Abstract
A high rate of aerobic glycolysis is a hallmark of malignant transformation. Accumulating evidence suggests that diverse regulatory mechanisms mediate this cancer-associated metabolic change seen in a wide spectrum of cancer. The echinoderm microtubule associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) fusion protein is found in approximately 3-7% of non-small cell lung carcinomas (NSCLC). Molecular evidence and therapeutic effectiveness of FDA-approved ALK inhibitors indicated that EML4-ALK is a driving factor of lung tumorigenesis. A recent clinical study showed that NSCLC harboring EML4-ALK rearrangements displayed higher glucose metabolism compared to EML4-ALK-negative NSCLC. In the current work, we presented evidence that EML4-ALK is coupled to overexpression of hexokinase II (HK2), one of the rate-limiting enzymes of the glycolytic pathway. The link from EML4-ALK to HK2 upregulation is essential for a high rate of glycolysis and proliferation of EML4-ALK-rearranged NSCLC cells. We identified hypoxia-inducible factor 1α (HIF1α) as a key transcription factor to drive HK2 gene expression in normoxia in these cells. EML4-ALK induced hypoxia-independent but glucose-dependent accumulation of HIF1α protein via both transcriptional activation of HIF1α mRNA and the PI3K-AKT pathway to enhance HIF1α protein synthesis. The EML4-ALK-mediated upregulation of HIF1α, HK2 and glycolytic metabolism was also highly active in vivo as demonstrated by FDG-PET imaging of xenografts grown from EML4-ALK-positive NSCLC cells. Our data reveal a novel EML4-ALK-HIF1α-HK2 cascade to enhance glucose metabolism in EML4-ALK-positive NSCLC.
Keywords: EML4-ALK, lung cancer, HK2, HIF1α, cancer metabolism, glycolysis
INTRODUCTION
Cancer cells are metabolically transformed to respond to environmental changes. The most prominent aspect of metabolic transformation is the glycolytic phenotype or the Warburg effect, whereby cancer cells sustain a high rate of aerobic glycolysis.1 Active glycolysis not only provides quick ATP but also serves as a primary route for carbon source required for biosynthesis of complex macromolecules in actively proliferating cancer cells.2 The metabolic switch from oxidative phosphorylation to hyperactive glycolysis is mediated by overexpression or hyperactivity of glucose transporters and/or key glycolytic enzymes as a result of activation of oncogene and/or inactivation of tumor suppressors.3 The oncogenes Ras, AKT, and c-Myc have been reported to promote expression of various glycolytic enzymes.4, 5 In contrast, loss of the p53 tumor suppressor inhibits the mitochondrial respiratory chain by suppression of SCO2 (the synthesis of cytochrome c oxidase protein) and promotes glycolysis via TIGAR, a p53-inducible regulator of glycolysis and apoptosis.6
The echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) fusion oncogene arises from an inversion on the short arm of chromosome 2 that joins the N-terminus of EML4 to the intracellular catalytic domain of ALK in patients with non-small cell lung cancer (NSCLC). First described in 2007 by two independent groups,7, 8 the fusion protein was now found in 3-7% of NSCLC patients.8, 9 The fusion oncogene is transforming both in vitro and in vivo.8 Molecular evidence and therapeutic effectiveness of FDA-approved ALK inhibitors demonstrates that EML4-ALK is a driving factor of lung tumorigenesis.10, 11 NSCLC harboring the EML4-ALK fusion oncogene is associated with specific clinical features, such as never or light smoking history and younger age.12-14 In addition, EML4-ALK rearrangement does not overlap with mutations of epidermal growth factor receptor (EGFR) or K-Ras.12 Therefore, the EML4-ALK fusion oncogene defines a distinct clinicopathologic subset of NSCLC.
There have been few studies of metabolic characteristics of EML4-ALK-positive NSCLC although this molecularly defined subtype of lung cancer represents an ideal model to investigate the role of oncogenes in metabolic transformation. Through analysis of fluorodeoxyglucose-positron emission tomography (FDG-PET) images of NSCLC patients, Choi et al. recently reported that EML4-ALK-rearranged NSCLC exhibited higher glucose metabolism than NSCLC patients negative for EML4-ALK, implying a potential role of EML4-ALK in reprogramming glucose metabolism.15 In the present work, we showed that EML4-ALK is causally linked to HK2 overexpression and the glycolytic phenotype of EML4-ALK-positive NSCLC. The ALK kinase activity induced hypoxia-independent accumulation of HIF1α via both transcriptional and post-transcriptional mechanisms. Abundant HIF1α in turn acts as a primary transcription factor to stimulate HK2 expression, contributing to active glycolysis in EML4-ALK-rearranged NSCLC. Suppression of HK2 expression mimics the effects of EML4-ALK inactivation on glycolytic metabolism and proliferation of these cells. The EML4-ALK-mediated upregulation of HIF1α, HK2 and glucose catabolism was also fully reproducible in vivo as demonstrated by FDG-PET imaging of xenografts of EML4-ALK-positive lung cancer cells. These results revealed a novel EML4-ALK-HIF1α-HK2 pathway to drive glycolytic metabolism in EML4-ALK-rearranged NSCLC.
RESULTS
EML4-ALK activates aerobic glycolysis
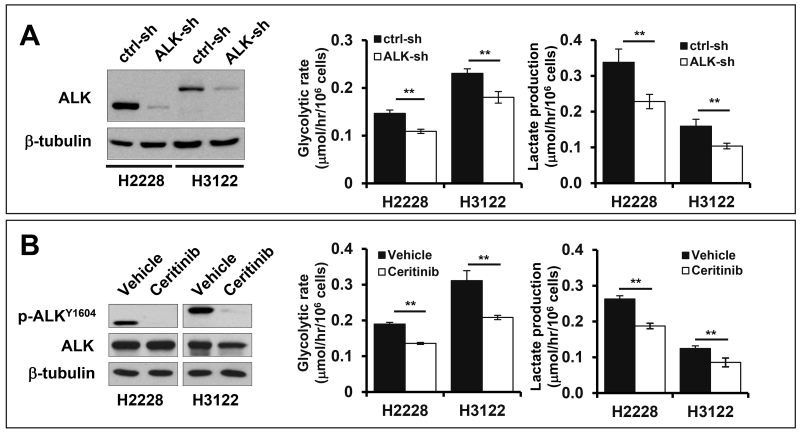
A recent clinical study demonstrated that NSCLCs with EML4-ALK rearrangement have higher FDG uptake than those carrying EGFR mutation or lacking known genetic aberrations, implying a potential role of EML4-ALK in glucose metabolic reprogramming.15 NPM-ALK, another form of ALK arrangement in anaplastic large cell lymphoma (ALCL), has been recently shown to phosphorylate pyruvate kinase M2 (PKM2), leading to inhibition of the enzyme activity and prevention of oxidative phosphorylation.16 Therefore we examined whether EML4-ALK is linked to upregulation of glucose metabolism in ALK-rearranged NSCLC. To this end, we took advantage of two well-defined NSCLC cell lines H3122 and H2228 known to harbor variant 1 and variant 3 of EML4-ALK, respectively.12 Variant 1 and variant 3 are generated by exon 13 and 6 of EML4 joined to exon 20 of ALK, respectively,17 and account for more than 60% of all EML4-ALK rearrangements in NSCLC.18 Lentivirally-mediated shRNA targeting ALK efficiently knocked down EML4-ALK expression in both cell lines (Fig. 1A). Downregulation of EML4-ALK significantly decreased glycolysis and lactate production in these cells. Similar effects were observed when H2228 and H3122 cells were treated with ceritinib, a second-generation ALK inhibitor for the treatment of ALK-positive NSCLC11, 19 (Fig. 1B). It should be pointed out that the expression of wide type ALK was undetectable in these cell lines, consistent with the fact that ALK expression was extremely low in adult tissues.20
Figure 1. EML4-ALK drives glycolysis in EML4-ALK-positive lung cancer cells.
(A) EML4-ALK expression in H2228 and H3122 cells was knocked down by ALK shRNA. The effects of ALK knockdown on glycolysis and lactate production were determined as detailed in Materials and Methods. (B) The effect of the ALK inhibitor cerinitib (200 nM, 48 hours) on glycolysis and lactate production was measured as in (A).
Expression of EML4-ALK is associated with upregulation of HK2
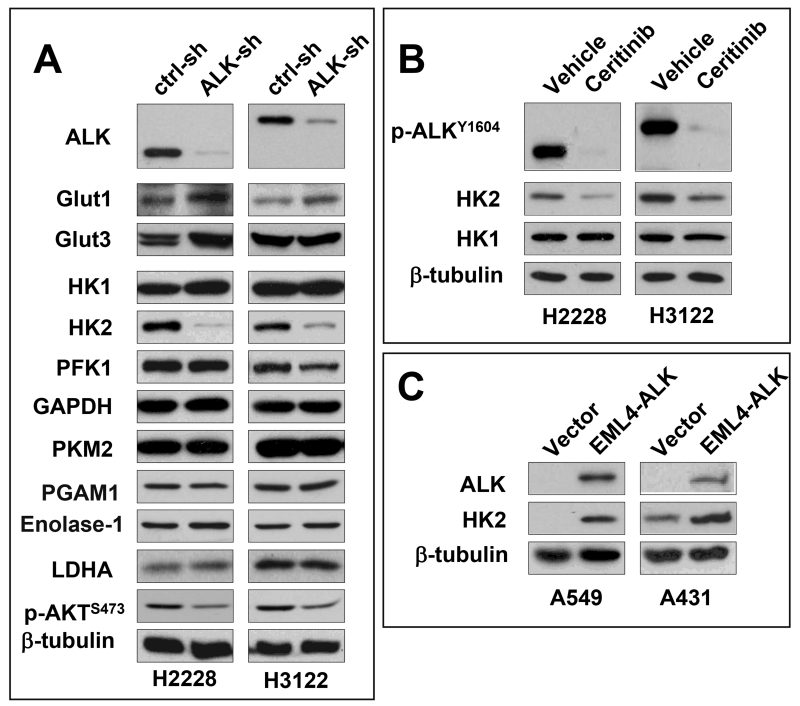
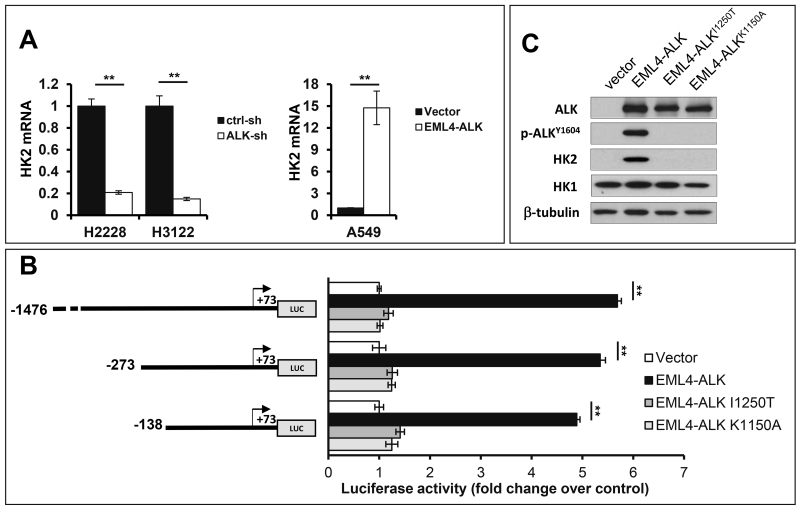
The elevated glucose utilization in tumors has been attributed primarily to upregulation of either glucose transporters or rate-limiting glycolytic enzymes.21 To elucidate the molecular mediator sustaining the glycolytic rate in EML4-ALK-positive cells, we examined the expression of glucose transporters and major glycolytic enzymes. Knockdown of EML4-ALK moderately increased expression of GLUT1 but did not affect that of GLUT3 (Fig. 2A), two major glucose transporters in lung cancer.22 Among the multiple glycolytic enzymes HK1, HK2, PFK1, PKM2, LDHA, PGAM1, Enolase-1 and GAPDH we examined, only HK2 protein was substantially decreased in response to shRNA silencing of EML4-ALK (Fig. 2A). Inhibition of HK2 expression was also observed in H2228 and H3122 cells treated with ceritinib (Fig. 2B). Thus the presence of the EML4-ALK fusion protein is concurrent with selective upregulation of HK2 in EML4-ALK-positive lung cancer cells.
Figure 2. HK2 is upregulated in EML4-ALK-positive lung cancer cells.
ALK was silenced in H2228 and H3122 by ALK shRNA. The expression of glucose transporters and multiple glycolytic enzymes was analyzed by immunoblotting (A). The selective inhibition of HK2 protein by EML4-ALK silencing was confirmed by the use of ALK inhibitor cerinitib (200 nM, 48 hours) (B). In (C), EML4-ALK was ectopically expressed in A549 and A431 cells. EML4-ALK induction of HK2 in the transduced cells compared to the vector control was analyzed by immunoblotting.
To confirm a causal role for EML4-ALK in upregulation of HK2, we used the pCDH-CMV-MCS-EF1-Puro lentivirus system to express EML4-ALK variant 1 in two EML4-ALK-negative cancer cell lines A549 and A431. Indeed, introduction of EML4-ALK dramatically enhanced HK2 protein expression in both cell lines (Fig. 2C).
HK2 mediates the metabolic effects of EML4-ALK
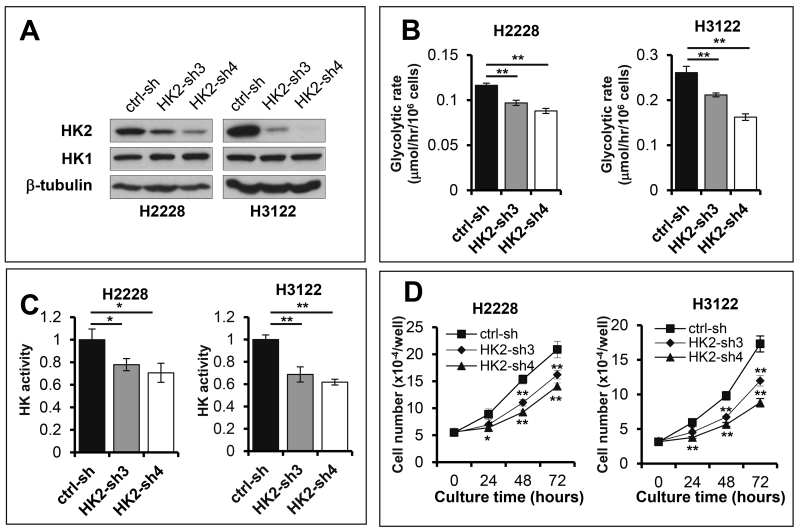
To further understand the role of upregulated HK2 in EML4-ALK-positive NSCLC cells, we examined the effect of HK2 silencing on glycolysis in H2228 and H3122 cells. As shown in Fig. 3, shRNA knockdown of HK2 expression suppressed glycolytic flux and HK activity in these cells. The inhibitory effects were directly related to the knockdown efficiencies of HK2 shRNAs with HK2-sh4 causing more dramatic decrease in glycolysis and HK activity (Fig. 3B and Fig. 3C). Likewise, HK2 silencing also attenuated proliferation of H2228 and H3122 cells (Fig. 3D), suggesting that EML4-ALK-mediated induction of HK2 is functionally important for sustaining active glycolysis and proliferation of EML4-ALK-positive NSCLC cells.
Figure 3. HK2 overexpression is a crucial mediator of glycolysis and proliferation in EML4-ALK positive lung cancer cells.
Expression of HK2 in H2228 and H3122 was knocked down by HK2 shRNA (A). Glycolytic rates (B), HK activity (C) and growth curves (D) of HK2 knockdown cells were determined and compared with the control cells.
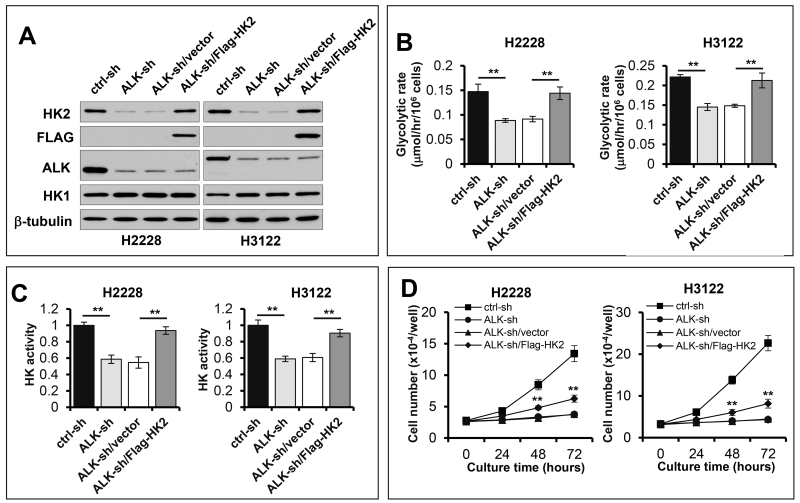
To confirm that the hyperactive glycolysis in EML4-ALK-positive NSCLC cells is truly due to ALK-driven HK2 overexpression, we first knocked down EML4-ALK to bring down cellular HK2 level and then introduced a flag-tagged HK2 under the control of the CMV promoter independent of ALK regulation. In these cells, exogenous HK2 reversed the inhibitory effects of ALK knockdown on glycolysis and HK activity (Fig. 4). However, ectopic expression of HK2 only partially rescued the growth rate of ALK knockdown cells (Fig. 4D), indicating that the ALK-HK2-glycolysis pathway is required but insufficient for ALK-driven cell proliferation. Other effectors downstream of ALK must act cooperatively to promote cell growth.
Figure 4. Exogenous HK2 rescues glycolysis in EML4-ALK knockdown cells.
The Flag-HK2 under the CMV promoter in lentivirus was transduced into EML4-ALK-silenced H2228 and H3122 cells (A). Glycolytic rates (B), HK activity (C) and growth curves (D) of ctrl-sh, EML4-ALK-sh, HK2 reconstituted and the vector control cells were determined as in Figure 3. The statistical significances marked in (D) refer to comparisons between the HK2 reconstituted cells and the vector control cells.
EML4-ALK drives HK2 expression through transcriptional activation
We next explored the molecular mechanism underlying the EML4-ALK-dependent induction of HK2. Quantitative PCR (qPCR) was performed to measure the steady state levels of HK2 mRNA in EML4-ALK-depleted H2228 and H3122 and the corresponding control cells. As shown in Fig. 5A, shRNA knockdown of EML4-ALK resulted in marked decrease in HK2 mRNA, indicating that EML4-ALK stimulates HK2 expression via upregulation of HK2 transcripts. In further support of this, introduction of EML4-ALK into A549 cells led to a multifold increase in HK2 mRNA.
Figure 5. EML4-ALK induces transcription of HK2 in an ALK kinase activity-dependent manner.
(A) EML4-ALK was knocked down in H2228 and H3122 cells or ectopically expressed in A549 cells. The effects of these approaches on HK2 mRNA levels were determined by qPCR. The results were normalized to 18S rRNA and presented as values relative to the corresponding control cells (defined as 1 fold). (B) The HK2 luciferase reporter (pGL2-1476HK2-Luc, pGL2-273HK2-Luc, or pGL2-138HK2-Luc) was co-transfected into A549 cells along with pCDNA3.1/Zeo(+) (vector), pCDNA3.1/Zeo(+)-EML4-ALK, pCDNA3.1/Zeo(+)-EML4-ALKI1250T or pCDNA3.1/Zeo(+)-EML4-ALKK1150A. The transactivation of the HK2 promoter by co-transfected ALK was measured by luciferase assays. The luciferase activities were presented as fold changes relative to the value of the vehicle control cells (defined as 1 fold). (C) Expression of co-transfected wild type and mutant forms of EML4-ALK in A549 cells was confirmed by immunoblotting.
We took advantage of the highly responsive A549 cells to explore the transcriptional mechanism leading to HK2 overexpression. The HK2 gene promoter (from −1476 to +73) was cloned into the pGL2-Basic-Luc reporter to construct pGL2-1476HK2-Luc. Cotransfection of pGL-1476HK2-Luc with pcDNA3.1/Zeo(+)-EML4-ALK, a variant 1 EML4-ALK expression vector, in A549 cells resulted in strong induction of luciferase activity from the HK2 promoter (Fig. 5B). A series of nested 5’ deletions of the HK2 promoter (pGL2-273HK2-Luc, pGL2-138HK2-Luc) were constructed and tested. The major luciferase activity remained intact when HK2 promoter was shortened to −138 bp (Fig. 5B). The transactivation of the HK2 promoter by EML4-ALK depended entirely on ALK kinase activity. Two kinase-dead mutants of EML4-ALK (I1250T, K1150A)23, 24 failed to transactivate the HK2 promoter (Fig. 5B), although they were almost equally expressed in transfected cells (Fig. 5C).
HIF1α is a key transcription factor mediating HK2 upregulation in EML4-ALK-expressing cells
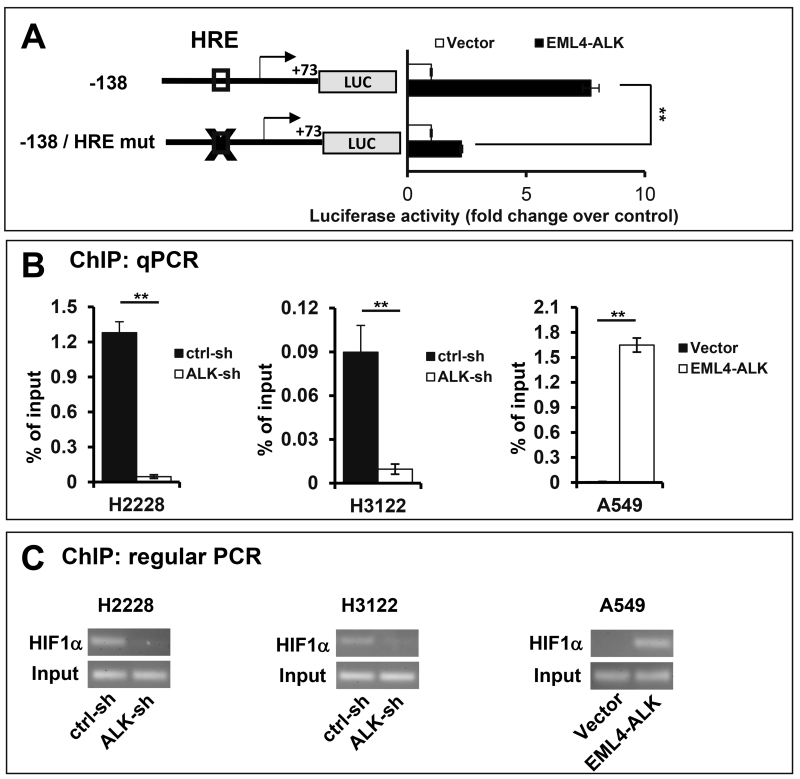
The DNA sequences (−138 to +73) of the HK2 gene promoter responsive to ALK contain potential binding sites for multiple transcription factors, including those for NF-Y, NF-1, SP-1, CREB, AP-2, GCF, and HIF1, as predicted from PROMO analysis.25 Among these regulatory elements, the putative hypoxia response element (HRE) for HIF1 is of particular interest. A functional HRE is usually composed of a pair of contiguous sites, at least one of which contains the core sequence 5′-RCGTG-3′,26, 27 which is exactly the case with the HRE located at −60/−49 upstream of the HK2 transcription start site. This prompted us to test whether HIF1 was involved in activation of the HK2 gene. Interestingly, mutation of the HRE site located at −60/−49 of the HK2 promoter inhibited EML4-ALK activation of the HK2 gene promoter as reflected by loss of more than 75% luciferase activity in A549 cells transfected with the HRE mutant reporter (Fig. 6A and Fig. S1). The weak remaining activity could be due to the existence of other minor yet HIF1-independent factors downstream of EML4-ALK.
Figure 6. HIF1α is a key transcription factor upregulating HK2 expression in EML4-ALK-expressing lung cancer cells.
(A) The HK2 luciferase reporter pGL2-138HK2-Luc or the HRE mutant form was co-transfected into A549 cells along with pCDNA3.1/Zeo(+) (vector) or pCDNA3.1/Zeo(+)-EML4-ALK. The transactivation of the HK2 promoter by co-transfected ALK was measured by luciferase assays. The luciferase activities were presented as fold changes relative to the value of the vector control cells (defined as 1 fold). ChIP assay was performed to assess the binding of HIF1α to the HK2 promoter in ALK knockdown and control cells of H2228 and H3122 and in A549 cells transduced to express EML4-ALK. The HK2 promoter DNA sequence co-precipitated with HIF1α was analyzed by qPCR using SYBR Green (B) and regular PCR (C) using the primers listed in Table 1. Fractions of immunoprecipitates and total cellular DNA were also analyzed with regular PCR amplification of the HK2 promoter..
To gain direct evidence that HIF1α binds to the −60/−49 HRE region of the native HK2 promoter to activate its transcription, we performed chromatin immunoprecipitation (ChIP) assay in H2228 and H3122 cells, and in EML4-ALK-transduced A549 cells. The qPCR analysis of HIF1α immunoprecipitates showed that EML4-ALK knockdown resulted in 28- and 9-fold decreases in the binding of HIF1α to the −60/−49 HRE in H2228 and H3122 cells, respectively (Fig. 6B). In EML4-ALK-transduced A549 cells, there was a 173-fold increase in HIF1α binding to the HRE region over the control cells lacking EML4-ALK (Fig. 6B). EML4-ALK-dependent induction of HIF1α binding to the HK2 promoter was also readily detected by regular PCR analysis of HIF1α immunoprecipitates (Fig. 6C). Hence EML4-ALK expression led to robust increases in HIF1α binding to the HK2 promoter at the −60/−49 HRE.
EML4-ALK induces hypoxia-independent but glucose-dependent accumulation of HIF1α
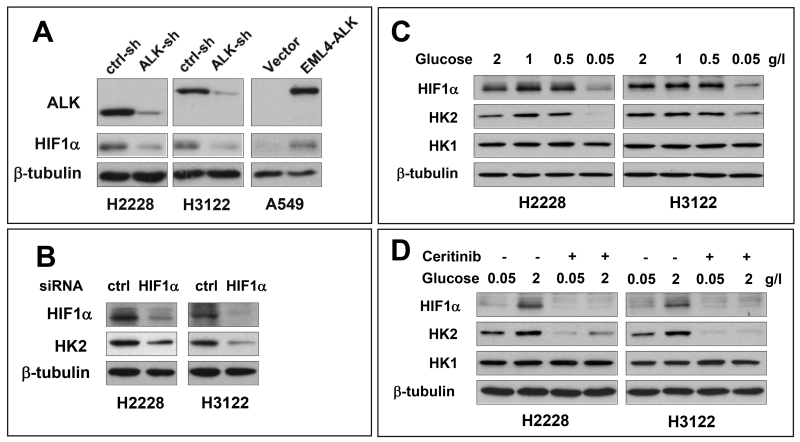
Consistent with the luciferase reporter and ChIP assays, basal HIF1α protein, but not HIF2α, was readily detectable in both H2228 and H3122 cells (Fig. 7A and Fig. S2), in contrast to absent or low expression of HIF1α in most other cancer cell lines in normoxic conditions. The inactivation of EML4-ALK by shRNA silencing or ceritinib was accompanied by diminished HIF1α protein in these cells. Conversely, ectopic expression of EML4-ALK in A549 cells increased HIF1α protein (Fig. 7A), indicating that EML4-ALK is sufficient to enhance HIF1α protein abundance independently of hypoxia. To confirm that the abundantly present HIF1α is responsible for HK2 overexpression in H2228 and H3122 cells, we used HIF1α siRNA to transiently knockdown its expression. This led to significant decreases in protein levels of not only HIF1α but also HK2 (Fig. 7B).
Figure 7. EML4-ALK induces hypoxia-independent but glucose-dependent accumulation of HIF1α.
(A) The effects of ALK knockdown in H2228 and H3122 or ALK ectopic expression in A549 on HIF1α protein levels were analyzed by immunoblotting. (B) H2228 and H3122 were transfected with HIF1α siRNA or non-target control siRNA. HK2 and HIF1α protein expression was analyzed by immunoblotting. (C) H2228 and H3122 were cultured in RPMI 1640 with indicated concentrations of glucose for 24 hours before analysis of HIF1α protein. (D) After incubation with 0.05 g/L glucose overnight, H2228 and H3122 cells were cultured with indicated concentrations of glucose in the presence and absence of 200 nM ceritinib for 24 hours. Expression of HIF1α and HK2 protein was analyzed by immunoblotting.
In addition to oxygen shortage, other environmental or nutritional factors including glucose availability have been shown to protect HIF1α protein through inhibition of lysosomal degradation of HIF1α, an oxygen and pVHL-independent arm of control of HIF1α protein.28, 29 We therefore asked whether ALK-dependent accumulation of HIF1α is influenced by glucose availability. When shifted from high to low concentrations of glucose (from 2 to 0.05 g/L), H2228 and H3122 cells exhibited much reduced HIF1α protein (Fig. 7C). The abundance of HIF1α was recovered by addition of glucose back to culture in an ALK activity-dependent manner (Fig. 7D). In further support of a crucial role for HIF1α in mediating HK2 transcription, HK2 protein was also similarly up and down-regulated under these high and low glucose conditions (Fig. 7C, 7D).
If EML4-ALK is causally linked to elevated HIF1α protein, one may expect that other known HIF1α target genes such as GLUT1 and LDHA, in addition to HK2, should be similarly upregulated in EML4-ALK-expressing cells. As shown earlier, we did not detect significant changes in protein expression of GLUT1 and LDHA in EML4-ALK stable knockdown cells (Fig. 2). However, when we examined mRNA of GLUT1 and LDHA, we found that depletion of EML4-ALK caused obvious decreases in GLUT1 and LDHA mRNA (Fig. S3). Consistent with this, GLUT1 and LDHA mRNAs but not their proteins were elevated in EML4-ALK stably transduced A549 cells (Fig. S3). Such discrepancy suggests that HIF1α-mediated transcriptional activation is not necessarily sufficient to enhance protein level expression of other HIF1α-targeted genes.
The PI3K-AKT pathway is required for ALK upregulation of HIF1α protein synthesis
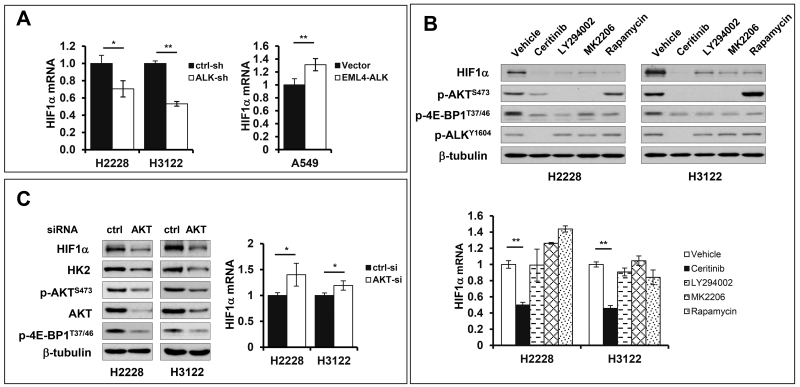
Previous studies showed that the NPM-ALK fusion protein in T-cell lymphoma cell lines and EML4-ALK in NSCLC cells stimulated transcription of HIF1α mRNA through signal transducer and activator of transcription 3 (STAT3)30 or through cooperation of STAT3 with CEBP/β.31 In both cases, however, enhanced HIF1α protein was seen only in hypoxia, consistent with the concept that O2-dependent proteasomal degradation of HIF1α plays a dominant role in the control of HIF1α homeostasis.32 In our hands, HIF1α mRNA was moderately but significantly increased in EML4-ALK-expressing cells. HIF1α mRNA levels were up or down-regulated in accordance with overexpression of EML4-ALK in A549 cells or inactivation of ALK by shRNA knockdown in H2228 and H3122 cells (Fig. 8A). Thus our data confirmed that ALK-mediated accumulation of HIF1α protein involves transcriptional activation of HIF1α.
Figure 8. The PI3K-AKT-mTOR pathway is involved in EML4-ALK-mediated upregulation of HIF1α protein.
(A) The effects of ALK knockdown in H2228 and H3122 or ALK ectopic expression in A549 on levels of HIF1α mRNA were analyzed by qPCR. (B) H2228 and H3122 cells were treated with vehicle, ceritinib (200 nM), LY294002 (10 μM), MK2206 (4 μM), or rapamycin (20 nM) for 8 hours. HIF1α protein and mRNA levels in control and treated cells were analyzed by immunoblotting and qPCR, respectively. (C) AKT1/2 expression in H2228 and H3122 cells was knocked down simultaneously by siRNA targeting both isoforms. The effects on HIF1α, HK2 and other proteins as well as HIF1α mRNA were analyzed as in (B). In all panels, qPCR results were normalized to 18S rRNA and presented as values relative to the corresponding control cells (defined as 1 fold).
Since the transcriptional regulation is usually insufficient to maintain HIF1α abundances under normoxia, we asked whether other post-transcriptional processes are implicated in upregulation of HIF1α protein. AKT has been reported to enhance HIF1α abundance in cells that overexpress oncogenic tyrosine kinases such as ErbB2 or activating mutant of AKT.33, 34 The PI3K-AKT pathway is particularly relevant as EML4-ALK constitutively activates PI3K.20, 35 Interestingly, pharmacological inhibition of PI3K, AKT and mTOR with LY294002, MK2206 and rapamycin, respectively, dramatically decreased levels of HIF1α protein in H2228 and H3122 cells (Fig. 8B). None of these inhibitors of the PI3K-AKT-mTOR pathway, however, negatively affected HIF1α mRNA (Fig. 8B). Only the ALK inhibitor ceritinib reduced both protein and mRNA expression of HIF1α. Consistent with an essential role of the PI3K-AKT-mTOR pathway in promotion of HIF1α protein synthesis, treatment with ceritinib, LY294002, MK2206 or rapamycin led to dephosphorylation of 4E-BP1, a biochemical process associated with functional activation of the translational repressor. These results together indicate that the PI3K-AKT-mTOR pathway is important for EML4-ALK upregulation of HIF1α protein synthesis.
To obtain molecular evidence for involvement of AKT, we took advantage of siRNA that was designed to silence both AKT1 and AKT2 isoforms simultaneously. Knockdown of AKT1/2 in H2228 and H3122 cells brought down HIF1α protein level with no concomitant decrease in HIF1α mRNA (Fig. 8C). Instead, HIF1α mRNA was moderately increased in AKT1/2 knockdown cells. Similar to the inhibitors of ALK, PI3K, AKT and mTOR, AKT1/2 siRNA also triggered dephosphorylation of 4E-BP1. Consistent with HIF1α being a primary transcription activator of HK2, AKT1/2 siRNA also decreased HK2 expression in these cells.
The EML4-ALK-HIF1α-HK2 pathway drives glycolytic metabolism in vivo
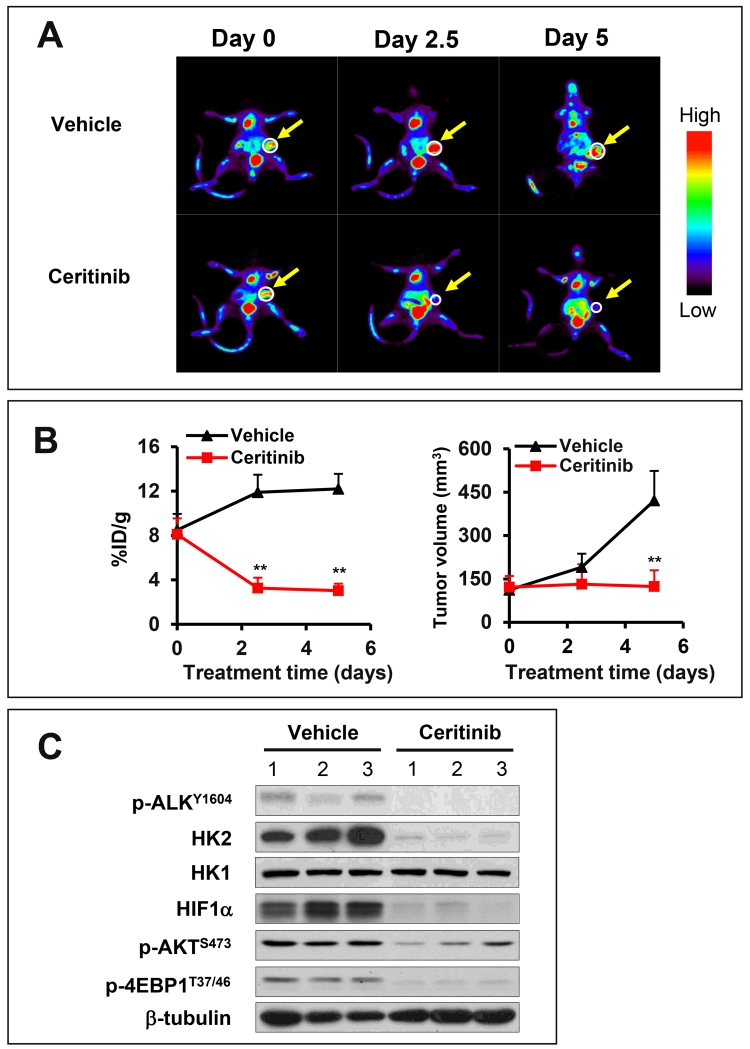
The Warburg effect of aerobic glycolysis constitutes the principle for tumor imaging in vivo following the administration of the labeled glucose analog FDG. The excess uptake and HK2-mediated phosphorylation of the labeled FDG in cancer cells can then be detected by PET scanning. Since our data suggests that the high rate of glycolysis in the EML4-ALK-positive subset of NSCLC is attributed to the kinase activity of EML4-ALK, we next examined whether inhibition of EML4-ALK is sufficient to limit glucose catabolism in xenografts of H3122 cells in immunodeficient SCID mice. H3122 cells were subcutaneously injected in SCID mice. Ten days after cell injection, the mice carrying palpable xenografts were randomly divided into experiment and control groups for treatment with ceritinib (25mg/kg, daily) or vehicle for 5 days. The FDG-PET scanning was conducted before treatment, and then in the middle (2.5 days) and at the end (5 days) of the treatment.
As shown in Fig. 9A, ceritinib resulted in a remarkable decrease in values of the percent injected dose/gram of tissue (%ID/g) in the xenografts. The difference between treatment and control groups was strikingly significant after treatment with cerinitib for only 2.5 days when tumors showed little reduction in volume (Fig. 9A-9B). We also examined the effects of ceritinib on expression of HIF1α, HK2, p-AKT, and p-4E-BP1 in the xenografts after 5 days of treatment when the animals were euthanized. As expected, expression HIF1α, HK2, p-AKT, and p-4E-BP1 proteins was reduced in the xenografts from mice treated with ceritinib (Fig. 9C). These results indicate that the EML4-ALK-HIF1α-HK2 axis to regulate glycolysis is fully operational in vivo. Inhibition of EML4-ALK was sufficient to disrupt the signaling from the ALK oncogene to glucose metabolism in mice.
Figure 9. Inhibition of ALK kinase activity blocks ALK-HIF1α-HK2 signaling and glycolysis in vivo.
(A) Shown were FDG-PET images of one representative xenograft from vehicle control group and from experimental group treated with cerinitib for 0, 2.5 and 5 days. (B) 18F-FDG uptake (% ID per g) (left) and tumor volumes (right) from 8 mice of each group were quantified as detailed in Materials and Methods. (C) Lysates from xenografts treated with vehicle or ceritinib for 5 days were analyzed for expression of indicated proteins by immunoblotting.
DISCUSSION
EML4-ALK rearranged lung cancer represents a molecularly defined subset of NSCLC, wherein ALK is clearly one of the oncogenic drivers.36 A recent clinical study found that this subset of lung cancer exhibited higher glucose metabolism and metastatic potential than EML4-ALK-negative NSCLC.15 We examined the hypothesis that oncogenic ALK is causally linked to the hyperactive glycolysis in EML4-ALK-positive NSCLC. Our results identified a novel regulatory pathway EML4-ALK-HIF1α-HK2 that upregulates glycolysis specifically in EML4-ALK-positive NSCLC cells. Brief exposure to an ALK inhibitor is sufficient to decrease glycolytic metabolism in EML4-ALK-positive xenografts, which could be monitored by FDG-PET scans in live animals.
Hexokinases catalyze ATP-dependent conversion of glucose to glucose-6-phosphate (G-6-P), the irreversible first rate-limiting step of glucose metabolism.37 G-6-P then serves as a co-substrate for both glycolysis and the pentose phosphate pathway (PPP). While a high rate of glycolysis is essential for providing quick ATP and carbon source for biosynthesis, PPP is a critical route for producing nucleotides required by actively replicating cancer cells.38 Among the four isoforms of hexokinase, HK2 is aberrantly overexpressed in cancer.39, 40 The higher affinity for substrate, lower sensitivity to negative feedback from product and greater catalytic power of HK2 compared to other hexokinase isoforms are likely the metabolic reasons for the propensity of tumors to selectively overexpress HK2.41 The N-terminal hydrophobic domain allows HK2 to interact with voltage-dependent anion channel at the mitochondria and to gain preferential access to mitochondrially generated ATP to facilitate HK reaction.41 Substantial studies have shown the essentiality of HK2 in growth, survival and tumorigenesis of different types of cancer including glioblastoma multiforme (GBM), medulloblastoma, K-Ras-driven lung cancer, ErbB2-driven breast cancer, and PTEN-/p53-deficiency-induced prostate cancer.42-45 It is noteworthy that HK2 is expressed only in a limited number of adult tissues.37 There are no adverse physiological consequences with systemic HK2 deletion in a mouse model.43 Therefore, HK2-selective inhibitors could be promising anti-cancer agents.46
Cancer cells upregulate HK2 expression and activity via diverse molecular mechanisms including gene amplification, transcriptional activation by p53 deficiency, Ras, AKT, c-Myc or HIF, downregulation of miR-143, and promoter demethylation.47-53 HIF1α is a crucial transcription factor to drive expression of HK2 and other glycolytic enzymes in the adaptive response to hypoxia.54 However, hypoxia is not the causal determinant of the glycolytic phenotype that occurs in both hypoxic and oxygenated regions of a tumor. Tumor cells in vitro also rely on glycolysis even when cultured in normoxic conditions. Involvement of HIF1α in hypoxia-independent overexpression of HK2 has not been yet demonstrated. Our study suggests that oncogenic ALK mediates transcriptional and translational upregulation of HIF1α to increase HIF1α protein synthesis. In the presence of appropriate glucose concentrations to prevent chaperone-mediated lysosomal degradation, HIF1α protein is sufficiently accumulated to activate the downstream HK2 gene expression to promote glucose metabolism in normoxia.α. Another interesting aspect of our observation is that the ALK-HIF1α signaling induced mRNA expression of other known HIF1α target genes such as GLUT1 and LDHA in addition to HK2. At the protein level, however, only HK2 was elevated. The selectivity suggests that gene expression in response to HIF1α activation is more complicated than transcriptional impact. Other post-transcriptional processes likely play critical roles in delivering functional outcomes of HIF1α activation in a gene-specific manner.
In addition to NSCLC, ALK rearrangements through chromosomal translocations are found in anaplastic large-cell lymphomas, large B-cell lymphoma, neuroblastoma, inflammatory myofibroblastic tumors, renal cell carcinoma, and ovarian cancer.20, 55 It is unlikely that the effect of ALK on HIF1α, HK2 and glycolysis is restricted to EML4-ALK positive NSCLC.
The currently available methods to diagnose EML4-ALK-positive NSCLC are FISH, IHC and RT-PCR analyses of tumor biopsies.56 Priorities are given to more prevalent EGFR mutation followed by that of Ras.57 If they are negative, EML4-ALK will be considered. This is a lengthy process and a significant proportion of EML4-ALK-positive NSCLC are never identified or only diagnosed after the commencement of conventional chemotherapies. In the present study, we showed that the FDA-approved ceritinib strongly reduced glycolytic rate in xenografts grown from EML4-ALK-positive NSCLC cells. A dramatic decrease was observed after treatment for only 2.5 days. The decrease occurred before a significant reduction in tumor volume. The causal link from ALK to glycolysis could be exploited to distinguish EML4-ALK-positive NSCLC from EML4-ALK-negative NSCLC with FDG-PET scans following 2-3 days administration of ceritinib. This could provide a quick and non-invasive approach to determining ALK status, which is critical for timely and effective treatment of ALK+ NSCLC with appropriate ALK-targeted inhibitors. The application of the current findings could be more useful in many parts of the world where routine examination of ALK status is not available to NSCLC patients.
MATERIALS AND METHODS
Cell culture and reagents
The cell lines H2228, A549, and A431 were originally obtained from ATCC (Manassas, VA). H3122 was obtained from NCI (Bethesda, MD). All the cell lines were authenticated in July of 2015 by STR DNA fingerprinting at the Characterized Cell Line Core Facility of MD Anderson Cancer Center (Houston, TX). H2228, H3122 and A549 were cultured in RPMI-1640 with 10% fetal bovine serum (FBS). A431 was cultured in DMEM with 10% FBS. D-[5-3H(N)]-glucose was obtained from Perkin Elmer (Boston, MA). Ceritinib, MK2206, and rapamycin were purchased from Selleckchem (Houston, TX). LY294002 was from Cell Signaling (Danvers, MA).
Plasmids and lentivirus
The shRNA target sequences for ALK43 and HK258 (Table 1) were cloned into the shRNA lentiviral vector pGreenPuro (System Biosciences, Mountain View, CA). EML4-ALK and HK2 with Flag-tag at the C-terminus were cloned into the lentiviral vectors pCDH-CMV-MCS-EF1-Puro and pCDH-CMV-MCS-EF1-neo (System Biosciences), respectively. Recombinant lentivirus was produced by co-transfection into 293TN cells with lentiviral vectors and packaging plasmids using lipofectamine 2000 (Life Technologies). Culture supernatants were collected 48 hours after transfection and used to infect target cells. The infected cells were selected with puromycin or G418 to establish stably transduced cells.
Table 1. Oligos used in the work.
| shRNA | |
| ALK-sh | 5’-GGGCGAGCTACTATAGAAA-3’ |
| HK2-sh3 | 5’-GCAGAAGGTTGACCAGTATCT-3’ |
| HK2-sh4 | 5’-CCAAAGACATCTCAGACATTG-3’ |
| Ctrl-sh | 5’-CCTAAGGTTAAGTCGCCCTCG-3’ |
| Luciferase assay | |
| HRE mut | 5’- CGCGCAGCCAATGGGAAAGCGCTTTTCACTGATCCGGAGGC-3’ (forward) |
| 5’- GCCTCCGGATCAGTGAAAAGCGCTTTCCCATTGGCTGCGCG-3’ (reverse) | |
| ChIP assay | |
| HK2 promoter | 5’-GTGCGCACGTCACTGATC-3’ (forward) |
| 5’-CTGCGCCGGAGTTTCATG-3’ (reverse) | |
| EML4-ALK expression vector | |
| EML4-ALK cloning | 5’-TAGAGAACGAGCGGGTCA-3’ (forward) |
| 5’-AGAAGCAGCTAATTCTGACTACAT-3’ (reverse) | |
| EML4-ALKK1150A | 5’-CTGCAAGTGGCTGTGGCGACGCTGC-3’ (forward) |
| 5’-GCAGCGTCGCCACAGCCACTTGCAG-3’ (reverse) | |
| EML4-ALKI1250T | 5’-ACCGAGACACTGCTGCCAGAAACTG-3’ (forward) |
| 5’-CAGTTTCTGGCAGCAGTGTCTCGGT-3’ (reverse) | |
| HK2 expression vector | |
| HK2 cloning | 5‘CGCGGATCCCAACTCTGCGCCGTCGGGCCGCGGCAGGATGATTGCCTCGCA-3’ (forward) |
| 5’-AAGGAAAAAAGCGGCCGCCTACTTGTCGTCATCGTCTTTGTAGTCTCGCTGTCCAGC-3’ (reverse) | |
siRNA
The siGENOME siRNAs for the human HIF1α were obtained from Dharmacon (Lafayette, CO). AKT1/2 siRNA were purchased from Cell Signaling. H2228 or H3122 were transfected with siRNAs using DharmaFECT 1 according to the manufacturer (Dharmacon). Twenty-four hours after transfection, cells were incubated with fresh complete medium for another 48 hours before protein and mRNA analysis.
Cell growth
Cells were plated and cultured in 12-well plates. Cell numbers were determined after indicated treatments using a Z1 Coulter counter (Beckman Coulter, Sykesville, MD).
Glycolysis
The glycolytic rate was measured by quantification of the conversion of 5-3H-glucose to 3H2O as we described previously.58 The glycolytic rate was presented as μmol glucose/hour/106 cells.
Lactate production
Lactate levels in culture supernatants were measured with a colorimetric lactate quantification kit according to the manufacturer (Eton Bioscience, San Diego, CA). The concentrations were normalized on cell numbers.
HK activity
HK activity in cell homogenates was measured as we described recently.58 The results were normalized on protein contents of samples.
qPCR
Total cellular RNA was extracted using Trizol reagent (Life Technologies) and used to synthesize cDNA with High-Capacity cDNA Reverse Transcription Kit (Life Technologies) after DNase treatment. The qPCR probes for HIF1α, HK2, GLUT1, LDHA and 18S rRNA were obtained from Life Technologies. The levels of mRNA expression were quantified with the CFX96 real-time PCR detection system (BioRad, Hercules, CA).
Western blot analysis
Lysates from cells or xenografts were prepared in RIPA buffer supplemented with protease/phosphatase inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Proteins were analyzed by immunoblotting as previously described.58 The antibodies for ALK, β-tubulin, phospho-ALKY1604, HK1, HK2, PFK1, GAPDH, PKM2, LDHA, PGAM1, Enolase-1, AKT, phospho-AKTS473 and phospho-4E-BP1T37/46 were purchased from Cell Signaling. The antibodies for GLUT1 and GLUT3 were obtained from Abcam (Cambridge, MA). The HIF1α antibody was from BD Biosciences (San Jose, CA).
Luciferase assay
The human HK2 gene promoter sequences (−1476 to +73, −273 to +73, or −138 to +73) were cloned into pGL2-Basic-Luc as we described recently.58 The HIF1α binding consensus within the −138 to +73 fragment was converted into inactive sequences (Table 1) by QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). For co-transfection along with luciferase vectors, EML4-ALK cDNA was subcloned into pcDNA3.1/Zeo(+). The two ALK kinase dead mutants pCDNA3.1/Zeo(+)-EML4-ALKI1250T and pCDNA3.1/Zeo(+)-EML4-ALKK1150A 23, 24 were generated by site-directed mutagenesis (Table 1). DNA sequences in all these plasmids were authenticated by automatic sequencing.
A549 cells were co-transfected with the luciferase reporter vectors along with EML4-ALK expression plasmids using TransIT-LT1 (Mirus Bio, Madison, WI). Forty-eight hours after transfection, cell extracts were prepared and assayed with the luciferase assay kit from Promega (Madison, WI).
ChIP assay
ChIP assay was performed as described previously.59 Briefly, nuclear fractions were extracted and chromatin was sonicated to achieve an average length of 200-1000 bp after cells were cross-linked with formaldehyde. Ten percent of the starting material was used as input controls. DNA-protein complexes were immunoprecipitated with HIF1α antibody or control IgG. The immunoprecipitates and input samples were reversed at 65°C overnight and then digested with Proteinase K. DNA samples were purified with the QIAquick Spin Columns (Qiagen, Valencia, CA) and quantified by qPCR with the QuantiNova SYBR Green PCR Kit (Qiagen) using primers in Table 1.
Animal study
Eight-week-old female NOD/SCID mice (Jackson Labs, Bar Harbor, ME) were implanted subcutaneously at the right flank with 5 × 106 H3122 cells. Tumor volumes were determined using a caliper and calculated with the formula: 0.5 × width2 × length. Once the tumor volume reached 100-150 mm3, mice were divided randomly to be treated with ceritinib at 25 mg/kg or vehicle by oral gavage (8 mice/group). Before drug treatment, mice with H3122 xenografts were scanned with the Inveon preclinical small-animal PET/computerized tomography (CT) system (Siemens Healthcare, Malvern, PA) one hour after intravenous injection of 300 μCi of 18F-FDG (IBA Molecular Inc., Sterling, VA). The mice underwent repeat PET/CT imaging after 2.5 and 5 days of daily treatment with ceritinib or vehicle. The 18F-FDG uptake was expressed as the percentage injected dose per gram tissue (%ID/g). All animal experiments were conducted in compliance with the policies and regulations of VCU IACUC.
Statistical analysis
All numerical data were presented as mean ± standard deviation of triplicate assays. The statistical significances were determined using Student’s two-tail t test, where p<0.05 was considered statistically significant. In all figures, the statistical significances were indicated with * if p<0.05 or ** if p<0.01.
Supplementary Material
ACKNOWLEDGEMENTS
The work was supported in part by Astar Biotech research award (XF), the Jeffress Memorial Fund award (XF), and the NIH grant P30 CA16059 to Massey Cancer Center of VCU School of Medicine.
Footnotes
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
REFERENCES CITED
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 4.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 6.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 9.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida A, Tsuta K, Nakamura H, Kohno T, Takahashi F, Asamura H, et al. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol. 2011;35:1226–1234. doi: 10.1097/PAS.0b013e3182233e06. [DOI] [PubMed] [Google Scholar]
- 14.Nishino M, Klepeis VE, Yeap BY, Bergethon K, Morales-Oyarvide V, Dias-Santagata D, et al. Histologic and cytomorphologic features of ALK-rearranged lung adenocarcinomas. Mod Pathol. 2012;25:1462–1472. doi: 10.1038/modpathol.2012.109. [DOI] [PubMed] [Google Scholar]
- 15.Choi H, Paeng JC, Kim DW, Lee JK, Park CM, Kang KW, et al. Metabolic and metastatic characteristics of ALK-rearranged lung adenocarcinoma on FDG PET/CT. Lung Cancer. 2013;79:242–247. doi: 10.1016/j.lungcan.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 16.McDonnell SR, Hwang SR, Rolland D, Murga-Zamalloa C, Basrur V, Conlon KP, et al. Integrated phosphoproteomic and metabolomic profiling reveals NPM-ALK-mediated phosphorylation of PKM2 and metabolic reprogramming in anaplastic large cell lymphoma. Blood. 2013;122:958–968. doi: 10.1182/blood-2013-01-482026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki T, Rodig SJ, Chirieac LR, Janne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010;46:1773–1780. doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–673. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 21.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- 22.Younes M, Brown RW, Stephenson M, Gondo M, Cagle PT. Overexpression of Glut1 and Glut3 in stage I nonsmall cell lung carcinoma is associated with poor survival. Cancer. 1997;80:1046–1051. doi: 10.1002/(sici)1097-0142(19970915)80:6<1046::aid-cncr6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Schonherr C, Ruuth K, Eriksson T, Yamazaki Y, Ottmann C, Combaret V, et al. The neuroblastoma ALK(I1250T) mutation is a kinase-dead RTK in vitro and in vivo. Transl Oncol. 2011;4:258–265. doi: 10.1593/tlo.11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo AH, Stoica GE, Riegel AT, Wellstein A. Recruitment of insulin receptor substrate-1 and activation of NF-kappaB essential for midkine growth signaling through anaplastic lymphoma kinase. Oncogene. 2007;26:859–869. doi: 10.1038/sj.onc.1209840. [DOI] [PubMed] [Google Scholar]
- 25.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 26.Riddle SR, Ahmad A, Ahmad S, Deeb SS, Malkki M, Schneider BK, et al. Hypoxia induces hexokinase II gene expression in human lung cell line A549. Am J Physiol Lung Cell Mol Physiol. 2000;278:L407–416. doi: 10.1152/ajplung.2000.278.2.L407. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 28.Hubbi ME, Hu H, Kshitiz Ahmed I, Levchenko A, Semenza GL. Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. J Biol Chem. 2013;288:10703–10714. doi: 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira JV, Fofo H, Bejarano E, Bento CF, Ramalho JS, Girao H, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349–1366. doi: 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- 30.Marzec M, Liu X, Wong W, Yang Y, Pasha T, Kantekure K, et al. Oncogenic kinase NPM/ALK induces expression of HIF1alpha mRNA. Oncogene. 2011;30:1372–1378. doi: 10.1038/onc.2010.505. [DOI] [PubMed] [Google Scholar]
- 31.Martinengo C, Poggio T, Menotti M, Scalzo MS, Mastini C, Ambrogio C, et al. ALK-dependent control of hypoxia-inducible factors mediates tumor growth and metastasis. Cancer Res. 2014;74:6094–6106. doi: 10.1158/0008-5472.CAN-14-0268. [DOI] [PubMed] [Google Scholar]
- 32.Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A. Akt1 activation can augment hypoxia-inducible factor-1alpha expression by increasing protein translation through a mammalian target of rapamycin-independent pathway. Molecular cancer research. 2006;4:471–479. doi: 10.1158/1541-7786.MCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 34.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1alpha (HIF-1alpha) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clinical cancer research. 2015;21:4014–4021. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- 36.Mano H. The EML4-ALK oncogene: targeting an essential growth driver in human cancer. Proc Jpn Acad Ser B Phys Biol Sci. 2015;91:193–201. doi: 10.2183/pjab.91.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- 38.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 39.Sato S, Matsushima T, Sugimura T. Hexokinase isozyme patterns of experimental hepatomas of rats. Cancer Res. 1969;29:1437–1446. [PubMed] [Google Scholar]
- 40.Mathupala SP, Rempel A, Pedersen PL. Aberrant glycolytic metabolism of cancer cells: a remarkable coordination of genetic, transcriptional, post-translational, and mutational events that lead to a critical role for type II hexokinase. J Bioenerg Biomembr. 1997;29:339–343. doi: 10.1023/a:1022494613613. [DOI] [PubMed] [Google Scholar]
- 41.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208:313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gershon TR, Crowther AJ, Tikunov A, Garcia I, Annis R, Yuan H, et al. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer Metab. 2013;1:2. doi: 10.1186/2049-3002-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, Xiong H, Wu F, Zhang Y, Wang J, Zhao L, et al. Hexokinase 2-mediated Warburg effect is required for PTEN- and p53-deficiency-driven prostate cancer growth. Cell Rep. 2014;8:1461–1474. doi: 10.1016/j.celrep.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganapathy-Kanniappan S, Vali M, Kunjithapatham R, Buijs M, Syed LH, Rao PP, et al. 3-bromopyruvate: a new targeted antiglycolytic agent and a promise for cancer therapy. Curr Pharm Biotechnol. 2010;11:510–517. doi: 10.2174/138920110791591427. [DOI] [PubMed] [Google Scholar]
- 47.Rempel A, Mathupala SP, Griffin CA, Hawkins AL, Pedersen PL. Glucose catabolism in cancer cells: amplification of the gene encoding type II hexokinase. Cancer Res. 1996;56:2468–2471. [PubMed] [Google Scholar]
- 48.Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776–22780. doi: 10.1074/jbc.272.36.22776. [DOI] [PubMed] [Google Scholar]
- 49.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto S, Murphy AN, Brown JH. Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ. 2008;15:521–529. doi: 10.1038/sj.cdd.4402285. [DOI] [PubMed] [Google Scholar]
- 51.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang R, Xiao T, Fang Z, Sun Y, Li F, Gao Y, et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J Biol Chem. 2012;287:23227–23235. doi: 10.1074/jbc.M112.373084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goel A, Mathupala SP, Pedersen PL. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J Biol Chem. 2003;278:15333–15340. doi: 10.1074/jbc.M300608200. [DOI] [PubMed] [Google Scholar]
- 54.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 55.Ren H, Tan ZP, Zhu X, Crosby K, Haack H, Ren JM, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–3323. doi: 10.1158/0008-5472.CAN-11-3931. [DOI] [PubMed] [Google Scholar]
- 56.Soda M, Isobe K, Inoue A, Maemondo M, Oizumi S, Fujita Y, et al. A prospective PCR-based screening for the EML4-ALK oncogene in non-small cell lung cancer. Clin Cancer Res. 2012;18:5682–5689. doi: 10.1158/1078-0432.CCR-11-2947. [DOI] [PubMed] [Google Scholar]
- 57.Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin Oncol. 2009;27:4232–4235. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukherjee A, Ma Y, Yuan F, Gong Y, Mohamed E, Fang Z, et al. Lysophosphatidic Acid Stimulates Proliferation of Ovarian Cancer Cells through Upregulation of Hexokinase 2 and Glycolytic Metabolism. Neoplasia. 2015;17:723–734. doi: 10.1016/j.neo.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, Mukherjee A, Lebman DA, Fang X. Gene expression of the lysophosphatidic acid receptor 1 is a target of transforming growth factor beta. Oncogene. 2013;32:3198–3206. doi: 10.1038/onc.2012.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.