63 binding sites, while NF-κB family member cREL either resides in the cytoplasm or in the nucleus with binding on AP-1 sites, and ΔNp63α is found in the nucleus unbound to DNA (upper left). TNF-α, a major inflammatory cytokine produced in the tumor microenvironment, can promote cREL nuclear translocation, and complexes with ΔNp63α, to occupy TP53/p63 binding sites. TNF-α also induces nuclear displacement or reprogramming of TAp73 to bind neighboring AP-1 sites (upper and bottom right). These dynamic alterations diminish TAp73 tumor suppressor activity, while enhancing cREL/ΔNp63α and TAp73 mediated inflammatory and cancer gene programs (lower left). This model helps explains how TNF-α modulates NF-κB and AP-1 signaling while altering tumor suppressor activity, to promote gene programs implicated in inflammation, survival, and metastasis.
After cross comparison of 12 different cancer types, TCGA PanCancer 12 project uncovered several unique signatures that differentiate squamous cancers from other cancer types, such as high rates of TP53 mutation and TP63 amplification (2). In depth genetic analyses of the gene signatures of squamous cancers, revealed attenuated TP53 and increased expression of compensatory TP63/TP73 gene signatures. These were not strictly linked with the relatively infrequent loss of heterozygosity (LOH) or TP53-truncating loss-of-function mutations, but with TP63 amplification and TP63/73 gene expression by PARADIGM-SHIFT analysis (23).
Interestingly, our previous experimental results supported a relationship between attenuated TP53 target gene expression and partial compensatory function of TP53 family members TP63 and/or TP73 in SCC (6, 8). HNSCC with mtTP53 displayed higher ΔNp63 and TAp73, and intermediate “compensatory” basal expression of several TP53/TP63/TP73-regulated apoptotic genes. Expression of these genes was further attenuated by TNF-α, DNA binding interaction of cREL with ΔNp63, and displacement of TAp73 (6). In the present study, ChIPseq, ChIP PCR, and binding analyses provide evidence for genome-wide reprogramming of TAp73 binding to AP-1RE containing promoters, and varying regulation of target genes of TP53 as well as AP-1 and REL/NF-κB. Such TP63/TP73 compensatory TP53 function has been associated with higher sensitivity to cisplatin chemotherapy in both HNSCC (29, 30) and in BRCA1-related triple-negative breast cancer (31), but together with promotion of opposing inflammatory and oncogenic gene programs, is apparently insufficient to prevent development of these cancers.
TP53 is highly mutated in HNSCC, ~80-90% in HPV negative tumors, as evident by recent TCGA data (1). As we previously reported in the manuscript by Lu et al (6), the increased expression of TAp73 is predominantly observed in mtTP53 cell lines, and reduced TAp73 occupancy is associated with replacement by cREL, not mtTP53 in response to TNF-α. The mtTP53 does not co-IP with the cREL/ΔNp63 complex, and is not recruited to CDKN1A (p21), NOXA or PUMA promoters by ChIP assay following TNF-α treatment. In addition, dependence on cREL and ΔNp63 for TAp73 binding to p21 promoter was shown, whereby cREL inversely modulated TAp73 binding. Similar results for TNF-α treatment or cREL modulation of TAp73 were obtained in three UM-SCC lines (6). Furthermore, our prior data are also consistent with other's evidence for a strong protein interaction is between p63 and p73, but not with TP53, because TP53 is lacking the SAM domain at the C-terminal, which mediates heterotetramerization between these family members (32, 33).
Our prior and current findings help further clarify the apparent function of TP63/TP73 in compensating for TP53 and promoting oncogene expression. First, such compensatory activity is associated with enhanced expression of typical TP53 target genes, such as CDKN1A (p21) and GADDA45, which control cell cycle (9, 30), as observed in our cell line model (Figure 3-5), broader microarray studies of HNSCC tissues (Figure 6B), and PanCancer 12 TCGA project (Figure 7). However, TNF-α, cREL, ΔNp63 modulate TAp73, attenuating its TP53 compensatory function, while also enhancing a much broader oncogene program implicated in cancer promotion (6, 8). This includes growth factor mediated signaling, inflammatory cytokines, prosurvival transcription factors such as AP-1 and RELs, and anti-apoptotic genes (e.g. CFLAR) (Figure 4-6), which can potentially counter and limit compensatory TP53 tumor suppressor function. Secondly, as shown by our and other laboratories, as well as in PanCancer 12 project, ΔNp63α is the dominant isoform expressed in squamous cancers (2). We and others have shown the strong oncogenic activity of ΔNp63α in squamous cancers, including promoting tumor cell proliferation, migration, colony formation, and inflammatory responses (8). Thirdly, as our and other laboratories previously showed and supported by HNSCC and Pancancer 12 TCGA project, a subset of squamous cancers exhibited inflammatory signatures concurrent with TP63/TP73 compensatory activity (2). Such inflammatory signaling mediated by TNF-α could reprogram TP73 tumor suppressor to oncogenic activity, through binding to AP-1 sites, as supported by ChIP seq, ChIP RT-PCR, and binding assays (Figure 1-4) in this study. This novel finding is also supported by the PanCancer 12 data indicating that inflammatory gene signatures linked by cytokine TNF-α and NF-κB transcription factors are connected to TP63 gene signatures through JUN/FOS mediated pathway and network (Figure 7C and Figure S10).
The novel finding from this study of physical interaction among these TFs with genomic DNA is supported, among other evidence, by a narrow binding peak within ~200 bp surrounding TP53/p63 consensus sites, and a broader peak of ~400-500 bp within which nearby AP-1 motifs are distributed. Interestingly, a ~500 bp distance between p73 binding and AP-1 sites was independently identified in ChIP-seq experiments in osteosarcoma cells, where anti-TAp73α and c-Jun antibodies detected co-binding by ChIP-re-ChIP assay (34). In addition, Pietenpol and colleagues identified overlapping AP-1 and p73 binding sites in ChIP-seq experiments, and confirmed c-Jun binding within 500 bp of p73 binding sites in rhabdomyosarcoma cells (35). Furthermore, we identified palindromic binding sequences for TP53/p63 and AP-1 (Figure 2), which are similar to the sequences of TP53/TP63/TP73 and nearby AP-1 binding motifs observed in previous studies (34-39). Our study reveals that in the absence of TNF-α, TAp73 was mainly bound to TP53/TP63 consensus motifs, consistent with that found for growth arrest and pro-apoptotic genes in multiple HNSCC lines including UM-SCC46 (6), where TAp73 exhibited a partial compensatory function for mutated TP53 (9). However, previous studies have not observed how TNF-α induced dynamic alterations of these TF binding activities. After TNF-α treatment, ΔNp63α bound at TP53/TP63 sites previously occupied by TAp73, while TAp73 bound to AP-1 sites (Figure 2B and D). These dynamic alterations of binding activities reversed TAp73 tumor suppressor activity, while increased cREL/ΔNp63α and TAp73 modulated expression of an inflammatory and cancer related gene program.
Our previous study demonstrated that TNF-α modulates reciprocal and mutually exclusive protein-protein interactions between endogenous nuclear ΔNp63α/TAp73 and cREL/ΔNp63α complexes (6, 8). Our prior ChIP studies did not resolve if cREL or TAp73 can individually bind the same minimal TP53RE, or if TP73 can bind to AP-1 REs independent of ΔNp63α or other cofactors. Our sedimentation velocity centrifugation experiments revealed that both cREL and TP73 bind these DNA REs and can form dimers and/or tetramers upon DNA binding. These results provide biophysical support to the model of co-localized binding of cREL and TP73 suggested by the ChIP-seq and bioinformatics analyses. Collectively, the different experimental approaches provide mechanistic details of how TFs can occupy binding sites at the same as well as very close promoter sequences.
Our reporter assays provide evidence for functional overlap in transcriptional activities in addition to binding, which demonstrate differing effects of the TFs and TNF-α on modulation of transcription (Figure 4A). The reporters contained TP53, AP-1 or NF-κB REs. ΔNp63α exhibited minimal effects on TP53, decreased CDKN1A (p21), and significantly induced IL-6 expression. This induction was significantly reduced by mutation of the NF-κB and AP-1 REs, which is consistent with our and others’ publications (8, 40). Significantly, TAp73α strongly induced CDKN1A (p21) and SERPINE1 reporter activity, consistent with its compensatory function for mutant TP53 (9, 31, 35, 41, 42), as well as induced specific AP-1 promoter activity (43, 44). In addition, reporters with specific TP53 or AP-1 REs demonstrated the ability of TNF-α to repress TAp73-induced TP53 RE, or enhance TAp73-mediated AP-1 RE transactivation, supporting a reciprocal role for TNF-α in modulating these response elements.
TNF-α, an inflammatory cytokine produced in the tumor microenvironment of many cancers, was previously shown to activate both NF-κB and AP-1 (45, 46), the latter through MEK/ERK signaling. These findings are also consistent with the Pan-TCGA data, where the Squamous-like cancer type exhibited elevated activity of MAPK signaling pathways (2). Here we propose an expanded model, wherein TNF-α modulated TAp73 also appears capable of reprogrammed binding of AP1-containing REs, while inducing co-expression of AP-1 family member FOSL1/Fra1 and reporters containing AP-1 REs. In addition, FOSL1 and JUNB expression were significantly associated with p63 and p73 expression in human HNSCC cells and ΔNp63α transgenic mice. This is consistent with our finding that FOSL/Fra1, cJUN and JUNB are the major AP-1 protein family members expressed and bound to AP-1 sites, and which mediate cell proliferation and migration in HNSCC cell lines (47, 48). Our data are supported by others’ observations that p73 and AP-1 family members can enhance each other's binding and transcription activities (35, 43, 44). Our present study adds new evidence that TNF-α is a key factor that induces binding of TAp73 to AP-1 sites and increases AP-1 family member transcription, and suggests a new model whereby TNF-α can co-modulate AP-1 and reprogram TAp73 to cooperatively activate AP-1 target genes.
Thus, our and others’ studies help explain why when TP53 is mutated or defective in function, the TAp73 compensatory function for TP53 is repressed. First, overexpressed ΔNp63α is able to form a complex with TAp73 to block its tumor suppressor function (6, 8, 31). Secondly, TNF-α promotes displacement of TAp73 from pro-apoptotic genes and reprograms its binding and activation of AP-1 sites (Figure 2A, 2D, 2E, 3C). Thirdly, nuclear TAp73 binding is displaced from TP53/p63 sites by the cREL/ΔNp63α complex as supported by experimental validations (Figure 2, 3) (6). Instead, TAp73 function is converted to potentiate AP-1 activity and promote survival and inflammatory gene expression (Figure 5), as supported by other studies (34, 43). Furthermore, in an independent study of a group of genes predicted to be controlled by cJUN/NF-κB (49), 20% of genes overlapped with the genes differentially regulated by TAp73 (34).
Together, our current and recent studies (6, 8, 48) reveal a complex, genome-wide transcriptional regulatory mechanism whereby TP53, NF-κB and AP-1 family members interact to promote a broad signaling network favoring tumor survival and inflammation. Our current study helps explain the global mechanisms underlying the newly discovered deregulated TP53/p63 network and inflammation, presented recently by TCGA pan-cancer project (2), and a study in neuroblastoma (50). The transcriptional regulatory mechanisms mediated by TP53, NF-κB and AP-1 family members genome-wide are far more complex and nuanced than originally anticipated, and understanding the mechanisms will shed light on more effective means for targeting of cancer therapeutics.
Materials and Methods
ChIP and next-generation sequencing (ChIP-seq)
DNA fragments bound by antibodies were prepared following the ChIP protocol using sheared DNA (~100-400 bp). The DNA samples were sequenced using a next-generation GAIIx sequencer from Illumina following the manufacturer's protocol (San Diego, CA). Sequence reads were mapped to the UCSC human genome Hg18 assembly using the Eland algorithm (Illumina), permitting up to 2 mismatches and no gaps. Only unique mapped reads were used in the binding peak calling analysis. To identify binding peaks, we employed MACS (51), in which a dynamic Poisson distribution model was used to detect the statistically significant binding peaks in ChIP samples compared to DNA input controls. MACS is a widely used algorithm for detection of protein-DNA binding sites when analyzing ChIP-seq data. This method is specifically designed to detect the promoter transcription factor binding sites, which are typically located within a few hundred DNA base pairs. In our study, the default setting was selected, which is commonly utilized and suitable to our datasets. Detected peak regions were visualized mainly by using the CisGenome browser (52) together with gene structure, DNA sequences and conservation scores. We compared the fraction of peaks residing in various genomic features with the corresponding genome background using the CEAS program (53). A binomial test was used in this method to obtain the p value of each comparison. Gibbs Sampler (54) was used for the de novo motif search for binding site sequences and MEME-ChIP suit (55) was used to process the found motifs and compare the results to known motifs in database. Peak location, distance to transcriptional start sites (TSSs), peak intersection, motif location and intersection analysis were performed using customized python/R scripts. ChIP-seq experiments were repeated in UM-SCC46 cell lines, and a representative experiment was analyzed and presented in Supplemental Table S1. Minimal background peaks were observed in the samples of input DNA or DNA from isotype control antibodies. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Supplementary Material
Acknowledgments
The authors thank Dr. Fan Yang (NCI/NIH), Dr. Kairong Cui (NHLBI/NIH), Dr. Bingmei Zhu (NIDDK/NIH), Mr. Jeffery Burnett (NIDCD/NIH), Ms. Jamie Coupar (NIDCD/NIH), Ms. Guanmei Liang (Thomas Jefferson High School for Science and Technology, Alexandria, Virginia), and Mr. Eric Nicolson (Ithaca High School, Ithaca, New York NY) for their technical assistance and suggestions. The authors express appreciation to Drs. James W. Rocco and Leif W. Ellisen (Harvard University) for providing ΔNp63 and TAp63 expression vectors, Dr. Thomas Gilmore (Boston University) for cRel expression plasmids, Professor Gerry Melino (University of Leicester) for TAp73α expression plasmids, Dr. J. Silvio Gutkind (NIDCR/NIH) for IL-6 promoter reporter plasmids, Dr. Gourisankar Ghosh (UCSD) for the cRel expression plasmid, and Drs. Michal Karin (University of California, San Diego), Cheng-Ming Chiang (University of Texas, Southwestern) and Xuan Liu (University of California, Riverside) for critique of and helpful suggestions for the manuscript.
HL, HS, XY, AM, MJ, YB, CVW, and ZC are supported by intramural projects ZIA-DC-000073, ZIA-DC-000074, and RAR and SS are supported by a grant from NIH (R01AR049238). JMS acknowledges support from NCI (R01-CA180778 and U24-CA143858), Stand Up to Cancer, Prostate Cancer Foundation, and the Movember Foundation.
Footnotes
Supplemental information
For information on cell lines, reagents and antibodies, chromatin immunoprecipitation (ChIP) analyses, quantitative PCR and RT-PCR, plasmid and siRNA transfection, reporter gene assays, oligo-based binding assays, protein expression and purification, Analytical ultracentrifugation, microarray data collection and analysis, immunohistochemistry, the ΔNp63α transgenic mouse model, CircleMap, and PARADIGM SuperPathway subnetwork, sequence data storage, see the Supplemental Materials and Methods and related references.
References
- 1.Cancer Genome Atlas N Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12(4):303–12. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 2010;17(6):901–11. doi: 10.1038/cdd.2010.35. [DOI] [PubMed] [Google Scholar]
- 5.Leslie M. Brothers in arms against cancer. Science. 2011;331(6024):1551–2. doi: 10.1126/science.331.6024.1551. [DOI] [PubMed] [Google Scholar]
- 6.Lu H, Yang X, Duggal P, Allen CT, Yan B, Cohen J, et al. TNF-alpha promotes c-REL/DeltaNp63alpha interaction and TAp73 dissociation from key genes that mediate growth arrest and apoptosis in head and neck cancer. Cancer Res. 2011;71(21):6867–77. doi: 10.1158/0008-5472.CAN-11-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, et al. Biological functions of p53 isoforms through evolution: lessons from animal and cellular models. Cell Death Differ. 2011;18(12):1815–24. doi: 10.1038/cdd.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71(10):3688–700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26(36):5169–83. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 10.Perez CA, Pietenpol JA. Transcriptional programs regulated by p63 in normal epithelium and tumors. Cell Cycle. 2007;6(3):246–54. doi: 10.4161/cc.6.3.3801. [DOI] [PubMed] [Google Scholar]
- 11.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21(3):632–41. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunting K, Rao S, Hardy K, Woltring D, Denyer GS, Wang J, et al. Genome-wide analysis of gene expression in T cells to identify targets of the NF-kappa B transcription factor c-Rel. J Immunol. 2007;178(11):7097–109. doi: 10.4049/jimmunol.178.11.7097. [DOI] [PubMed] [Google Scholar]
- 13.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71(4):587–97. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 14.Dong G, Chen Z, Li ZY, Yeh NT, Bancroft CC, Van Waes C. Hepatocyte growth factor/scatter factor-induced activation of MEK and PI3K signal pathways contributes to expression of proangiogenic cytokines interleukin-8 and vascular endothelial growth factor in head and neck squamous cell carcinoma. Cancer Res. 2001;61(15):5911–8. [PubMed] [Google Scholar]
- 15.Dong G, Lee TL, Yeh NT, Geoghegan J, Van Waes C, Chen Z. Metastatic squamous cell carcinoma cells that overexpress c-Met exhibit enhanced angiogenesis factor expression, scattering and metastasis in response to hepatocyte growth factor. Oncogene. 2004;23(37):6199–208. doi: 10.1038/sj.onc.1207851. [DOI] [PubMed] [Google Scholar]
- 16.Andreasen PA. PAI-1 - a potential therapeutic target in cancer. Current drug targets. 2007;8(9):1030–41. doi: 10.2174/138945007781662346. [DOI] [PubMed] [Google Scholar]
- 17.Boccaccio C, Sabatino G, Medico E, Girolami F, Follenzi A, Reato G, et al. The MET oncogene drives a genetic programme linking cancer to haemostasis. Nature. 2005;434(7031):396–400. doi: 10.1038/nature03357. [DOI] [PubMed] [Google Scholar]
- 18.Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23(14):2484–98. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 19.O'Donnell RK, Kupferman M, Wei SJ, Singhal S, Weber R, O'Malley B, et al. Gene expression signature predicts lymphatic metastasis in squamous cell carcinoma of the oral cavity. Oncogene. 2005;24(7):1244–51. doi: 10.1038/sj.onc.1208285. [DOI] [PubMed] [Google Scholar]
- 20.Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, et al. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27(51):6607–22. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 21.Romano RA, Ortt K, Birkaya B, Smalley K, Sinha S. An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PLoS One. 2009;4(5):e5623. doi: 10.1371/journal.pone.0005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano RA, Sinha S. Dynamic life of a skin keratinocyte: an intimate tryst with the master regulator p63. Indian J Exp Biol. 2011;49(10):721–31. [PubMed] [Google Scholar]
- 23.Ng S, Collisson EA, Sokolov A, Goldstein T, Gonzalez-Perez A, Lopez-Bigas N, et al. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics. 2012;28(18):i640–i6. doi: 10.1093/bioinformatics/bts402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26(12):i237–45. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sethi G, Shanmugam MK, Ramachandran L, Kumar AP, Tergaonkar V. Multifaceted link between cancer and inflammation. Biosci Rep. 2012;32(1):1–15. doi: 10.1042/BSR20100136. [DOI] [PubMed] [Google Scholar]
- 26.Wong CK, Vaske CJ, Ng S, Sanborn JZ, Benz SC, Haussler D, et al. The UCSC Interaction Browser: multidimensional data views in pathway context. Nucleic Acids Res. 2013;41(Web Server issue):W218–24. doi: 10.1093/nar/gkt473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du J, Romano RA, Si H, Mattox A, Bian Y, Yang X, et al. Epidermal overexpression of transgenic DeltaNp63 promotes type 2 immune and myeloid inflammatory responses and hyperplasia via NF-kappaB activation. J Pathol. 2014;232(3):356–68. doi: 10.1002/path.4302. [DOI] [PubMed] [Google Scholar]
- 28.King KE, Ponnamperuma RM, Allen C, Lu H, Duggal P, Chen Z, et al. The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 2008;68(13):5122–31. doi: 10.1158/0008-5472.CAN-07-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee A, Chang X, Sen T, Ravi R, Bedi A, Sidransky D. Regulation of p53 family member isoform DeltaNp63alpha by the nuclear factor-kappaB targeting kinase IkappaB kinase beta. Cancer Res. 2010;70(4):1419–29. doi: 10.1158/0008-5472.CAN-09-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9(1):45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117(5):1370–80. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natan E, Joerger AC. Structure and kinetic stability of the p63 tetramerization domain. Journal of molecular biology. 2012;415(3):503–13. doi: 10.1016/j.jmb.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belyi VA, Levine AJ. One billion years of p53/p63/p73 evolution. Proc Natl Acad Sci U S A. 2009;106(42):17609–10. doi: 10.1073/pnas.0910634106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koeppel M, van Heeringen SJ, Kramer D, Smeenk L, Janssen-Megens E, Hartmann M, et al. Crosstalk between c-Jun and TAp73alpha/beta contributes to the apoptosis-survival balance. Nucleic Acids Res. 2011;39(14):6069–85. doi: 10.1093/nar/gkr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbluth JM, Mays DJ, Jiang A, Shyr Y, Pietenpol JA. Differential regulation of the p73 cistrome by mammalian target of rapamycin reveals transcriptional programs of mesenchymal differentiation and tumorigenesis. Proc Natl Acad Sci U S A. 2010;108(5):2076–81. doi: 10.1073/pnas.1011936108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–12. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 37.Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36(11):3639–54. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang A, Zhu Z, Kettenbach A, Kapranov P, McKeon F, Gingeras TR, et al. Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar dna-binding profiles in vivo. PLoS One. 2010;5(7):e11572. doi: 10.1371/journal.pone.0011572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortt K, Sinha S. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 2006;580(18):4544–50. doi: 10.1016/j.febslet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Koshiba S, Ichimiya S, Nagashima T, Tonooka A, Kubo T, Kikuchi T, et al. Tonsillar crypt epithelium of palmoplantar pustulosis secretes interleukin-6 to support B-cell development via p63/p73 transcription factors. J Pathol. 2008;214(1):75–84. doi: 10.1002/path.2266. [DOI] [PubMed] [Google Scholar]
- 41.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 42.Vayssade M, Haddada H, Faridoni-Laurens L, Tourpin S, Valent A, Benard J, et al. P73 functionally replaces p53 in Adriamycin-treated, p53-deficient breast cancer cells. Int J Cancer. 2005;116(6):860–9. doi: 10.1002/ijc.21033. [DOI] [PubMed] [Google Scholar]
- 43.Vikhanskaya F, Toh WH, Dulloo I, Wu Q, Boominathan L, Ng HH, et al. p73 supports cellular growth through c-Jun-dependent AP-1 transactivation. Nat Cell Biol. 2007;9(6):698–705. doi: 10.1038/ncb1598. [DOI] [PubMed] [Google Scholar]
- 44.Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J Biol Chem. 2004;279(43):44713–22. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- 45.Bancroft CC, Chen Z, Dong G, Sunwoo JB, Yeh N, Park C, et al. Coexpression of proangiogenic factors IL-8 and VEGF by human head and neck squamous cell carcinoma involves coactivation by MEK-MAPK and IKK-NF-kappaB signal pathways. Clin Cancer Res. 2001;7(2):435–42. [PubMed] [Google Scholar]
- 46.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 47.Ondrey FG, Dong G, Sunwoo J, Chen Z, Wolf JS, Crowl-Bancroft CV, et al. Constitutive activation of transcription factors NF-(kappa)B, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and pro-angiogenic cytokines. Mol Carcinog. 1999;26(2):119–29. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 48.Nottingham LK, Yan CH, Yang X, Si H, Coupar J, Bian Y, et al. Aberrant IKKalpha and IKKbeta cooperatively activate NF-kappaB and induce EGFR/AP1 signaling to promote survival and migration of head and neck cancer. Oncogene. 2014;33(9):1135–47. doi: 10.1038/onc.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barenco M, Brewer D, Papouli E, Tomescu D, Callard R, Stark J, et al. Dissection of a complex transcriptional response using genome-wide transcriptional modelling. Mol Syst Biol. 2009;5:327. doi: 10.1038/msb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bid HK, Roberts RD, Cam M, Audino A, Kurmasheva RT, Lin J, et al. DeltaNp63 promotes pediatric neuroblastoma and osteosarcoma by regulating tumor angiogenesis. Cancer Res. 2014;74(1):320–9. doi: 10.1158/0008-5472.CAN-13-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang H, Wang F, Dyer NP, Wong WH. CisGenome Browser: a flexible tool for genomic data visualization. Bioinformatics. 2010;26(14):1781–2. doi: 10.1093/bioinformatics/btq286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–6. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 54.Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262(5131):208–14. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 55.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–7. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.