Abstract
Slowing of replication in response to DNA damage is a universal response to DNA damage during S-phase. Originally discovered to be defective in checkpoint mutant cells in metazoans, this S-phase DNA damage checkpoint response has been extensively studied in yeast. Unlike other checkpoints that completely arrest cell cycle, the S-phase DNA damage checkpoint slows but does not completely halt replication in response to DNA damage. An analysis of mutants defective in the slowing response requires a sensitive assay to measure this quantitative effect. The use of centrifugal elutriation to synchronize cells and improved techniques in preparing cells for flow cytometry allow for more sensitive and accurate measurement of cells’ ability to slow replication in the presence of DNA damage. This chapter describes the use of transient cdc10-M17 temperature sensitive allele arrest and release combined with centrifugal elutriation to synchronize cells in G1. The S-phase progression of these cells is then assayed by flow cytometry of isolated nuclei, which allows sensitive determination of replication kinetics.
Keywords: S-phase DNA damage checkpoint, DNA replication, fission yeast, Schizosaccharomyces pombe, flow cytometry, centrifugal elutriation, cell division cycle
1. Introduction
Slowing of DNA replication is a hallmark of the S-phase DNA damage checkpoint response. Failure to reduce new synthesis in the presence of DNA damage is correlated with genomic instability and cancer development (1). In metazoans, new DNA synthesis may be measured in asynchronous cultures by the incorporation of radio-labeled nucleotides (2). However, in fission yeast only about 10% of cells in an asynchronous population are in S-phase, so kinetic measurement of replication in asynchronous cultures is not currently practical. Accurate measurement of replication progression requires the synchronization of cells, allowing one to follow the population through S-phase.
Many methods are available to synchronize yeast. Cells may be synchronized using cell division cycle (cdc) mutants, nutrient starvation or drug-induced arrest. G1 synchronization may be achieved using temperature sensitive alleles of the transcriptional regulator cdc10 or starvation in nitrogen-free media (3, 4). These techniques allow the manipulation of multiple cultures and the uses of large quantities of cells. However, these methods suffer from various disadvantages including checkpoint activation, inefficient release from starvation, and physiological disruption. In particular, we have observed that block-and-release protocols can prevent wild-type cells from properly responding to DNA damage (Kommajosyula and Rhind, 2006).
To avoid synchronization artifacts, we combine cdc10-ts arrest with centrifugal elutriation (Figure 3.1A). As mentioned above, prolonged arrest of cdc10-ts cells can compromise the S-phase DNA damage checkpoint. However, due to a peculiarity of the fission yeast cell cycle, G1 is cryptic, with cells beginning DNA replication before they finish cytokinesis. Therefore, in order treat collect cells in G1, one must arrest cells in one way or another. Fortunately, one can briefly arrest cdc10-ts cells and then select the smallest, G1 arrested cells by centrifugal elutriation. This approach allows for the preparation of a synchronous G1 population that retains robust S-phase checkpoint response to DNA damage.
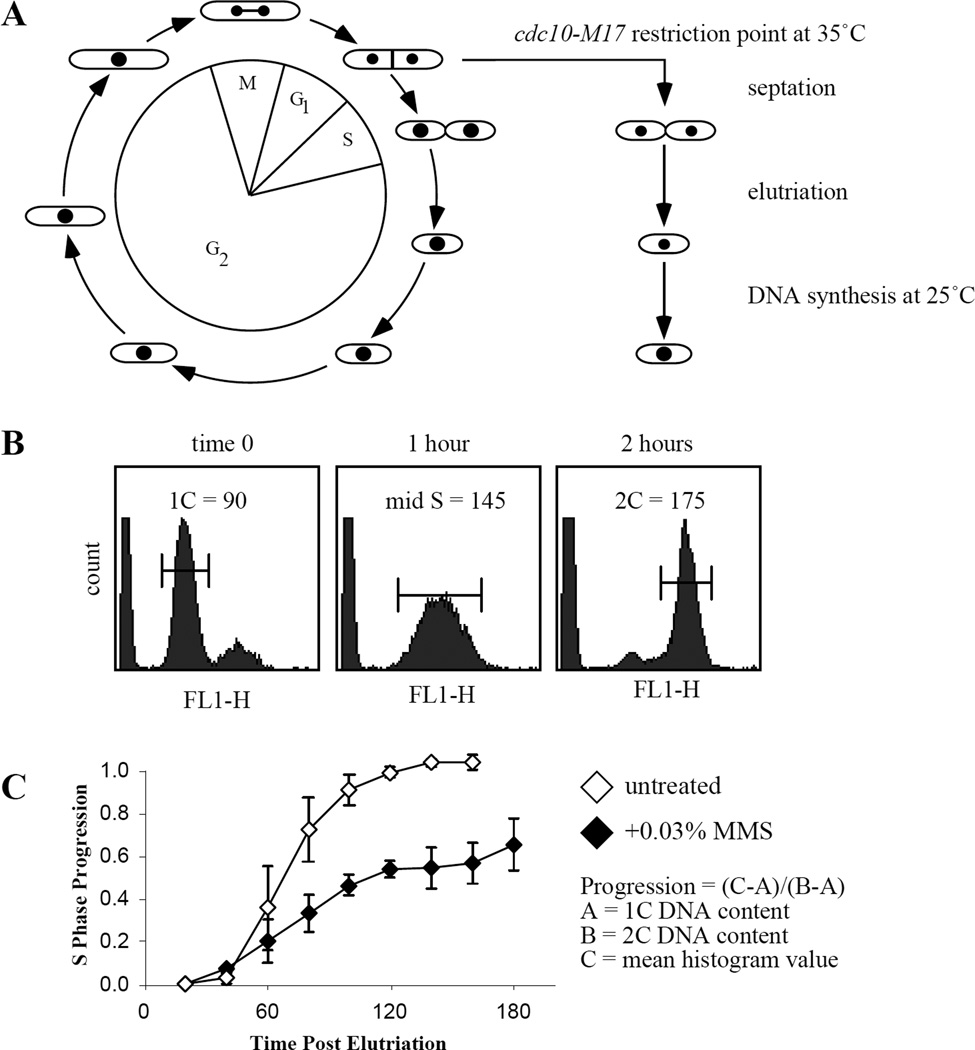
Figure 3.1. S-phase DNA Damage Response.
(A) Cytokinesis and replication normally occur almost simultaneously. However, use of the cdc10-M17 temperature sensitive allele prevents replication while allowing cells to complete cytokinesis. Transient arrest of an asynchronous culture will produce small cells arrested in G1 containing an unreplicated genome. Centrifugal elutriation allows the harvesting of these cells. (B) Examples of flow cytometry profiles for 0, 1 and 2 hours after release. Cells display unreplicated, mid-S phase, and replicated nuclear DNA profiles, respectively. Brackets indicate sample portion measured to determine mean histogram peak value used for S-phase progression plots. (C) Example of S phase progression plot in wild-type strain untreated or exposed to 0.03% MMS. The profile shown is an average of three experiments and error bar represent the standard error of the mean.
To accurately measure progression through S-phase and assess strains’ ability to slow replication in response to DNA damage, we employ flow cytometry of isolated nuclei. Cytoplasmic background leads to an overestimation of nuclear DNA content due to the contribution of RNA and mitochondrial DNA to overall cellular nucleic acid content (5). Isolating nuclei reduces background due the contribution of mitochondrial DNA and cell size, greatly increasing sensitivity and simplifying data interpretation (6, 7).
Synchronization of cells in G1 is performed using the following scheme. Cells are arrested for 2 hours at non-permissive temperature 35ûC, meanwhile the elutriator is setup and temperature set to 35ûC. The semi-synchronous culture is then loaded and the smallest and transiently arrested G1 cells are harvested. Harvested cells are released from the arrest to 25ûC in the absence or presence of DNA damage. Samples are fixed at regular intervals for analysis by flow cytometry or frozen in liquid nitrogen for isolation of DNA, RNA or protein.
2. Materials
2.1 G1 synchronization
-
1.
cdc10-M17 fission yeast strains.
-
2.
YES rich media (Yeast Extract + Supplements): 5 g/l yeast extract, 30 g/l glucose, 75 mg/l leucine, 75 mg/l uracil, 75 mg/l adenine, 75 mg/l histidine, autoclaved.
-
3.
Beckman J-20 with a JE-5 series rotor and 4 ml elutriation chamber, Beckman Instruments.
-
4.
Two shaking water baths set to 25°C and 35°C, 200 rpm.
-
5.
70% ethanol.
-
6.
methyl methane sulfonate, Sigma, cat # M4016.
2.2 Nuclei Flow Cytometry
-
7.
0.6 M KCl.
-
8.
0.1 M KCl, 0.1% SDS.
-
9.
20 mM Tris pH 8.0, 1mM EDTA pH 8.0.
-
10.
10 mg/ml RNAse A, Sigma, cat # R5503.
-
11.
Branson 450 Analog Sonifier, #101-063-200, VWR, cat # 33995-320, or equivalent.
-
12.
Branson Sonifier 3mm tapered tip, VWR cat # 33996-163.
-
13.
BD FACScan flow cytometer, BD Sciences, or equivalent.
-
14.
BD Sciences Cellquest software version 3.3.
-
15.
Lysing enzymes from Trichoderma harzianium, Sigma, cat # L1412.
-
16.
Zymolyase 20T, US Biological, cat# Z1000.
-
17.
FACS sheath fluid, Diluent 2 (1x PBS), VWR, cat # 45001-012.
-
18.
Sytox Green nucleic acid stain diluted to 2 µm in FACS sheath fluid, Invitrogen, cat # S7020.
-
19.
Falcon 100 × 17mm 2054 tubes, Fischer, cat # 149595.
3. Methods
3.1 G1 Cell Elutriation
-
1.
Grow cells to a final volume of 500 ml, mid-log phase (OD600 ~1) (See Note 1).
-
2.
Preheat shaking water baths to 25°C and 35°C, set to 200 rpm (See Note 2).
-
3.
Pellet 0.5 OD cells in a 1.7ml eppendorf tube, 16000x g 2 minutes at room temperature and resuspend in 1 ml 70% ethanol to serve as the asynchronous control for flow cytometry (See Note 3).
-
4.
Incubate 500 ml culture at 35°C in the shaking water bath for 2 hours (See Note 4).
-
5.
Setup the elutriator as described in Chapter II, and set the chamber temperature to 35°C (See Note 5).
-
6.
Load cells as described for G2 elutriation in Chapter II.
-
7.
Pellet and save 0.5 OD cells while loading to serve as the transiently arrested control (See Note 6).
-
8.
Perform elutriation and harvest 150 ml cells. Visually check cells, homogenous small size with no divided pairs or septated cells is ideal (See Notes 7 and 8).
-
9.
Sample cells as in #3 to serve as the time 0 samples.
-
10.Divide cells into three 50 ml cultures and treat as follows
- mock treatment.
- add MMS to 0.03%.
- add hydroyurea to 10 mM (See Note 9).
-
11.
Place cultures in 25°C shaking water bath (See Note 10).
-
12.
Sample cells as in #3 at 20 minute intervals for analysis by flow cytometry.
-
13.
Store samples in 70% alcohol overnight at +4°C (See Note 11).
3.2 Preparation of Isolated Nuclei for Flow Cytometry
-
14.
Pellet fixed cells at 5000x g for 3 minutes at room temperature, decant supernatant (See Note 12).
-
15.
Wash once with 1 ml 0.6 M KCl.
-
16.
Resuspend cells in 1 ml 0.6 M KCl, 1 mg/ml lysing enzyme, 0.5 mg/ml Zymolyase 20T (See Note 13).
-
17.
Incubate cells for 30 minutes at 37°C.
-
18.
Pellet cells and decant supernatant.
-
19.
Resuspend cells in 1 ml 0.1M KCl 0.1% triton-x-100, vortex well.
-
20.
Pellet cells and decant supernatant.
-
21.
Wash cells in 1 ml 20 mM Tris-HCl, 5 mM EDTA pH 8.0.
-
22.
Resuspend cells in 1 ml 20 mM Tris-HCl, 5 mM EDTA pH 8.0 (See Note 14).
-
23.
Add 25 µl 10 mg/ml RNase A to each sample (final conc = 200 µg/ml), vortex well.
-
24.
Incubate cells overnight at 37°C.
-
25.
Chill cells at −20°C for 5–10 minutes (See Notes 15, 16 and 17).
-
26.
Sonicate samples using a Branson 450 Sonifier and a 3 mm tapered tip, maximum micro-tip power for 10 seconds (See Note 18).
-
27.
Mix 300 µl of prepared nuclei with 300 µl of 2 µM Sytox in a Falcon 2054 tube. Briefly vortex samples.
3.3 Analysis by Flow Cytometry
-
28.
Measure nuclear DNA content by FL1 signal for each sample according to flow cytometer instructions (See Note 19).
-
29.
Calculate the position within S-phase for each sample taken using the following equation (Figure 3.1B and see Note 20).
% replicated = (C - A)/(B - A)
A = 1C, B = 2C, and C = mean histogram value.
- 30.
4. Notes
4.1 G1 Elutriation
-
1.
Grow 500 ml culture of cdc10-M17 in YES at 25°C, 200 rpm to an optical density at 600 nm (OD600) of about 1, starting from a mid-log 50 ml starter culture. OD600 is used to follow yeast cell proliferation. Optical density is also used approximate cell number; 1 OD unit (abbreviated OD) is the number of cells required to give 1 ml of culture an optical density of 1. Therefore, a 100 ml culture at OD600 of 0.1, a 10 ml culture at OD600 of 1 and a 1 ml culture at OD600 of 10 all contain 10 ODs of cells. 1 OD is approximately 2 × 107 cells. The 4 ml and 40 ml chambers hold approximately 400 and 4000 OD yeast cells, respectively. For the large chamber, several liters of cells may be grown to an optical density of 2.0. Sick strains may require more cells to fill the chamber, in excess of 800 or 8000 ODs of cells for the small and large chambers respectively. For cultures that are extremely sick or flocculate at low OD, sonicate the culture using 10–20 50 msec pulses at maximum power. Sonication also helps to break up divided pairs, thus increasing enrichment of small cells for any strain.
-
2.
Water baths are required for rapid temperature shifts used for arrest and release. Air incubators are not suitable for quick temperature shifts.
-
3.
The asynchronous sample serves as a control with mostly 2C nuclear DNA content. Keep the quantity of cells fixed for all timepoints approximately equal. Subsequent processing steps are sensitive to the total number of cells in each sample.
-
4.
Culture density should increase by 50–100% depending on strain doubling time.
-
5.
Allow about 40 minutes prior to cell loading to heat the chamber and recirculating media to 35°C.
-
6.
Asynchronous cultures arrested for 2 hours display two populations of cells, those containing 1C and 2C genomic DNA content. The 1C cells have encountered and arrested at the restriction point while the 2C cells have not.
-
7.
At most, ten percent of cells loaded may be synchronously harvested. Harvested culture density should be between 0.1 and 0.2 OD600. Greater than 0.4 OD600 results in far less synchronous population. For S-phase progression, 50 ml volume for each condition is sufficient.
-
8.
The time zero control should contain mostly cells with 1C nuclear DNA content. Harvested cells should all appear small and homogenous in size. Sick, slow growing cultures may look far worse than wild-type control. Cultures may be sonicated to help separate cells. The larger 40 ml chamber may be employed to help select a healthy minority from sick cultures.
-
9.
MMS induced replication slowing is dose dependent; 0.015% MMS causes very little slowing, 0.03% causes robust, checkpoint-dependent slowing, and 0.06% and greater concentrations induce checkpoint-independent slowing. Alternatives to 0.03% MMS include incubation with 0.5 uM 4NQO or exposure to 100–200 joule/m2 UV radiation. See Chapter II for details on UV exposure.
-
10.
Multiple consecutive elutriations may be staggered approximately 30 minutes apart after sufficient experience operating the elutriator. Time courses may be synchronized by maintaining freshly harvested cells at 35ûC for 5–15 minutes until release with the next timepoint of a previous and ongoing timecourse.
-
11.
Fixed cells may be stored at +4ûC for days before processing. Overnight fixation in ethanol is ideal. Samples processed the same day as the timecourse do not look as tight by flow cytometry as cells processed after an overnight fixation. Additionally, the longer cells are stored after 24 hours, the worse the raw flow cytometry data will look.
-
12.
G1 cells synchronized by centrifugal elutriation begin replication approximately 40 minutes after release and complete replication by 100 minutes. Most replication occurs about 80 minutes after release.
4.2 Nuclei Preparation and Flow Cytometry
-
13.
Cell pellets are often soft and lost during supernatant aspiration. Simply decant supernatant by hand and do not attempt to remove more than 90% of the supernatant.
-
14.
Properly digested cells appear dark by phase microscopy when mixed with 1% SDS. Lysing enzymes and zymolyase may be increased by 50% if needed. A titration of the enzymes should be done if using an alternative vendor or the purified 100T fraction of zymolyase. Setup digests using 0.5x, 1x, 1.5x and 2x concentration of enzymes used and digest as normal. Process all the samples in parallel for flow cytometry and compare. Select the enzyme concentration that displays sharp G1 and G2 peaks; a sub-G1 shoulder indicates over digestion and nuclear breakdown.
-
15.
Be careful since the pellets at this step are extremely fragile.
-
16.
Anecdotally, sonicating pelleted cells improves signal to noise of samples by flow cytometry. If necessary, pellet cells for 5 minutes at 5000 xg prior to chilling.
-
17.
Cooling of the samples prevents over heating during sonication. If the samples freeze, allow them to thaw on the benchtop before sonication.
-
18.
Do not touch the side of the tube to the microtip, gently move the tip up and down in the tube while sonicating. Do not allow samples to froth up during sonication. This leads to a very poor sample flow profile. This nuclei preparation may be stored at 4ûC for days.
4.3 Calculate and Plot Population S-phase Progression
-
19.
BD Sciences Cellquest software is used to measure the mean peak values of histograms displayed by populations exiting G1 and progressing through S phase. For this analysis, all detectors are set to linear scale. Calibrate the FSC and SSC (forward and side-scatter) so that the samples analyzed are well within the cytometer detection range. Calibrate FL1 voltage and amperage gain to allow visualization of both the 1C and the 2C DNA populations prior to data collection. The FL1 signal representing the 1C population should be set to appear at around the 400th channel on a 1024 channel detection scale. Flow cytometry plots show a sub-1C peak, which is due to cell fragments lacking nuclei.
-
20.
For calculation of the percent replicated, the 1C unreplicated value, A, is determined from the DNA content of the freshly elutriated, time 0 sample; the 2C fully replicated value B is determined from the DNA content of the later time points of the untreated sample and the 2C peaks of the asynchronous and transiently arrested samples (Figure 1).
-
21.
If sample signal drifts to the right for later timepoints, after cells have reached 2C DNA content, try resonicating the samples and collect data again and sonicate subsequent samples for longer than 10 seconds.
-
22.
G1 cells synchronized by centrifugal elutriation begin replication approximately 40 minutes after release and complete replication by 100 minutes. Most replication occurs about 80 minutes after release.
References
- 1.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 2.Painter RB, Young BR. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci U S A. 1980;77:7315–7317. doi: 10.1073/pnas.77.12.7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- 4.Aves SJ, Durkacz BW, Carr A, Nurse P. Cloning, sequencing and transcriptional control of the Schizosaccharomyces pombe cdc10 'start' gene. EMBO J. 1985;4:457–463. doi: 10.1002/j.1460-2075.1985.tb03651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- 6.Carlson CR, Grallert B, Bernander R, Stokke T, Boye E. Measurement of nuclear DNA content in fission yeast by flow cytometry. Yeast. 1997;13:1329–1335. doi: 10.1002/(SICI)1097-0061(199711)13:14<1329::AID-YEA185>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 7.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]