Abstract
Acidithiobacillus caldus (A. caldus) is a common bioleaching bacterium that possesses a sophisticated and highly efficient inorganic sulfur compound metabolism network. Thiosulfate, a central intermediate in the sulfur metabolism network of A. caldus and other sulfur-oxidizing microorganisms, can be metabolized via the tetrathionate intermediate (S4I) pathway catalyzed by thiosulfate:quinol oxidoreductase (Tqo or DoxDA) and tetrathionate hydrolase (TetH). In A. caldus, there is an additional two-component system called RsrS-RsrR. Since rsrS and rsrR are arranged as an operon with doxDA and tetH in the genome, we suggest that the regulation of the S4I pathway may occur via the RsrS-RsrR system. To examine the regulatory role of the two-component system RsrS-RsrR on the S4I pathway, ΔrsrR and ΔrsrS strains were constructed in A. caldus using a newly developed markerless gene knockout method. Transcriptional analysis of the tetH cluster in the wild type and mutant strains revealed positive regulation of the S4I pathway by the RsrS-RsrR system. A 19 bp inverted repeat sequence (IRS, AACACCTGTTACACCTGTT) located upstream of the tetH promoter was identified as the binding site for RsrR by using electrophoretic mobility shift assays (EMSAs) in vitro and promoter-probe vectors in vivo. In addition, ΔrsrR, and ΔrsrS strains cultivated in K2S4O6-medium exhibited significant growth differences when compared with the wild type. Transcriptional analysis indicated that the absence of rsrS or rsrR had different effects on the expression of genes involved in sulfur metabolism and signaling systems. Finally, a model of tetrathionate sensing by RsrS, signal transduction via RsrR, and transcriptional activation of tetH-doxDA was proposed to provide insights toward the understanding of sulfur metabolism in A. caldus. This study also provided a powerful genetic tool for studies in A. caldus.
Keywords: RsrS-RsrR, two-component system, Acidithiobacillus caldus, sulfur metabolism, thiosulfate oxidation, S4I pathway, transcriptional regulation, cis regulatory element
Introduction
Sulfur oxidizing microorganisms, widely distributed within the chemoautotrophic bacteria and archaea (Goebel and Stackebrandt, 1994; Friedrich, 1997; Suzuki, 1999; Kletzin et al., 2004; Friedrich et al., 2005; Frigaard and Dahl, 2008; Ghosh and Dam, 2009), have evolved a variety of sulfur redox enzymes to metabolize elemental sulfur and various reduced inorganic sulfur compounds (RISCs). Thiosulfate, a central intermediate, plays a key role in inorganic sulfur metabolism in these sulfur oxidizers (Friedrich et al., 2005; Ghosh and Dam, 2009). It is metabolized mainly through the sulfur oxidizing (Sox) enzyme system and the tetrathionate intermediate (S4I) pathway. The Sox system, composed of SoxYZ, SoxAX, SoxB, and Sox(CD)2 (Friedrich et al., 2000, 2005), completely decomposes thiosulfate to sulfate without generating any sulfur intermediates. Many acidophiles (Friedrich et al., 2005; Ghosh and Dam, 2009; Williams and Kelly, 2013) have a truncated Sox system without Sox(CD)2 (Dahl and Prange, 2006). The alternate S4I pathway is widely found in chemoautotrophic genera including Acidithiobacillus, Thermithiobacillus, Halothiobacillus, and Tetrathiobacter (Dam et al., 2007; Ghosh and Dam, 2009). This pathway is made up of a thiosulfate:quinol oxidoreductase (Tqo or DoxDA) and a tetrathionate hydrolase (TetH). DoxDA oxidizes thiosulfate to tetrathionate, while TetH hydrolyzes tetrathionate to thiosulfate and other products (Hallberg et al., 1996; Ghosh and Dam, 2009). Thus, the Sox and S4I pathways play important roles in the metabolism of RISCs in sulfur-oxidizing microorganisms.
Acidithiobacillus caldus (A. caldus) is an obligate chemoautotrophic sulfur-oxidizing bacterium and one of the most abundant microorganisms in industrial bioleaching systems (Hallberg and Lindström, 1994, 1996; Rawlings, 1998; Dopson and Lindström, 1999). A. caldus possesses a truncated Sox system encoded by two sox clusters (sox-I and sox-II) and also has a typical S4I pathway encoded by a tetH cluster (Valdés et al., 2008, 2009; Chen et al., 2012). Furthermore, sulfur metabolism also occurs by other enzymes in this organism. A sulfur quinone oxidoreductase enzyme (SQR) is responsible for oxidation of hydrogen sulfide (Wakai et al., 2004). A sulfur oxygenase reductase (SOR) catalyzes the disproportionation of elemental sulfur to produce sulfite, thiosulfate, and sulfide (Kletzin, 1989, 1992). A sulfur dioxygenase (SDO) can oxidize the thiol-bound sulfane sulfur atoms (R-S-SH) which is activated from S8 (Rohwerder and Sand, 2003, 2007). It was proposed that the disulfide reductase complex (HdrABC) could catalyze sulfane sulfate (RSSH) to produce sulfite and regenerate RSH, following donation of electrons to the quinone pool (Quatrini et al., 2009). The Rhodanese (RHD) enzyme can transfer a sulfur atom from thiosulfate to sulfur acceptors such as cyanide and thiol compounds (Schlesinger and Westley, 1974; Gardner and Rawlings, 2000). Furthermore, two thiosulfate-transferring proteins, DsrE and TusA, react with tetrathionate to yield protein Cys-S-thiosulfonates, and trigger an irreversible transfer of thiosulfate from DsrE to TusA. This indicates that both these proteins are important players in the dissimilatory sulfur and tetrathionate metabolism (Liu et al., 2014). The tetH cluster of A. caldus includes ISac1, rsrR, rsrS, tetH, and doxD, which encode a transposase, a RsrS-RsrR two-component system (TCS), a tetrathionate hydrolase and a thiosulfate:quinol oxidoreduetase subunit, respectively (Rzhepishevska et al., 2007). The differences in the expression of TetH with different sulfur substrates and the location of the RsrS-RsrR system upstream of the tetH gene imply that a regulatory mechanism exists at the transcriptional level (Bugaytsova and Lindström, 2004; Rzhepishevska et al., 2007). However, up to now nothing is known about this potential mechanism. Additionally, we also found a σ54-dependent two-component system (named TspS-TspR), upstream of the sox-I cluster of A. caldus (unpublished data). The discoveries of TCSs in tetH and sox clusters of A. caldus indicated that TCSs are potentially involved in signal transduction from substrate sensing to subsequent transcriptional regulation of the sulfur-oxidizing genes. These TCS-dependent regulatory systems possibly allow A. caldus to adapt to a variety of sulfur energy sources in different growth environments.
TCSs are predominant signal transduction components used by prokaryotic microorganisms to convert rapid environmental changes into specific adaptive responses (Bourret and Silversmith, 2010; Capra and Laub, 2012; Lehman et al., 2015). They typically consist of a membrane-bound sensor histidine kinase (HK), which senses a specific environmental stimulus and undergoes autophosphorylation, and a cognate response regulator (RR), which receives the phosphoryl group via various phosphotransfer pathways and modulates gene transcription by binding to cis regulatory elements in the promoter region (Forst et al., 1989; Huang et al., 1997; Bilwes et al., 1999; Stock et al., 2000; Bourret and Silversmith, 2010).
The most effective way to study gene function in vivo is mutagenesis of the gene of interest. Gene transfer methods, conjugation, and electroporation, have been developed for A. caldus, and mutants were constructed by a marker replacement knockout method (Liu et al., 2007; Zyl et al., 2008; Chen et al., 2010, 2012). However, previously reported gene knockout methods are extremely difficult and not reproducible. Moreover, the antibiotic marker gene introduced into the mutants makes it difficult for creating multiple mutations and may lead to polar effects on downstream genes as well as cause potential biological safety issues in industrial applications. Therefore, the development of a reliable and markerless gene knockout method is of great significance for performing molecular biology research and genetic engineering in A. caldus.
In order to detect the regulatory mechanism of the S4I pathway, we analyzed the tetH cluster of the sulfur oxidation system in A. caldus. We developed an efficient markerless gene knockout system for A. caldus and successfully obtained knockout mutants of rsrR and rsrS. Physiological and transcriptional analyses of the mutants were carried out to uncover the regulatory mechanism of RsrS-RsrR on the S4I pathway.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The strain A. caldus MTH-04 has been deposited in the China General Microbiological Culture Collection Center (CGMCC) with the accession number CGMCC 1.15711. Liquid Starkey-S0 and -K2S4O6 inorganic media and solid Starkey-Na2S2O3 plates for A. caldus MTH-04 culture were prepared as reported previously (Jin et al., 1992). Elemental sulfur (S0; boiling sterilized, 8g/L) or K2S4O6 (membrane filtration, 3 g/L) were added prior to inoculation. Chloromycetin, kanamycin, and streptomycin were added to a final concentration of 34, 100, and 100 μg/mL in LB media, and at 60, 100, and 100 μg/mL in liquid and solid Starkey media. The culture conditions were 37°C, 200 r/min for Escherichia coli (E. coli), and 40°C, 150 r/min for A. caldus MTH-04 (Chen et al., 2012).
Table 1.
Strains and plasmids used in the study.
| Strains or plasmids | Genotype or description | Source or references |
|---|---|---|
| STRAINS | ||
| A. caldus MTH-04 | Isolated from Tengchong area, Yunnan province, China | Our lab |
| A. caldus MTH-04 ΔrsrR mutant | ΔrsrR | This study |
| A. caldus MTH-04 ΔrsrS mutant | ΔrsrS | This study |
| Escherichia coli DH5α | 80d lacZΔM15Δ(lacZYA-argF) U169 end A1 recA1 hsdR17(rk−,mk+) supE44λ-thi-1 gyr96 relA1 phoA | TransGen Biotech Corp. China |
| Escherichia coli SM10 | Thr leu hsd recA KmrRP4-2-Tc::Mu | Simon et al., 1983 |
| Escherichia coli BL21 (DE3) | F−ompT hdsSB(Rb−mB−) gal dgmmet(DE3) | TransGen Biotech Corp. China |
| PLASMIDS | ||
| pMD19-T | Apr, ColE1 replicon | TaKaRa Cor. |
| pSDUDI | suicide plasmid; Apr; Kmr; oriTRP4; multi-cloning sites | This study |
| pSDUDI::rsrR(UHA + DHA) | suicide plasmid for rsrR deletion | This study |
| pSDUDI::rsrS(UHA + DHA) | suicide plasmid for rsrS deletion | This study |
| pSDU1-tac | Cmr; IncQ, mob+, tac promoter | Our lab |
| pSDU1-I-SceI | Cmr; mob+; Ptac; containing I-SceI gene | This study |
| pET-22b | Ampr | Novagen Cor. |
| pJRD215 | Smr, Kmr; IncQ, Mob+ | Davison et al., 1987 |
| pJRD-P360IRS | Smr, Kmr; IncQ, Mob+; PtetH(360 bp); gusA | This study |
| pJRD-P148IRS | Smr, Kmr; IncQ, Mob+; PtetH(148 bp); gusA | This study |
| pJRD-P90 | Smr, Kmr; IncQ, Mob+; PtetH(90 bp); gusA | This study |
| RP4 | Apr, Tcr, Kmr; IncP, tra+ | Datta et al., 1971 |
| pACBSR | Cmr; I-Sce I, λ-Red recombination system | Herring et al., 2003 |
The wild type and mutant strains of A. caldus were initially grown on a solid Starkey-Na2S2O3 plate. One colony from each plate was inoculated into 10 mL Starkey-S0 liquid medium and grown to stationary phase. The saturated 10 mL culture was then transferred to 150 mL Starkey-S0 liquid medium and allowed to grow to stationary phase. Finally, cells in the 150 mL culture were collected by centrifugation at 12000 × g for 5 min and diluted with Starkey liquid medium to a final concentration of OD600 = 1.0. In order to measure growth or extract RNA, 1 mL of this culture was inoculated into 150 mL Starkey-S0 or -K2S4O6 liquid medium. Cell growth in Starkey-S0 medium was measured at OD600 after removal of elemental sulfur in the sample by low-speed centrifugation at 400 × g for 5 min (Yu et al., 2014). Only a small amount of cells (<1.5 %) were found attached to the sulfur particles, which were neglected in the cell growth measurement. All experiments were performed in triplicate.
Construction of plasmids pSDUDI and pSDU1-I-Sce I
To construct the basic suicide plasmid vector pSDUDI, the oriT region was initially amplified from the plasmid RP4 using primers oriT EcoR sen and oriT Sal ant and digested with EcoR I/Sal I. A ColE1-AmpR fragment was amplified by PCR from pMD19-T vector using primers pMD19 Sal sen and pMD19 EcoR ant and digested with EcoR I/Sal I. The two PCR products were ligated together to generate plasmid pMD-oriT. The plasmid pMD-oriT was then amplified by PCR using primers pMD19 Nde sen and oriT Apa ant to generate a linearized plasmid. This was digested with Nde I/Apa I, and ligated to Nde I/Apa I digested Km resistant gene amplified from plasmid pJRD215 using primers Km Apa sen and Km Nde ant. The resulting plasmid was the basic suicide plasmid pSDUDI. An I-Sce I recognition site (5′-TAGGGATAACAGGGTAAT-3′) and multiple cloning sites (MCS) were introduced into pSDUDI by PCR using primers Km Apa sen and pMD19 Nde sen/Km Nde ant, respectively. All primers used are listed in Table 2.
Table 2.
Primers used in construction of suicide plasmid and I-Sce I-expressing plasmid.
| Primer name | Primer Sequence (5′ → 3′) |
|---|---|
| oriT EcoR sen | ATTCCGGAATTCGCTCGTCCTGCTTCTCTTCG |
| oriT Sal ant | TCACGCGTCGACCGGGATTCAACCCACTCG |
| pMD19 Sal sen | TCACGCGTCGACGCGGTAATACGGTTATCCACAG |
| pMD19 EcoR ant | ATTCCGGAATTCAATGGTTTCTTAGACGTCAGGT |
| pMD19 Nde sen | TTATCATATGCAATTGAAGCTTGGTACCGCGGCTAGCGG |
| CCGCGCGGTAATACGGTTATCCACAG | |
| oriT Apa ant | TATAGGGCCCCGACCGGGATTCAACCCA |
| Km Apa sen | GTGAGGGCCCATAGGGATAACAGGGTAATAT GAATGTCA |
| GCTACTGG | |
| Km Nde ant | TTATCATATGACGCGTGGATCCGAGCTCTAGACTAG |
| TCGACAATCGAAATCTCGTGATGG | |
| pSDU1 Xba sen | CTTCATGCATTCTAGATCATGTTTGACAGCTTATC |
| tac Bam ant | TCGCGGATCCTCCTGCAGTGTTTCCTGTGTGAAATTG |
| I-Sce Bam sen | TTACGCGGATCCAGGAGGGTACCTATATGCATATGAAAAA |
| CATCAAAAAAAACC | |
| I-Sce Xba ant | TTCTTCTCTAGAACGTCGGGCCCTTATTTCAGGAAAGTTT |
| CGGAGGAG |
Restriction sites were indicated with underline.
The I-Sce I-expressing plasmid pSDU1-I-Sce I was constructed by generating a linearized form of the vector pSUD1-tac by PCR amplification with pSDU1 Xba sen and tac Bam ant. The I-Sce I gene was then amplified from plasmid pACBSR using primers I-Sce Bam sen and I-Sce Xba ant, digested with BamH I and Xba I, and ligated into the BamH I/Xba I treated linearized vector pSUD1-tac. The resulting plasmid was designated as pSDU1-I-Sce I.
Generation of knockout mutants
To generate the suicide plasmid for rsrR, the upstream and downstream homologous arms (UHA and DHA) of this gene were amplified using the primer pairs R1F/R1R and R2F/R2R, respectively. The two homologous arms were then linked using fusion PCR (Yon and Fried, 1989). Finally, the fused fragments were digested with Spe I and Not I, and ligated to Spe I/Not I digested plasmid pSDUDI, thus generating the suicide plasmid pSDUDI::rsrR (UHA + DHA).
The suicide plasmid for rsrS was constructed in a similar manner by amplifying the two homologous arms (UHA and DHA) of rsrS using S1F/S1R and S2F/S2R. The UHA, DHA, and plasmid pSDUDI were then digested with Spe I/Hind III, Hind III/Kpn I, and Spe I/Kpn I, respectively. The three digested fragments were finally ligated to each other, and transformed into E. coli DH5α and screened for the suicide plasmid pSDUDI::rsrS (UHA + DHA).
The suicide plasmids for both genes were verified by restriction enzyme digestion and sequencing.
The suicide plasmids were transformed into E. coli SM10, and the transformed E. coli SM10 were conjugated with A. caldus as described earlier (Liu et al., 2007). After the first homologous recombination, the single crossover mutants were selected for kanamycin resistance on Starkey-Na2S2O3 solid plates. Single recombination events were rapidly identified by PCR using R4F/R and S4F/R primers for rsrR and rsrS, respectively. The I-Sce I-expressing plasmid (pSDU1-I-Sce I) was then transferred into the single crossover mutants to induce a second homologous recombination, thereby generating the target mutants. The ΔrsrR and ΔrsrS strains were identified by PCR based screening using R4F/R and S4F/R primers. Finally, the PCR fragments amplified from ΔrsrR and ΔrsrS were sequenced using primers R5F/R and S5F/R, respectively to confirm their identity. All primers used in this section are listed in Table S1.
Elimination of the I-Sce I expression plasmid was achieved by spontaneous loss. The mutant containing I-Sce I expression plasmid was inoculated into the non-selective liquid Starkey-S0 medium and grown to stationary phase. An aliquot from the culture was diluted and plated on non-selective solid Starkey-Na2S2O3 medium. Colony PCR was carried out to screen for loss of the plasmid using the primers RepA sen and RepC, which are also listed in Table S1. About 3–5 consecutive transfers were carried out to obtain the ΔrsrR and ΔrsrS mutants without the I-Sce I expression plasmid.
Real-time quantitative PCR
For RNA isolation, the culture was filtered through filter papers with a pore size of 15–20 μm to remove the sulfur powder, after which the cells were harvested by centrifugation at 12000 × g under refrigerated conditions. RNAprotect Bacteria Reagent (Qiagen Cor.) was added to resuspend the cells and inhibit changes to RNA transcripts. The suspension was mixed immediately by vortexing for 5 s, incubated for 5 min at room temperature, and centrifuged for 10 min at 5000 × g. The supernatant was discarded and the harvested cells were stored at −70°C. The RNAs were extracted by using RNAiso Plus kit (TaKaRa Cor.) according to the manufacturer's instructions. Reverse transcription was performed using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa Cor.). One microgram of total RNA was used for every 20 μL reverse transcription reaction system to obtain cDNA under the following reaction conditions: 42°C for 2 min, 37°C for 15 min, and 85°C for 5 s. The cDNAs from various cultures were used with SYBR® Premix Ex TaqTM (TliRNaseH Plus; TaKaRa Cor.) for real-time quantitative PCR reactions, which were performed using LightCycler® 480 (Roche). Two-hundred nanograms of cDNA was used in a 20 μL RT-qPCR reaction. The conditions for qPCR were as follows: 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 60°C for 30 s, and a final cycle at 95°C for 5 s, 60°C for 1 min and 95°C with continuous mode. The data and fold change were calculated using the LightCycler® 480 software. The primers used in this assay were designed using PRIMER PREMIER 5 software (PREMIER Biosoft Int., Palo Alto, CA, USA) and are listed in Table S2. The gapdH gene of A. caldus, encoding glyceraldehyde-3-phosphate dehydrogenase, was used as the reference gene for normalization (Livak and Schmittgen, 2001). Relative expression was calculated using the comparative ΔΔCT method, and the values were expressed as 2−ΔΔCT (Livak and Schmittgen, 2001). Three independent replicates were performed for each experiment. The values shown in this study are the means of three independent replicates showing fold changes (FC). FC ≥ 2, P ≤ 0.05 and FC ≤ 0.5, P ≤ 0.05 were regarded as significant changes, designated as up-regulation and down-regulation, respectively. No significant change was inferred when 0.5 ≤ FC ≤ 2, P ≥ 0.05. The SD-value was calculated using Origin software with “descriptive statistics,” and the P-value was calculated by using GraphPad Prism software with “unpaired t-test.”
EMSA assays
The expression and purification of RsrR was performed as described. The rsrR gene of A. caldus MTH-04 was amplified by PCR using primers rsrR-F and rsrR-R listed in Table S3. The PCR product was cloned into plasmid pET-22b, generating the expression plasmid pET-22b-rsrR. A positive clone was verified by sequencing, and transformed into E. coli BL21 (DE3). RsrR was purified using HisTrap HP column (GE Healthcare), and the concentration of the purified protein was determined using the Bradford assay.
DNA fragments were obtained by PCR amplification using different sets of primers listed in Table S3. The G360, T360, T148, and T90 fragments were obtained using primer-pairs G360-F and G360-R, T360-F and T360/148/90-R, T148-F and T360/148/90-R, and T90-F and T360/148/90-R, respectively. The G360+58IRS fragment was obtained after two rounds of PCR. The first round of PCR was performed with G360 fragment as the template using the primer-pair G360-F and G360+58IRS–R1. In the second round of PCR, the PCR product from the first round was used as the template with the primer-pair G360-F and G360+58IRS–R2, to generate the G360+58IRS fragment. The T360Δ19 fragment was obtained by fusion PCR. Two fragments were generated by PCR from the T360 fragment using the primer-pairs T360-F and T360Δ19-R, and T360Δ19-F1 and T360/148/90-R. Fusion PCR was then performed using the two generated fragments as templates with the primer pair T360-F and T360/148/90-R to obtain the T360Δ19 fragment without the 19 bp IRS. After PCR amplification, the amplified fragments were purified using QIAquick Gel Extraction Kit (Qiagen Corp.), desalted, and concentrated using ultrafiltration. Ultrafiltration was carried out using Amicon Ultra-15 mL, 3 kDa Centrifugal Filter Unit (Millipore Corp.) at 5000 × g in a refrigerated centrifuge.
EMSA assays were performed as described in Pardo and Orejas (2014). Initially, a 15 μL reaction mixture containing 1.5 μL 10 × Binding Buffer, 1 μg of salmon sperm DNA, and 1 μg RsrR was mixed and used as the reaction solution. Reaction solution containing PBS (phosphate buffer saline) instead of RsrR was used as a negative control. Both solutions were prepared and incubated at room temperature for 20 min. Two-hundred nanogram of DNA fragments were then added to both mixtures and incubated at room temperature for another 20 min. The DNA-protein complexes in both reactions were separated on 6.5% nondenaturing polyacrylamide gels in 0.25 × TBE (22.25 mM Tris-Boric acid, 0.5 mM EDTA) on ice (150 volts for 1.5 h). Visualization of the bands was done using ethidium bromide staining as described earlier (Bidart et al., 2014).
The primers used for constructing the three IRS-probe vectors and the primers that were used to verify the function of the 19bp-IRS in vivo are listed in Table S4.
Gene sequences
The nucleotide sequences of rsrR and rsrS have been deposited with GenBank accession numbers KX161704 and KX161705, respectively. This Whole Genome Shotgun project for A.caldus MTH-04 has been deposited at DDBJ/ENA/GenBank under the accession LXQG00000000.
Results
Analysis of tetH clusters in A. caldus and other sulfur oxidizers
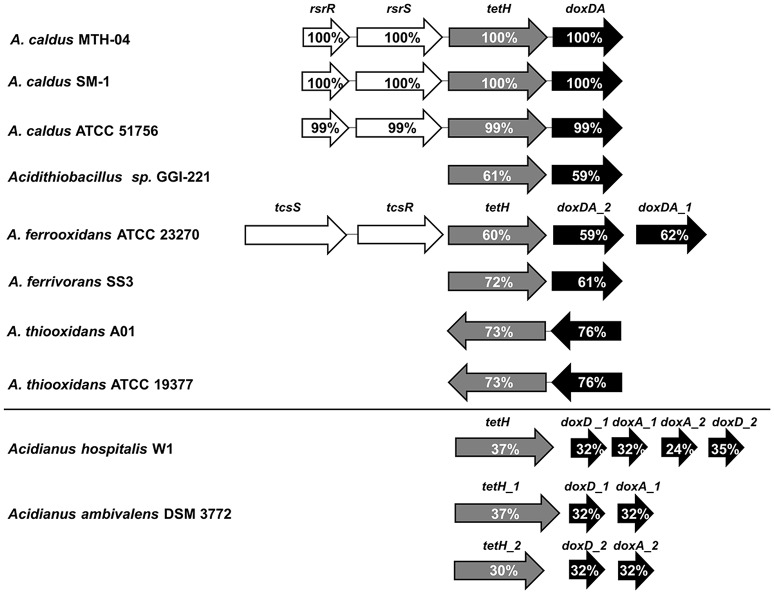
The tetH clusters in various bacteria and archaea were compared for analysis. As shown in Figure 1, the two functional genes (tetH and doxDA) of the S4I pathway were distributed in chemoautotrophic sulfur oxidizers including bacteria and archaea of the genera Acidithiobacillus and Acidianus, respectively. However, only A. caldus and Acidithiobacillus ferrooxidans have a specific two-component system (TCS) upstream the tetH gene. The cluster of A. caldus MTH-04 was identical to that of A. caldus SM-1 and shared 99% identity with that of A. caldus ATCC 51756. Furthermore, the tetH cluster in Acidithiobacillus sp. GGI-221 has certain components including tetH and doxDA with 61 and 59% identity, respectively. The 16S rRNA gene sequence of Acidithiobacillus sp. GGI-221 shows 99.7% identity to Acidithiobacillus ferrooxidans indicating that this is a strain of Acidithiobacillus ferrooxidans (Williams and Kelly, 2013). However, the TCS of A. ferrooxidans is σ54-dependent and has a different order from tcsS to tcsR encoding the sensor histidine kinase and the cognate response regulator when compared to A. caldus. The tetH and doxDA genes are arranged in a cluster in A. caldus and Acidithiobacillus thiooxidans, while they were separately distributed in the genomes of Acidianus and other Acidithiobacillus spp. Two copies of doxDA genes are located separately in the genomes of A. ferrooxidans and Acidianus hospitalis W1, while in Acidianus ambivalens DSM 3772 there are two copies of tetH. Moreover, doxDA in Acidithiobacillus spp. encodes a protein with two domains DoxD and DoxA, but DoxD and DoxA in Acidianus hospitalis W1 and Acidianus ambivalens DSM 3772 are two individual subunits encoded by two separate genes, doxD and doxA (Müller et al., 2004; Valenzuela et al., 2008; Valdés et al., 2009; Mangold et al., 2011). BLAST and multiple alignment (Figure S1) results demonstrated that the two subunits DoxD and DoxA are fused as one protein in the three A. caldus strains, MTH-04, SM-1, and ATCC 51756, indicating that the doxD gene in this tetH cluster encodes a protein that has two domains corresponding to DoxD and DoxA (Müller et al., 2004). Thus, we renamed the doxD gene in these strains as doxDA or tqo.
Figure 1.
Comparison of tetH clusters in various bacteria and archaea. rsrR and tcsR, response regulators of TCSs (two-component systems); rsrS and tcsS, histidine kinases of TCSs; tetH, tetrathionate hydrolase; doxDA, thiosulfate:quinone oxidoreductase. The percentages of amino acid similarity between proteins and their homologs are indicated. The database of National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/ was used in this study. NCBI Taxonomy IDs for Acidithiobacillus caldus SM-1, Acidithiobacillus caldus ATCC 51756, Acidithiobacillus sp. GGI-221, Acidithiobacillus ferrooxidans ATCC 23270, Acidithiobacillus ferrooxidans SS3, Acidithiobacillus thiooxidans A01, Acidithiobacillus thiooxidans ATCC 19377, Acidianus hospitalis W1, and Acidianus ambivalens DSM 3772 are 990288, 637389, 872330, 243159, 743299, 1432062,637390, 933801, and 2283, respectively. Accession numbers (GenBank) for these proteins are: A. caldus MTH-04, RsrR (ANJ65973.1), RsrS (ANJ65974.1), TetH (OAN03451.1), DoxDA (OAN03452.1); A. caldus SM-1, RsrR (AEK59530.1), RsrS (AEK58242.1), TetH (AEK58243.1), DoxDA (AEK58244.1); A. caldus ATCC 51756, RsrR (ABP38227.1), RsrS (ABP38226.1), TetH (ABP38225.1), DoxDA (ABP38224.1); Acidithiobacillus sp. GGI-221, TetH (EGQ60847.1), DoxDA (EGQ62792.1); A. ferrooxidans ATCC 23270, TcsS (WP_041646693.1), TcsR (WP_012535753.1), TetH (ACK80599.1), DoxDA_2 (ACK79881.1), DoxDA_1 (ACK78481.1); A. ferrooxidans SS3, TetH (AEM46280.1), DoxDA (AEM47534.1); A. thiooxidans A01, TetH (WP_024894935.1), DoxDA (WP_024894934.1); A. thiooxidans ATCC 19377, TetH (WP_029316048.1), DoxDA (WP_010638552.1); Acidianus hospitalis W1, TetH (AEE94548.1), DoxD_1 (AEE93006.1), DoxA_1 (AEE93005.1), DoxA_2 (AEE93131.1), DoxD_2 (AEE93130.1); Acidianus ambivalens DSM 3772, TetH_1 (CBY66038.1), DoxD_1 (CAA69986.1), DoxA_1 (CAA69987.1), TetH_2 (CBY66040.1), DoxD_2 (CAA70827.1), DoxA_2 (CAA70828.1).
Development of a markerless gene knockout system to construct ΔRsrS and ΔRsrR
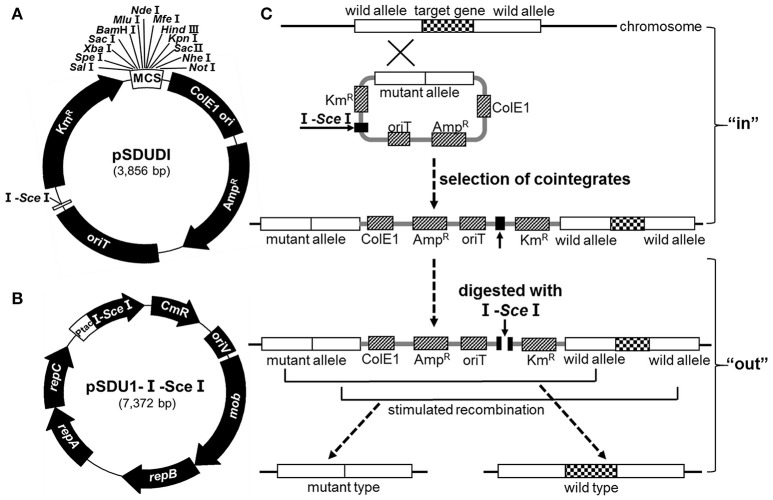
A mobile suicide vector (pSDUDI, Figure 2A) was employed to introduce homologous sequences of the target genes into A. caldus cells, and to integrate these homologous sequences into the genome by homologous recombination, thus generating cointegrates (Figure 2C). The backbone of the suicide plasmid was derived from pUC19 and therefore cannot replicate in A. caldus. Second, the origin of transfer (oriT) of plasmid RP4 was cloned into this plasmid, which allows it to be mobilized into A. caldus with high efficiency. Third, an 18 bp I-Sce I endonuclease recognition site (5′-TAGGGATAACAGGGTAAT-3′) was also introduced into this plasmid to facilitate cleavage of the cointegrate by I-Sce I endonuclease.
Figure 2.
The markerless gene knockout strategy in A. caldus. (A) Map of the suicide plasmid pSDUDI. ColE1, the replication origin; oriT, the origin of transfer; I-SceI, endonuclease recognition site. (B) Plasmid pSDU1-I-Sce I. I-Sce I endonuclease gene was under the control of Ptac promoter. (C) General scheme of the markerless gene knockout method. At the “in” step, the suicide plasmid carrying homologous sequences specific to the targeted gene was conjugated into A. caldus, and integrated into the chromosome with the help of Rec homologous recombination system in A. caldus. At the “out” step, the I-Sce I endonuclease encoded by plasmid pSDU1-I-Sce I created the double-stranded breaks (DSBs) on the chromosome, which acted as the signal to stimulate a second recombination event, generating either a wild type or a mutant type.
The I-Sce I-expressing plasmid (pSDU1-I-Sce I) shown in Figure 2B was derived from pJRD215, which can replicate and remain stable in A. caldus. The Ptac promoter was introduced into this plasmid to express I-Sce I in A. caldus. Mobilization of pSDU1-I-Sce I into the cointegrate of A. caldus would lead to the generation of double-stranded breaks (DSBs) at the I-Sce I site. The subsequent second homologous recombination event, would ultimately lead to generation of the mutant or wild type strains (Figure 2C).
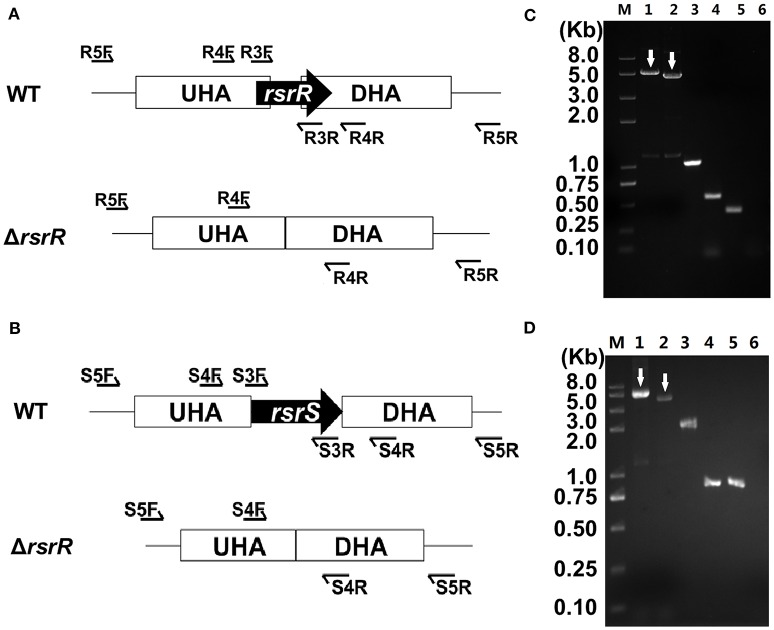
Verification of the ΔrsrR and ΔrsrS strains is shown in Figure 3. Smaller fragments were amplified from ΔrsrR and ΔrsrS strains compared to the wild type using primers R4F/R and S4F/R (lane 1, 5.1 kb; lane 2, 4.6 kb; lane 3, 1.0 kb, and lane 4, 551 bp), and R5F/R and S5F/R (lane 1, 5.0 kb; lane 2, 3.9 kb; lane 3, 2.1 kb, and lane 4, 910 bp; Figures 3C,D lanes 1–4). No band could be amplified from rsrR and rsrS knockout strains using primers R3F/R and S3F/R (Figures 3C,D lanes 6), while there were 413 and 850 bp bands for rsrR and rsrS genes, respectively in the wild type as shown in Figures 3C,D lanes 5. The sizes of the observed PCR bands were as expected in different strains. The precise sequences of the two mutants were confirmed by sequencing the PCR fragments (Figures 3C,D lane 2) derived from ΔrsrR and ΔrsrS.
Figure 3.
Confirmation of rsrR and rsrS mutants by PCR analyses. (A,B) Diagram of three sets of primers specific to target genes (rsrR and rsrS), the upstream and downstream homologous arms (UHA and DHA) and the sequences outside of homologous arms, respectively. (C,D) PCR analyses of the chromosomes of ΔrsrR and ΔrsrS. Lane 1 and 2, PCR amplifications from wild type and mutants using primers R5F/R or S5F/R, respectively; lane 3 and 4, PCR amplifications from wild type and mutants using primers R4F/R or S4F/R, respectively; lane 5 and 6, PCR amplifications from wild type and mutants using primers R3F/R or S3F/R, respectively. In (C,D), the arrows are added to indicate the target bands.
Transcriptional changes of the tetH cluster
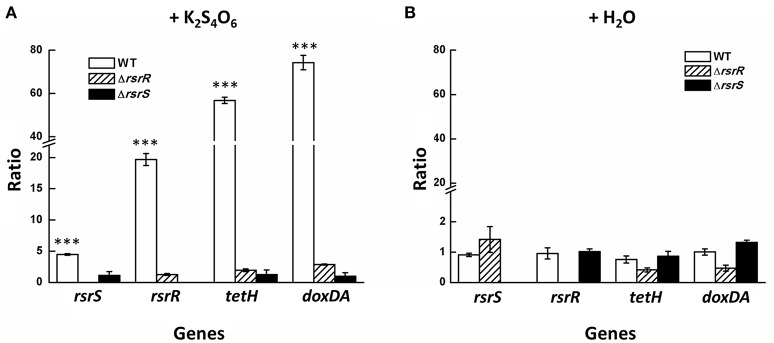
To verify the influence of the lack of RsrR or RsrS on the S4I pathway, relative RNA transcript levels of genes in the tetH cluster of ΔrsrR, ΔrsrS, and the wild type strains were tested by RT-qPCR. Strains were grown to mid-log phase in S0-medium (at the 4th day), followed by addition of equal volumes of either K2S4O6 or H2O to the stimulation and control groups, respectively. Cells at the 4th and 4.5th days were collected to purify total RNA for transcriptional analysis. The ratio of relative RNA transcripts levels of the genes at the two time points were calculated and are shown in Figure 4. After stimulation with exogenous K2S4O6 at a final concentration of 3 g/L, the relative RNA transcript levels of rsrS, rsrR, tetH, and doxDA in the wild type increased by about 5, 20, 60, and 80 fold, respectively (Figure 4A). However, the relative transcript levels of these genes in the two mutants did not show any obvious increase (0.5 ≤ ratio ≤ 2.0; Figure 4A). In the control group, addition of water did not result in a significant change in expression of any of the four genes in the three strains (0.5 ≤ ratio ≤ 2.0; Figure 4B). This result supported the notion that there is a K2S4O6-dependent positive regulation of RsrS-RsrR on tetH-doxDA.
Figure 4.
Relative RNA transcripts level changes of tetH cluster in ΔrsrR, ΔrsrS, and the wild type in S0-medium with the addition of K2S4O6 (A) and H2O (B). The addition of the same volume H2O was set as the control group. Asterisks denote statistically significant changes (***P ≤ 0.001).
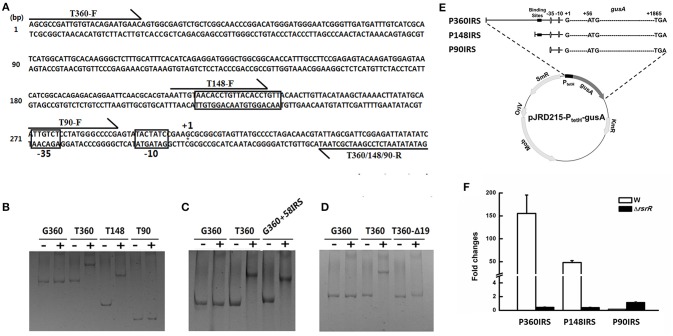
Determination of the Cis-regulatory element of RsrR
Positive control of the cotranscription of tetH and doxDA (Rzhepishevska et al., 2007) is likely achieved by binding of RsrR to a cis-regulatory element at the promoter region of tetH. To confirm the above hypothesis, three fragments about 360, 148, and 90 bp upstream of the “ATG” of tetH were amplified to test for their interaction with RsrR by electrophoretic mobility shift assays (EMSAs) (Figure 5A). A 360 bp fragment amplified from the gapdH gene of A. caldus was used as a negative control. The results showed that the binding region is located in a 58 bp region between 90 and 148 bp upstream of “ATG” (Figure 5B). When the 58 bp region was fused with the 360 bp gapdH fragment, the fusion could bind to RsrR (Figure 5C), indicating that this cis-regulatory element was located in the 58 bp region. To further narrow down the binding region of RsrR, the software package Repeat Around-2.1 was used to analyze this region and a 19 bp (AACACCTGTTACACCTGTT) inverted repeat sequence (IRS) within the 58 bp region was predicted to be the binding sequence (Figure 5A). Upon removal of the 19 bp-IRS from the 360 bp-fragment upstream of tetH, binding of RsrR could not be observed (Figure 5D), which suggested that the 19 bp-IRS was the key binding site of RsrR.
Figure 5.
The functional identification of a 19-bp inverted repeat sequence (IRS) upstream the tetH promoter. (A) The loci of IRS, −10/−35 regions, the transcriptional start site and primers on the sequence upstream tetH gene. (B–D) The results of EMSAs to determine the binding ability of RsrR to the regulatory sequence. −, the group containing the nucleotide fragments without RsrR; +, the group containing both the different nucleotide fragments and RsrR. G360, a 360 bp-fragment from the gapdH gene used as a control; T360, T148 and T90, a 360 bp-, 148 bp-, or 90 bp-fragment amplified from upstream region of tetH; G360+58IRS, a 58 bp sequence containing the IRS fused with the 360 bp-fragment from gapdH; T360Δ19: the T360 fragment with IRS removed. (E) Diagram of series of IRS-probe vectors containing various versions of the upstream region of tetH. (F) Transcriptional analysis of gusA on the IRS-probe vectors in ΔrsrR and the wild type.
To verify the function of the 19 bp-IRS on the transcription of tetH and doxDA in vivo, the 360, 148, and 90 bp fragments upstream of the “ATG” of tetH were fused to the reporter gene gusA to generate three IRS-probe vectors, as shown in Figure 5E. The resulting plasmids were designated as pJRD-P360IRS, pJRD-P148IRS, and pJRD-P90, respectively and were mobilized into wild type and ΔrsrR strains. All strains were grown in S0-medium, and the relative RNA transcript levels of gusA in each strain were measured after stimulation with K2S4O6. As shown in Figure 5F, a significantly low level of gusA transcript was observed in the wild type strain harvesting plasmid pJRD-P90 when compared with that with pJRD-P360IRS and pJRD-P148IRS, indicating that the 19 bp-IRS had a positive effect on the transcription of tetH promoter. The relative gusA RNA transcript levels from plasmids pJRD-P360IRS and pJRD-P148IRS in ΔrsrR were much lower than those in the wild type strain, indicating that IRS is needed for the positive effect of RsrR.
Structural simulation and protein sequence alignment between RsrS-RsrR and EnvZ-OmpR were also carried out in order to help understand the mechanism of regulation of the tetH cluster by RsrS-RsrR, which showed that the two TCSs are highly identical at the protein level (Figure S2).
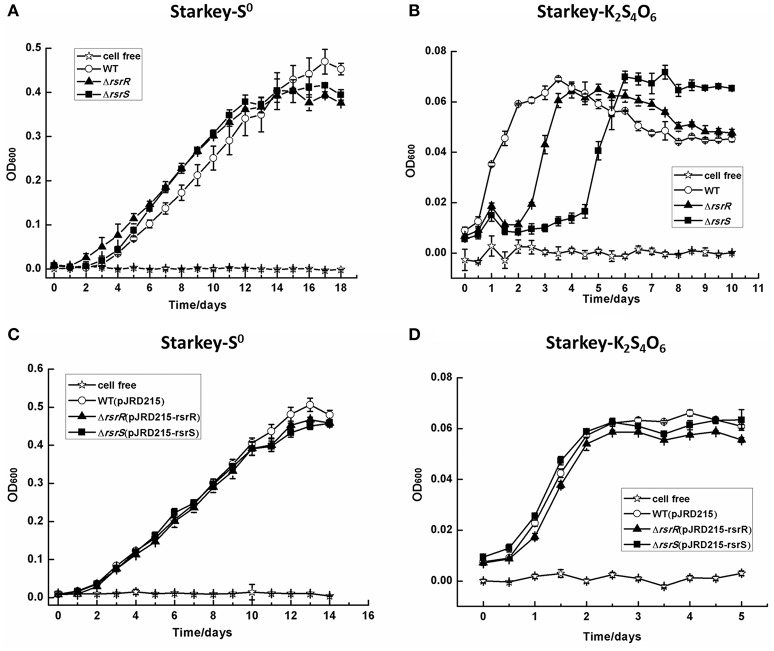
Growth analyses of the mutants in different sulfur-substrate media
As shown in Figure 6, the growth rates of the rsrR or rsrS knockout mutants were not similar to the wild type. When grown in S0-medium, both ΔrsrR and ΔrsrS had a slight growth advantage over the wild type strain from the 5th to the 11th day, which corresponds to the logarithmic growth phase prior to entering into the stationary phase. In contrast, the wild type strain grew slightly better than the mutants after the 15th day (Figure 6A). When K2S4O6 was used as the sole sulfur substrate, the three strains showed very slow overall growth (up to 0.06) compared to S0-medium due to differences in RISCs metabolism for different substrates (Zyl et al., 2008; Chen et al., 2012; Zhang et al., 2014). In K2S4O6-medium, ΔrsrR and ΔrsrS had a 2 and 4 day growth delay in lag phase, respectively when compared to the wild type. However, the three tested strains reached approximately the same growth when they reached stationary phase (Figure 6B). In addition, complementation of the mutations with wild type alleles resulted in a growth pattern similar to the wild type in both media (Figures 6C,D). Therefore, the recovery of growth of the mutants in K2S4O6-medium after a delay of several days suggested that RsrS-RsrR might be the primary, but not the sole signal transduction pathway that regulates the metabolism of tetrathionate.
Figure 6.
The growth curves of the wild type, ΔrsrR, and ΔrsrS mutants, and complemented strains of A. caldus MTH-04 grown in Starkey medium to which were added different energy sources. All the measurements were performed in triplicate. The values for OD600 are the mean of the three independent replicates. The SD-values are shown in the figure with short bars on the top of the columns, and they were calculated by using the Origin software with “descriptive statistics.” (A,C) Growth in Starkey-S0 medium; (B,D) Growth in Starkey-K2S4O6 medium. WT, ΔrsrR, and ΔrsrS, WT(pJRD215), ΔrsrR(pJRD215-rsrR), and ΔrsrS(pJRD215-rsrS) represent the wild type, mutants, wild type carrying plasmid pJRD215, and complemented strains of A. caldus MTH-04, respectively.
Influence of RsrR and RsrS on sulfur metabolism and signaling systems
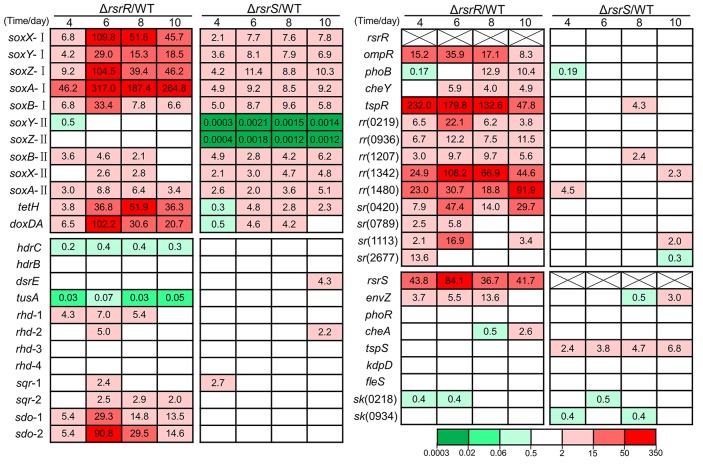
In A. caldus, the sulfur-oxidizing mechanisms include the periplasmic Sox system, the S4I pathway, and the RISCs oxidation enzymes such as SOR, SDO, SQR, HDR, RHD, DsrE, and TusA (mentioned in the Introduction Section). To investigate the effect of the absence of RsrS and RsrR on the S4I pathway, we analyzed the relative RNA transcript levels of genes attributed to play a role in the A. caldus RISCs metabolism. Cross-talk often occurs between a sensor kinase and a non-cognate response regulator in the TCSs (Procaccini et al., 2011), so genes encoding single- and two-component systems (SCSs and TCSs) were also analyzed for their relative RNA transcript levels. The transcriptional analysis of these sulfur-oxidizing and regulatory genes during cultivation on S0 was carried out by RT-qPCR. All data were calculated and the statistically valid data were showed as the mean value of three independent replicates in Figure 7 (original data with values for standard deviation and P-value are listed in Supplementary Text S1). As shown, several genes in the mutants had significant changes in expression (FC ≥ 2, P ≤ 0.05, up-regulation; FC ≤ 0.5, P ≤ 0.05, down-regulation) when compared with the wild type. The deletion of rsrR resulted in a sharp up-regulation (FC ≥ 6, P ≤ 0.05) of tetH-doxDA, sox operon I, sdo-1, and sdo-2, clear down-regulation (FC ≤ 0.01, P ≤ 0.05) of tusA, and weak transcriptional changes (2 ≤ FC ≤ 6, P ≤ 0.05) in sox operon II, sqr, and rhd. The absence of rsrR also resulted in significant up-regulation (FC ≥ 2, P ≤ 0.05) of a majority of regulator genes in TCSs including ompR (A5904_2590), phoB (A5904_0374), cheY (A5904_1450), tspR (A5904_2485), A5904_0219, A5904_0936, A5904_1207, A5904_1342 and A5904_1480, and in SCSs including A5904_0420, A5904_0789 and A5904_1113. The sensor histidine kinase (HK) genes rsrS and envZ in TCSs are up-regulated significantly in the rsrR knockout strain. However, the HK genes phoR (A5904_0373), cheA (A5904_1448), tspS (A5904_2484), kdpD (A5904_1340), fleS (A5904_1479), A5904_0218 and A5904_0934 had no obvious transcriptional changes in this mutant. In the ΔrsrS strain, the genes of the Sox operon (except soxYZ in operon II), tetH, and doxDA showed significant changes in expression when compared to the wild type. The expression of soxYZ in operon II was reduced to almost undetectable levels. Almost all the other genes involved in the sulfur oxidation system, TCSs, and SCSs did not show significant changes. However, tspS showed a significant change (2 ≤ FC ≤ 7, P ≤ 0.05) in the ΔrsrS strain.
Figure 7.
The relative transcription levels of genes involved in sulfur metabolism and signaling systems during the S0-cultivating process. This is the valid mean value of three independent replicates showing fold changes (FC) determined by RT-qPCR analyses of the mutant against the wild-type. FC ≥ 2, P ≤ 0.05 and FC ≤ 0.5, P ≤ 0.05 were regarded as the significant changes. FC ≥ 2, P ≤ 0.05, up-regulation; FC ≤ 0.5, P ≤ 0.05, down-regulation; 0.5 ≤ FC ≤ 2, P ≥ 0.05, no change (data are not shown in the figure). FC-values are represented with different colors as indicated in the color bar. The data of standard deviation (SD) and P-value were shown in Supplementary Text S1. The SD-value was calculated by using the Origin software with “descriptive statistics.” The P-value was calculated by using the GraphPad Prism software with “unpaired t-test.” The original data with the values for standard deviation and P-value are listed in Supplementary Text S1. The putative function of proteins encoded by these genes: soxX-I (A5904_2486), soxX-II (A5904_2525), cytochrome c class I; soxY-I (A5904_2487), soxY-II (A5904_2520), sulfur covalently binding protein; soxZ-I (A5904_2488), soxZ-II (A5904_2521), sulfur compound chelating protein; soxA-I (A5904_2489), soxA-II (A5904_2526), cytochrome c (diheme); soxB-I (A5904_2491), soxB-II (A5904_2522), sulfate thiol esterase; hdrC (A5904_1042), hdrB (A5904_1043), heterodisulfide reductase subunit C and B; dsrE (A5904_2473), tusA (A5904_2474), sulfur transferase; rhd-1 (A5904_0894), rhd-2 (A5904_1407), rhd-3 (A5904_2860), rhd-4 (A5904_2475), rhodanese (sulfur transferase); sqr-1 (A5904_1436), sqr-2 (A5904_2678), sulfide quinone reductase; sdo-1 (A5904_0421), sdo-2 (A5904_0790), sulfur dioxygenase; envZ (A5904_2589), ompR (A5904_2590), osmolarity regulation; phoB (A5904_0374), phoR (A5904_0373), phosphate regulon; cheY (A5904_1450), cheA (A5904_1448), chemotaxis; tspR (A5904_2485), tspS (A5904_2484), regulation for Sox pathway; kdpD (A5904_1340), unknown; fleS (A5904_1479), flagellum associated; rr (A5904_0219, A5904_0936, A5904_1207, A5904_1342, A5904_1480), putative response regulators in TCSs; hk (A5904_0218, A5904_0934), putative sensor histidine kinases in TCSs; sr (A5904_0420, A5904_0789, A5904_1113, A5904_2677), putative regulators in SCSs.
Discussion
The lack of a reliable gene knockout method for A. caldus has been a significant obstacle in the progress of uncovering the sulfur oxidation mechanism and other important physiological functions in this organism (Valdés et al., 2009; You et al., 2011; Chen et al., 2012). In this study, we developed a markerless gene knockout technique using a suicide plasmid and an I-Sce I-expressing plasmid. There are two advantages of our markerless gene knockout technique. The first is that we used an endogenous Rec (RecA and RecBCD) system of A. caldus, rather than an exogenous λ-Red or RecET system, for homologous recombination to avoid the incompatibility of an exogenous recombination system. The second advantage is that we used conjugation as our gene transfer method, which is the optimal way to incorporate plasmids into cells because conjugation facilitates homologous recombination between the suicide plasmid and the host chromosome (Dillingham and Kowalczykowski, 2008). Furthermore, this knockout technique can be further optimized to integrate genes or other sequences into A. caldus.
A. caldus has a complex sulfur oxidation system that includes periplasmic sulfur-oxidizing pathways (Sox and S4I) and cytoplasmic sulfur-oxidizing enzymes (SDO, SOR, HDR etc.) (Chen et al., 2012). The sor gene in the wild type and both mutants of A. caldus MTH-04 was lost during the long period of subcultivation in S0-medium under the laboratory conditions (as confirmed by sequence analysis of PCR products). The loss of sor is probably caused by transposition of the transposon as has been reported for the strain A. caldus SM-1 (You et al., 2011). The sulfur oxidation pathways in the two cellular compartments are probably connected by tetrathionate, which can enter the cytoplasm and react with DsrE or TusA to generate protein Cys-S-thiosulfonates, thus initiating sulfur metabolism in the cytoplasm (Liu et al., 2014). Comparative analysis of the S4I pathway genes tetH and doxDA in Acidithiobacillus spp. and the archaea Acidianus hospitalis and Acidianus ambivalens, indicated that only A. caldus evolved an rsrRS-tetH-doxDA-like cluster (Figure 1). The combination of two functional genes (tetH and doxDA) and the TCS regulatory genes (rsrR and rsrS) in this cluster potentially allows A. caldus to regulate its S4I pathway via the RsrS-RsrR system. Thus, A. caldus can efficiently maintain the balance between thiosulfate and tetrathionate in the periplasm and modulate the periplasmic and cytoplasmic sulfur-oxidizing pathways.
The positive regulatory role of RsrS-RsrR on the S4I pathway was inferred from the transcriptional analysis of the tetH cluster in the rsrR and rsrS knockout mutants and the wild type upon stimulation with K2S4O6. The relative transcriptional levels of genes in the tetH cluster in the rsrR or rsrS knockouts were much lower as compared to that in the wild type when stimulated with K2S4O6 (Figure 4A), indicating a positive effect of RsrS-RsrR on the transcriptional regulation of tetH and doxDA. Thus, RsrS-RsrR linked the signal from tetrathionate to the transcription of tetH and doxDA, which allowed adjustment of the S4I pathway in A. caldus to utilize tetrathionate in the growth environment.
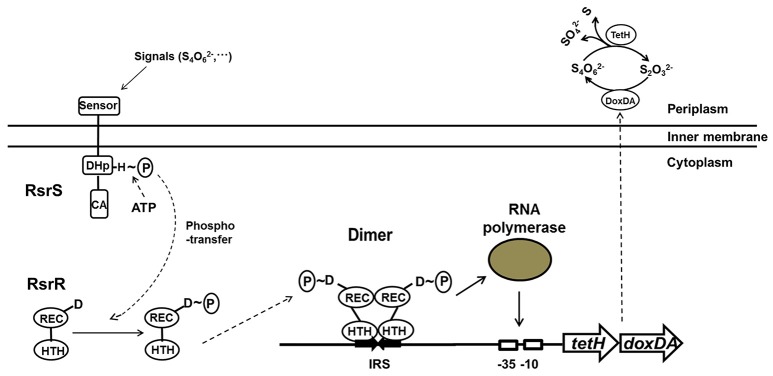
The determination of the positive regulation of RsrS-RsrR for S4I pathway, combined with simultaneous transcription of tetH-doxDA and the P1 promoter upstream of tetH (Rzhepishevska et al., 2007), indicated the existence of a cis-regulatory element in A. caldus. The data from the EMSA assays in vitro and the promoter-probe vector analysis in vivo revealed direct interaction between RsrR and the IRS upstream of the tetH promoter, along with the effects of the IRS on the transcriptional activity of the promoter. The 19 bp-IRS (AACACCTGTTACACCTGTT) is composed of two 9 bp complementary inverted half-sites (AACACCTGT and ACACCTGTT) with a 1-bp interval (T). The RsrS-RsrR in A. caldus is an EnvZ-OmpR like TCS (Rzhepishevska et al., 2007), in which the response regulators (RsrR and OmpR) share 42% identity and the sensor histidine kinases (RsrS and EnvZ) share 30% identity at the amino acid level. The high level of identity in the structures and protein sequences between RsrS-RsrR and EnvZ-OmpR indicate that RsrR is a typical regulator with a winged helix-turn-helix (HTH) DNA-binding domain, allowing the RsrR dimer to interact with the 19 bp IRS through the binding of two HTH domains to the two 9 bp half-sites of the IRS. RsrS has a predicted unique sensor domain, suggesting that it has the ability to detect the signal from tetrathionate. These results are consistent with the previously reported mechanism of TCS in translating environmental stimuli to specific adaptive responses (Martínez-Hackert and Stock, 1997; Mattison and Kenney, 2002; Wang, 2012). Therefore, we propose a tetrathionate-dependent transcriptional regulation model of the S4I pathway by RsrS-RsrR in A. caldus. As shown in Figure 8, the membrane-bound sensor of histidine kinase RsrS might sense the signal from tetrathionate in the periplasm, which may then autophosphorylate the conserved His site, and transfer the phosphoryl group to the conserved Asp site of RsrR, thus generating an active dimer. The RsrR dimer might recognize and bind to the 19 bp IRS with its HTH domains to promote the transcriptional activity of the tetH promoter by assisting the recruitment of the RNA polymerase or by strengthening the binding between the RNA polymerase and the DNA sequence (Hochschild and Dove, 1998; Kenney, 2002).
Figure 8.
The signal transduction and transcriptional regulation model of RsrS-RsrR on the S4I pathway in A. caldus.
Blocking of the signaling pathway from tetrathionate to S4I pathway caused changes in growth and transcription patterns. The absence of RsrS or RsrR led to several days delay of growth in K2S4O6-medium. The survival of mutants in K2S4O6-medium indicated that the RsrS-RsrR mediated signal pathway is redundant with other pathways in promoting the transcription of tetH-doxDA and decomposition of tetrathionate. Transcriptional analysis revealed that the knockout of rsrR had a much stronger impact on the transcription of these genes than that of rsrS, both in terms of the number of genes being affected and in the magnitude of changes in transcription levels. The relative change in the RNA transcript levels in the mutants during growth in S0-medium revealed that the knockout of either rsrS or rsrR not only caused significant up- or down-regulation of the majority of sulfur-oxidizing genes, but also resulted in significant changes in transcription of most regulatory genes. The RsrS-RsrR and EnvZ-OmpR like two-component systems share significant homology owing to their evolutionary relationship (Rzhepishevska et al., 2007). The high level of sequence similarity and close homologous relation between some TCSs raises the possibility of undesired cross-talk between a sensor kinase and a non-cognate response regulator (Howell et al., 2003; Siryaporn and Goulian, 2008; Procaccini et al., 2011; Guckes et al., 2013; Bielecki et al., 2015; Nguyen et al., 2015). The transcriptional changes of these genes of TCSs in ΔrsrS and ΔrsrR implied that cross-talk potentially occurs between RsrS-RsrR and other TCSs. Therefore, we propose that the absence of RsrS or RsrR in A. caldus might result in the remodulation of the signal transduction pathways and changes in the transcriptional regulatory mechanisms, and certain sulfur-oxidizing pathways may be adjusted to complete the sulfur metabolism. This may be a reasonable explanation for the differences in growth between the mutants and the wild type in S0-medium and the ability of ΔrsrS and ΔrsrR mutants to grow in K2S4O6-medium. In addition, sequences homologous to the 19-bp IRS were not found at any other loci across the whole genomes of three A. caldus strains MTH-04, SM-1 and ATCC51756 by using bioinformatics tools (BBS, MP3 and Fimo; Grant et al., 2011; Ma et al., 2013), implying that RsrS-RsrR was specific for regulation of the S4I pathway. The discovery of this sequence indicates an important role of the signaling and regulatory systems in efficient metabolism of various RISCs in A. caldus. Further, exploration of the two-component and other regulatory systems would provide novel insights to better understand the sulfur metabolism and regulation network in A. caldus.
Conclusion
In this study, we developed a reliable markerless gene knockout method for A. caldus and constructed RsrS-RsrR two-component system mutants. We illustrated the regulatory role of RsrS-RsrR on the S4I pathway and proposed a tetrathionate-dependent transcriptional regulation model for the two-component system in the S4I pathway. Our markerless gene knockout system has many potential applications both in investigations of molecular mechanisms as well as genetic engineering. The elucidation of the mechanism of regulation of the S4I pathway by the RsrS-RsrR system helps improve our understanding of molecular mechanisms in the regulation of sulfur metabolism network in A. caldus.
Author contributions
ZW, JQiL, and LC designed, conducted and composed the paper. ZW, YL, CZ, and YW conducted the experiments. BL and RW performed bioinformatics analysis. JQuL, XP, XL, BL, and RW analyzed the data and revised the paper.
Funding
This work was supported by grants from the Natural Science Foundation (Grant No. 31370138, 31570036), the National Basic Research Program (2010CB630902), the Natural Science Foundation (Grant No. 31400093, 31370084, 30800011), the China Postdoctoral Science Foundation (Grant No. 2015M580585) and the State Key Laboratory of Microbial Technology Foundation (M2015-03), People's Republic of China.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Qingsheng Qi and Prof. Lushan Wang from Shandong University for providing plasmid pACBSR and assisting protein homology modeling.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01755/full#supplementary-material
Multiple alignment of the combined DoxD and DoxA amino acid sequences with homologs. Conserved residues are shown with black shadow. Accession numbers (GenBank): Acidithiobacillus caldus SM-1, DoxD (fused DoxDA), AEK58244; Acidithiobacillus caldus MTH-04, fused DoxDA, OAN03452; Acidithiobacillus caldus ATCC 51756, DoxD (fused DoxDA), ABP38224; Acidithiobacillus thiooxidans, fused DoxDA, WP_024894934; Acidithiobacillus ferrooxidans, fused DoxDA, CDQ09967; Sulfolobus tokodaii, DoxD and DoxA, NP_377837 and NP_377838; Sulfolobus solfataricus, DoxD and DoxA, NP_343149 and NP_343148; Acidianus ambivalens, DoxD and DoxA, CAA70827 and CAA70828; Acidianus ambivalens, DoxD2 and DoxA2, CAC86936 and CAC86935; Bacteroides thetaiotaomicron, fused DoxDA, NP_809428.
Amino acid sequence and protein structure analysis of RsrR and RsrS. Every domain is shown in the figure. (A) Amino acid sequence alignment between RsrR and OmpR; (B) Amino acid sequence alignment between RsrS and EnvZ; (C) Protein structure fitting of RsrR and OmpR (blue for RsrR, red for OmpR); (D) Protein structure fitting of RsrS and EnvZ (gray for RsrS, red for EnvZ).
Primers used for constructing ΔrsrR and ΔrsrS.
Primers used for RT-qPCR.
Primers used for EMSA assays.
Primers used for constructing IRS-probe vectors.
The original data for the relative transcription levels of genes involved in sulfur metabolism and signaling systems during the S0-cultivating process.
References
- Bidart G. N., Rodríguez-Díaz J., Monedero V., Yebra M. J. (2014). A unique gene cluster for the utilization of the mucosal and human milk-associated glycans galacto-N-biose and lacto-N-biose in Lactobacillus casei. Mol. Microbiol. 93, 521–538. 10.1111/mmi.12678 [DOI] [PubMed] [Google Scholar]
- Bielecki P., Jensen V., Schulze W., Gödeke J., Strehmel J., Eckweiler D., et al. (2015). Cross talk between the response regulators PhoB and TctD allows for the integration of diverse environmental signals in Pseudomonas aeruginosa. Nucleic Acids Res. 43, 6413–6425. 10.1093/nar/gkv599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes A. M., Alex L. A., Crane B. R., Simon M. I. (1999). Structure of CheA, a signal-transducing histidine kinase. Cell 96, 131–141. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Silversmith R. E. (2010). Two-component signal transduction. Curr. Opin. Microbiol. 13, 113–115. 10.1016/j.mib.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaytsova Z., Lindström E. B. (2004). Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271, 272–280. 10.1046/j.1432-1033.2003.03926.x [DOI] [PubMed] [Google Scholar]
- Capra E. J., Laub M. T. (2012). The evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66, 325–347. 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lin J., Li B., Lin J., Liu X. (2010). Method development for electrotransformation of Acidithiobacillus caldus. J. Microbiol. Biotechnol. 20, 39–44. 10.4014/jmb.0905.05023 [DOI] [PubMed] [Google Scholar]
- Chen L., Ren Y., Lin J., Liu X., Pang X., Lin J. (2012). Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS ONE 7:e39470. 10.1371/journal.pone.0039470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl C., Prange A. (2006). Bacterial sulfur globules: occurrence, structure and metabolism in Inclusions in Prokaryotes, ed Shively J. M. (Heidelberg: Springer; ), 21–51. [Google Scholar]
- Dam B., Mandal S., Ghosh W., Das Gupta S. K., Roy P. (2007). The S4-intermediate pathway for the oxidation of thiosulfate by the chemolithoautotroph Tetrathiobacter kashmirensis and inhibition of tetrathionate oxidation by sulfite. Res. Microbiol. 158, 330–338. 10.1016/j.resmic.2006.12.013 [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. (1971). Properties of an R factor from Pseudomonas aeruginosa. J. Bacteriol. 108, 1244–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Chevalier N., Ha-Thi V., Brunei F. (1987). Vectors with restriction site banks V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51, 275–280. 10.1016/0378-1119(87)90316-7 [DOI] [PubMed] [Google Scholar]
- Dillingham M. S., Kowalczykowski S. C. (2008). RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671. 10.1128/MMBR.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopson M., Lindström E. B. (1999). Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl. Environ. Microbiol. 65, 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst S., Delgado J., Inouye M. (1989). Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 86, 6052–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich C. G. (1997). Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39, 235–289. 10.1016/S0065-2911(08)60018-1 [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Bardischewsky F., Rother D., Quentmeier A., Fischer J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259. 10.1016/j.mib.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Friedrich C. G., Quentmeier A., Bardischewsky F., Rother D., Kraft R., Kostka S., et al. (2000). Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182, 4677–4687. 10.1128/JB.182.17.4677-4687.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N. U., Dahl C. (2008). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200. 10.1016/S0065-2911(08)00002-7 [DOI] [PubMed] [Google Scholar]
- Gardner M. N., Rawlings D. E. (2000). Production of rhodanese by bacteria present in bio-oxidation plants used to recover gold from arsenopyrite concentrates. J. Appl. Microbiol. 89, 185–190. 10.1046/j.1365-2672.2000.01117.x [DOI] [PubMed] [Google Scholar]
- Ghosh W., Dam B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043. 10.1111/j.1574-6976.2009.00187.x [DOI] [PubMed] [Google Scholar]
- Goebel B. M., Stackebrandt E. (1994). Cultural and phylogenetic analysis of mixed microbial populations found in natural and commercial bioleaching environments. Appl. Environ. Microbiol. 60, 1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C. E., Bailey T. L., Noble W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckes K. R., Kostakioti M., Breland E. J., Gu A. P., Shaffer C. L., Charles R., et al. (2013). Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc. Natl. Acad. Sci. U.S.A. 110, 16592–16597. 10.1073/pnas.1315320110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg K. B., Dopson M., Lindström E. B. (1996). Reduced sulfur compound oxidation by Thiobacillus caldus. J. Bacteriol. 178, 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg K. B., Lindström E. B. (1994). Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140, 3451–3456. [DOI] [PubMed] [Google Scholar]
- Hallberg K. B., Lindström E. B. (1996). Multiple serotypes of the moderate thermophile Thiobacillus caldus, a limitation of immunological assays for biomining microorganisms. Appl. Environ. Microbiol. 62, 4243–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring C. D., Glasner J. D., Blattner F. R. (2003). Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 311, 153–163. 10.1016/S0378-1119(03)00585-7 [DOI] [PubMed] [Google Scholar]
- Hochschild A., Dove S. L. (1998). Protein-protein contacts that activate and repress prokaryotic transcription. Cell 92, 597–600. 10.1016/S0092-8674(00)81126-5 [DOI] [PubMed] [Google Scholar]
- Howell A., Dubrac S., Andersen K. K., Noone D., Fert J., Msadek T., et al. (2003). Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49, 1639–1655. 10.1046/j.1365-2958.2003.03661.x [DOI] [PubMed] [Google Scholar]
- Huang K.-J., Lan C.-Y., Igo M. M. (1997). Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. U.S.A. 94, 2828–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Yan W., Wang Z. (1992). Transfer of IncP plasmids to extremely acidophilic Thiobacillus thiooxidans. Appl. Environ. Microbiol. 58, 429–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney L. J. (2002). Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5, 135–141. 10.1016/S1369-5274(02)00310-7 [DOI] [PubMed] [Google Scholar]
- Kletzin A. (1989). Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J. Bacteriol. 171, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A. (1992). Molecular characterization of the sor gene, which encodes the sulfur oxygenase/reductase of the thermoacidophilic Archaeum Desulfurolobus ambivalens. J. Bacteriol. 174, 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kletzin A., Urich T., Müller F., Bandeiras T. M., Gomes C. M. (2004). Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36, 77–91. 10.1023/B:JOBB.0000019600.36757.8c [DOI] [PubMed] [Google Scholar]
- Lehman M. K., Bose J. L., Sharma-Kuinkel B. K., Moormeier D. E., Endres J. L., Sadykov M. R., et al. (2015). Identification of the amino acids essential for LytSR-mediated signal transduction in Staphylococcus aureus and their roles in biofilm-specific gene expression. Mol. Microbiol. 95, 723–737. 10.1111/mmi.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Stockdreher Y., Koch T., Sun S., Fan Z., Josten M., et al. (2014). Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur-oxidizing archaeon Metallosphaera cuprina. J. Biol. Chem. 289, 26949–26959. 10.1074/jbc.M114.591669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Lin J., Zhang Z., Bian J., Zhao Q., Liu Y. (2007). Construction of conjugative gene transfer system between E. coli and moderately thermophilic, extremely acidophilic Acidithiobacillus caldus MTH-04. J. Microbiol. Biotechnol. 17, 162–167. [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma Q., Liu B., Zhou C., Yin Y., Li G., Xu Y. (2013). An integrated toolkit for accurate prediction and analysis of cis-regulatory motifs at a genome scale. Bioinformatics 29, 2261–2268. 10.1093/bioinformatics/btt397 [DOI] [PubMed] [Google Scholar]
- Mangold S., Valdés J., Holmes D. S., Dopson M. (2011). Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbio. 2:17. 10.3389/fmicb.2011.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Hackert E., Stock A. M. (1997). The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5, 109–124. 10.1016/S0969-2126(97)00170-6 [DOI] [PubMed] [Google Scholar]
- Mattison K., Kenney L. J. (2002). Phosphorylation alters the interaction of the response regulator OmpR with its sensor kinase EnvZ. J. Biol. Chem. 277, 11143–11148. 10.1074/jbc.M111128200 [DOI] [PubMed] [Google Scholar]
- Müller F. H., Bandeiras T. M., Urich T., Teixeira M., Gomes C. M., Kletzin A. (2004). Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate:quinone oxidoreductase. Mol. Microbiol. 53, 1147–1160. 10.1111/j.1365-2958.2004.04193.x [DOI] [PubMed] [Google Scholar]
- Nguyen M. P., Yoon J. M., Cho M. H., Lee S. W. (2015). Prokaryotic 2-component systems and the OmpR/PhoB superfamily. Can. J. Microbiol. 61, 799–810. 10.1139/cjm-2015-0345 [DOI] [PubMed] [Google Scholar]
- Pardo E., Orejas M. (2014). The Aspergillus nidulans Zn(II)2Cys6 transcription factor AN5673/RhaR mediates L-rhamnose utilization and the production of α-L-rhamnosidases. Microb. Cell Fact. 13:161. 10.1186/s12934-014-0161-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccini A., Lunt B., Szurmant H., Hwa T., Weigt M. (2011). Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: Orphans and crosstalks. PLoS ONE 6:e19729. 10.1371/journal.pone.0019729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatrini R., Appia-Ayme C., Denis Y., Jedlicki E., Holmes D., Bonnefoy V. (2009). Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. 10.1186/1471-2164-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D. E. (1998). Industrial practice and the biology of leaching of metals from ores. J. Ind. Microbiol. Biotechnol. 20, 268–274. [Google Scholar]
- Rohwerder T., Sand W. (2003). The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149, 1699–1709. 10.1099/mic.0.26212-0 [DOI] [PubMed] [Google Scholar]
- Rohwerder T., Sand W. (2007). Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng. Life Sci. 7, 301–309. 10.1002/elsc.200720204 [DOI] [Google Scholar]
- Rzhepishevska O. I., Valdés J., Marcinkeviciene L., Gallardo C. A., Meskys R., Bonnefoy V., et al. (2007). Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 73, 7367–7372. 10.1128/AEM.01497-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger P., Westley J. (1974). An expanded mechanism for rhodanese catalysis. J. Biol. Chem. 249, 780–788. [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechology 1, 784–790. [Google Scholar]
- Siryaporn A., Goulian M. (2008). Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol. Microbiol. 70, 494–506. 10.1111/j.1365-2958.2008.06426.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Suzuki I. (1999). Oxidation of inorganic sulfur compounds: chemical and enzymatic reactions. Can. J. Microbiol. 45, 97–105. [Google Scholar]
- Valdés J., Pedroso I., Quatrini R., Holmes D. S. (2008). Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94, 180–184. 10.1016/j.hydromet.2008.05.039 [DOI] [Google Scholar]
- Valdés J., Quatrini R., Hallberg K., Dopson M., Valenzuela P., Holmes D. S. (2009). Genome announcement: draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 191, 5877–5878. 10.1128/JB.00843-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L., Chi A., Beard S., Shabanowitz J., Hunt D. F., Jerez C. A. (2008). Differential-expression proteomics for the study of sulfur metabolism in the chemolithoautotrophic Acidithiobacillus ferrooxidans, in Microbial Sulfur Metabolism, eds Dahl C., Friedrich C. G. (Berlin: Springer; ), 77–86. [Google Scholar]
- Zyl L. J. V., Munster J. M. V., Rawlings D. E. (2008). Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl. Environ. Microbiol. 74, 5686–5694. 10.1128/AEM.01235-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakai S., Kikumoto M., Kanao T., Kamimura K. (2004). Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 68, 2519–2528. 10.1271/bbb.68.2519 [DOI] [PubMed] [Google Scholar]
- Wang S. (2012). Bacterial two-component systems: structures and signaling mechanisms, in Protein Phosphorylation in Human Health, ed Huang C. (Rijeka: InTech Press; ), 439–466. [Google Scholar]
- Williams K. P., Kelly D. P. (2013). Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int. J. Syst. Evol Microbiol 63, 2901–2906. 10.1099/ijs.0.049270-0 [DOI] [PubMed] [Google Scholar]
- Yon J., Fried M. (1989). Precise gene fusion by PCR. Nucleic Acids Res. 17, 4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X., Guo X., Zheng H., Zhang M., Liu L., Zhu Y., et al. (2011). Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J. Genet. Genomics 38, 243–252. 10.1016/j.jgg.2011.04.006 [DOI] [PubMed] [Google Scholar]
- Yu Y., Liu X., Wang H., Li X., Lin J. (2014). Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J. Bacteriol. 196, 2255–2264. 10.1128/JB.01472-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Jiang C., You X., Liu S. (2014). Construction and application of an expression vector from the new plasmid pLAtc1 of Acidithiobacillus caldus. Appl. Microbiol. Biotechnol. 98, 4083–4094. 10.1007/s00253-014-5507-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignment of the combined DoxD and DoxA amino acid sequences with homologs. Conserved residues are shown with black shadow. Accession numbers (GenBank): Acidithiobacillus caldus SM-1, DoxD (fused DoxDA), AEK58244; Acidithiobacillus caldus MTH-04, fused DoxDA, OAN03452; Acidithiobacillus caldus ATCC 51756, DoxD (fused DoxDA), ABP38224; Acidithiobacillus thiooxidans, fused DoxDA, WP_024894934; Acidithiobacillus ferrooxidans, fused DoxDA, CDQ09967; Sulfolobus tokodaii, DoxD and DoxA, NP_377837 and NP_377838; Sulfolobus solfataricus, DoxD and DoxA, NP_343149 and NP_343148; Acidianus ambivalens, DoxD and DoxA, CAA70827 and CAA70828; Acidianus ambivalens, DoxD2 and DoxA2, CAC86936 and CAC86935; Bacteroides thetaiotaomicron, fused DoxDA, NP_809428.
Amino acid sequence and protein structure analysis of RsrR and RsrS. Every domain is shown in the figure. (A) Amino acid sequence alignment between RsrR and OmpR; (B) Amino acid sequence alignment between RsrS and EnvZ; (C) Protein structure fitting of RsrR and OmpR (blue for RsrR, red for OmpR); (D) Protein structure fitting of RsrS and EnvZ (gray for RsrS, red for EnvZ).
Primers used for constructing ΔrsrR and ΔrsrS.
Primers used for RT-qPCR.
Primers used for EMSA assays.
Primers used for constructing IRS-probe vectors.
The original data for the relative transcription levels of genes involved in sulfur metabolism and signaling systems during the S0-cultivating process.