Abstract
As a leading cause of cancer deaths worldwide, lung cancer is a collection of diseases with diverse etiologies which can be broadly classified into small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC). Lung cancer is characterized by genomic and epigenomic alterations; however, mechanisms underlying lung tumorigenesis remain to be elucidated. Long non-coding RNAs (lncRNAs) are a group of non-coding RNAs that consist of ⩾200 nucleotides but possess low or no protein-coding potential. Accumulating evidence indicates that abnormal expression of lncRNAs is associated with tumorigenesis of various cancers, including lung cancer, through multiple biological mechanisms involving epigenetic, transcriptional, and post-transcriptional alterations. In this review, we highlight the expression and roles of lncRNAs in NSCLC and discuss their potential clinical applications as diagnostic or prognostic biomarkers, as well as therapeutic targets.
Keywords: Long non-coding RNA, Non-small-cell lung cancer, Expression spectrum, Biomarker, Therapeutic resistance
Introduction
Lung cancer is the most common cancer and the leading cause of cancer deaths among men and women worldwide. Among all lung cancer cases, non-small-cell lung cancers (NSCLCs) account for approximately 85% [1], which are at locally advanced or metastatic stage at diagnosis [2]. Based on its pathological characteristics, NSCLC is subdivided into three subtypes, namely, lung adenocarcinoma (LAD), large cell carcinoma (LCC), and lung squamous cell carcinoma (LSCC). LAD and LSCC are the predominant types of NSCLCs, which constitute ∼50% and ∼40% of NSCLC cases, respectively [3]. Although the traditional therapeutic strategies have been tremendously improved and targeted therapies, such as tyrosine kinase inhibitors (TKIs) of the epidermal growth factor receptor (EGFR) [4] and immune checkpoint inhibitors, have been successfully used in clinical practice [5], the five-year overall survival rate of lung cancer of all stages combined remains as low as 15.9% [6]. Such unfavorable outcome could be at least partially attributed to the poor understanding of the pathogenesis of NSCLC, as well as lack of early diagnostic biomarkers and therapeutic targets.
Genetic and epigenetic alterations have been widely recognized as the driving events of cancer. A recent high-throughput transcriptome analysis showed that nearly 75% of the human genome is transcribed into RNAs and only ∼2% of the genome serves as blueprints for proteins with others as non-coding RNAs (ncRNAs) [7]. ncRNAs can be short or small (<200 bp) or long (⩾200 bp) in length. Small ncRNAs include microRNAs (miRNAs), small interfering RNA (siRNAs), PIWI-interacting RNAs (piRNAs), as well as classical housekeeping ncRNAs such as tRNAs, rRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs). miRNAs and piRNAs have been implicated in multiple cellular functions that are essential for physiological or pathological processes [8]. Linearized ncRNAs containing >200 nucleotides are termed as long ncRNAs (lncRNAs), which have attracted much attention recently. A wealth of compelling evidence has demonstrated that aberrantly expressed lncRNAs play important roles in cancer development, including cancer cell proliferation and metastasis, through distinct transcriptional, post-transcriptional, or epigenetic mechanisms [9], [10]. In this review, we focus on the roles of lncRNAs in lung tumorigenesis and briefly introduce the development of lncRNA-directed diagnostics, prognostics and therapeutics.
Discovery of lncRNAs
The discovery of lncRNAs is attributed to studies on the size, evolution, and function of the genome. Higher species were previously thought to need more genes than lower species [11]. However, developmental complexity of animals is not determined by the amount of DNA in the genome [12]. For example, the genome of salamander is 15 times larger than that of humans [13]. With the aid of DNA–RNA hybridization technique, scientists have come to realize that most parts of the genome do not encode proteins and these non-coding regions of the genome were considered as “junk DNA” [14]. On the other hand, some researchers reckoned that junk DNA was not completely useless [11]. Therefore, substantial interest has been focused on determining the functional roles of these non-coding sequences. As a result, heterogeneous nuclear RNAs and introns were discovered in the 1970s [15], [16], [17]. Subsequent studies demonstrated that snRNAs and snoRNAs play important roles in post-transcriptional RNA processing [18], thus pushing forward investigations on other non-coding sequences. In the early 1990s, the roles of some lncRNAs (e.g., H19 and Xist) in epigenetic regulation were uncovered [19], [20], [21]. However, research on lncRNAs was suspended due to the discovery of miRNAs in 1993 [22] and sustaining keen interests in microRNA studies. Notably, introduction of whole-transcriptome sequencing in the early 2000s led to the identification and annotation of many lncRNAs [23], [24], [25]. A small number of characterized human lncRNAs were then recognized as the central regulators of a variety of biological processes including gene expression, mRNA processing, and protein translation or transport [26]. Up till now, ten thousands of lncRNAs have been identified in different species. However, functional identification of lncRNAs remains a gigantic challenge.
Characteristics of lncRNAs
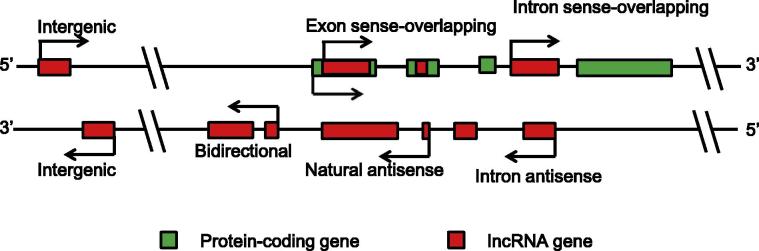
lncRNAs contain ⩾200 nucleotides and bear no or low translational potential [9]. Based on their relationships with protein-coding genes, lncRNAs are classified into six broad categories, namely, intergenic, bidirectional, intron sense-overlapping, exon sense-overlapping, intronic-antisense, and natural-antisense lncRNAs [27] (Figure 1). lncRNAs usually are transcribed by RNA polymerase II (RNAPII), but there are some exceptions. For instance, brain-associated BC200 is transcribed by RNAPIII [28]. Generally, lncRNAs are expressed at lower levels and are less conserved than protein-coding genes [29], [30], [31] and some lncRNAs exhibit cell-, tissue- and time-specific expression patterns [32]. A growing body of evidence has indicated that the expression of lncRNAs is tightly regulated through distinct mechanisms, such as chromatin state, transcription factors (TFs), and microRNAs [33]. And majority of lncRNAs are transcribed from antisense regions upstream of promoters, intragenic regions, intergenic regions distal to promoters, or gene bodies of protein-coding genes [7].
Figure 1.
A diagram of lncRNA categories
Intergenic: a lncRNA gene lies as an independent unit within the genomic interval between two genes. Bidirectional: expression of a lncRNA gene and its neighboring coding transcript on the opposite strand is initiated in close genomic proximity. Intron sense-overlapping: a lncRNA gene lies in the intron of a protein-coding gene on the same strand. Exon sense-overlapping: a lncRNA gene lies in the exons of protein-coding gene on the same strand. Intronic-antisense: a lncRNA lies in the introns of protein-coding gene on the opposite strand in the same region. Natural-antisense: a lncRNA gene lies in the exons of protein-coding gene on the opposite strand. lncRNA, long non-coding RNA.
Functions of lncRNAs
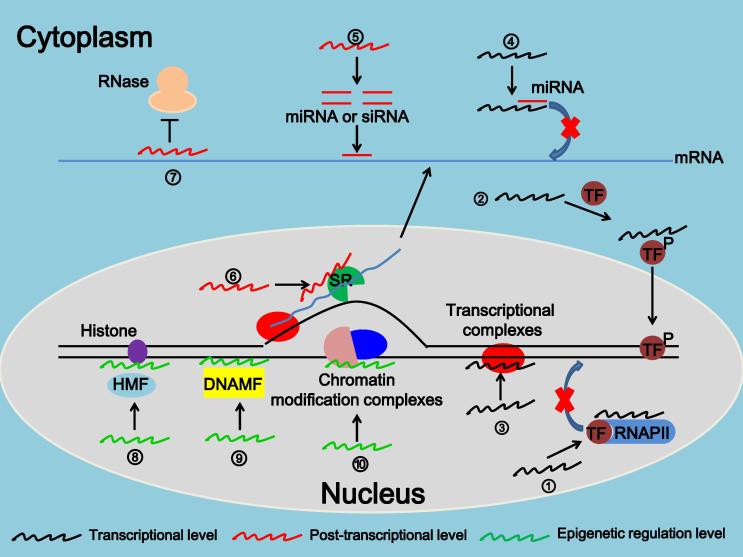
LncRNAs function in diverse biological processes by modulating the transcription and translation of protein-coding genes. Unlike miRNAs, which commonly participate in mRNA degradation or regulate mRNA translation [34], [35], [36], lncRNAs regulate the expression of target genes through multiple mechanisms at different levels (Figure 2). lncRNAs can interact directly with DNA, mRNA, or proteins to regulate chromatin modification or structure, transcription, splicing, and translation, so as to regulate a variety of physiological and pathological processes such as cell proliferation or differentiation, stem cell reprogramming, tumorigenesis, or drug resistance [10], [37], [38]. Functions of lncRNAs are summarized in Figure 2 and briefly described below.
Figure 2.
Molecular mechanisms for the functions of lncRNAs
① lncRNA acts as decoys for TFs or RNAPII; ② lncRNA alters the modification and location of transcription factors; ③ lncRNA interacts with DNA and forms triple helix structures, thereby recruiting transcriptional complex; ④ lncRNA acts as decoy for miRNA; ⑤ lncRNA acts as precursor for siRNAs or miRNAs; ⑥ lncRNA regulates the alternative splicing of pre-mRNAs through SR complex; ⑦ lncRNA protects mRNA from degradation through forming double-stranded RNA with mRNAs; ⑧ lncRNA regulates histone modification by interacting with modification factors; ⑨ lncRNA binds to DNA modification factors to modify the methylation of DNA; ⑩ lncRNA binds to chromatin modification complexes to regulate chromatin remodeling and structure. DNAMF: DNA modification factor; HMF: histone modification factor; miRNA, microRNA; siRNA, small-interfering RNA; TF, transcription factor; RNAPII, RNA polymerase II.
First, at the transcriptional level, lncRNAs (i) act as decoys for TFs or RNAPII to disrupt their binding to promoters/enhancers of target genes, thus promoting or suppressing gene expression [39]; (ii) interact directly with TFs and alter their modification or localization to regulate gene transcription [40]; (iii) interact with DNA and form scaffolds for TFs, thus affecting target gene transcription [41]; and (iv) act as competitive endogenous RNAs (ceRNAs) to control target gene transcription [42].
Second, at the post-transcriptional level, lncRNAs (i) act as precursors of siRNAs or miRNAs, leading to decreased expression of their target genes [43], (ii) form double-stranded RNA complexes with mRNAs and protect them from degradation [44], and (iii) regulate the alternative splicing of pre-mRNAs to produce different transcripts [45].
Lastly, at the epigenetic level, lncRNAs (i) interact with proteins associated with histone modifications to modify the methylation, acetylation or ubiquitination of histones [46]; (ii) get involved in gene silencing by regulating DNA methylation in the promoter region of target genes [47]; and (iii) get involved in chromatin remodeling or conformational alterations by binding to chromatin modification complexes, which is important for gene transcription [7].
Expression spectrum of lncRNAs in NSCLCs
Compelling evidence has demonstrated the important roles of lncRNAs in various diseases, particularly in cancer. Recent studies have reported lncRNA expression in NSCLCs. For instance, using high-throughput microarrays, Xu et al identified 2420 lncRNAs that were differentially expressed (fold change ⩾2) between LAD and normal tissue (NT) samples. Of these 2420 lncRNAs, the expression of 1213 lncRNAs was upregulated, whereas the expression of the remaining 1207 lncRNAs was downregulated [48]. As another example, Yang et al identified 47 lncRNAs (14 upregulated and 33 downregulated lncRNAs) from gene expression data of five NSCLC cohorts that were deposited in the Gene Expression Omnibus (GEO) database [49]. Interestingly, several novel lncRNAs were identified to be induced by established risk factors for NSCLC, such as cigarette smoking or exposure to a polycyclic aromatic hydrocarbon compound benzo(a)pyrene (BaP). These include cancer-associated lncRNA-1 (SCAL1), DQ786227, and LOC728228 [50], [51], [52]. We recently reported the screening for lncRNAs with abnormal expression in lung cancers that are associated with air pollution [53]. We found that the cancer samples of patients from high pollution region had much more dysregulated lncRNAs than patients from control regions when compared to their corresponding neighboring tissues. Among these, the expression of an lncRNA, CAR intergenic 10 (CAR10), was up-regulated in air pollution-related NSCLCs. Expression of CAR10 could be upregulated by the carcinogen dibenz[a,h]anthracene (DBA) through increasing expression of TF FoxF2. CAR10 binds to and stabilizes TF Y-box-binding protein 1 (YB-1), leading to up-regulation of EGFR and proliferation of lung cancer cells. Knockdown of CAR10 inhibited cell growth in vitro and in vivo, suggesting the role of lncRNAs in environmental lung carcinogenesis [53]. To gain new insights into the pathogenesis of NSCLCs, the molecular mechanisms underlying the roles of several lncRNAs such as MALAT1, HOTAIR, H19, and PVT1 have been extensively investigated. We list the majority of known NSCLC-associated lncRNAs and their functions in Table 1. Their potential application as early diagnostic or prognostic biomarkers and efficient therapeutic targets in patients with NSCLCs warrants further investigations.
Table 1.
NSCLC-associated lncRNAs
| lncRNA | Expression | Key factors | Functions | Ref. |
|---|---|---|---|---|
| CAR10 | Up | YB-1 | Promote cell proliferation | [53] |
| MALAT1 | Up | SR, PC2, hnRNP C | Promote cell proliferation, migration, and invasion | [54] |
| HOTAIR | Up | PRC2, LSD1 | Promote cell proliferation, invasion, and metastasis | [55] |
| H19 | Up | miR-675, c-MYC, p53 | Suppress apoptosis Promote cell growth |
[56] |
| RGMBAS1 | Up | RGMB | Promote cell metastasis | [57] |
| PVT1 | Up | Promote cell proliferation, migration, and invasion | [58] | |
| GHSROS | Up | Promote cell migration | [59] | |
| NKX2-AS1 | Up | EZH2, UTX | Promote cell growth, Regulate cell shape |
[60] |
| BCYRN1 | Up | c-MYC | Promote cell motility, migration, and invasion | [61] |
| DLX6-AS1 | Up | DLX6 | Carcinogenesis | [62] |
| AFAP1-AS1 | Up | Actin filament integrity | Promote cancer cell metastasis | [63] |
| SOX2-OT | Up | PRC2 | Promote cell proliferation | [64] |
| CARLo-5 | Up | Promote cell proliferation, migration, and invasion, | [65] | |
| Lnc_bc060912 | Up | PARP1, NPM1 | Repress cell apoptosis | [66] |
| MVIH | Up | Promote cell proliferation and invasion | [67] | |
| HNF1A-AS1 | Up | DNMT1 | Promote tumor proliferation and metastasis | [68] |
| CCAT2 | Up | Promote cell proliferation and invasion | [69] | |
| LUADT1 | Up | SUZ12, p27, LUAD | Regulate cell cycle | [70] |
| ZXF1 | Up | Promote cell invasion and metastasis | [71] | |
| ANRIL | Up | PRC2 | Correlate with TNM stages and tumor size | [72] |
| SCAL1 | Up | Nrf-2 | Mediate oxidative stress protection | [52] |
| NRG1 | Up | Carcinogenesis | [73] | |
| DQ786227 | Up | Mediate oxidative stress protection | [50] | |
| LOC728228 | Up | Mediate oxidative stress protection | [51] | |
| GAS5 | Down | p53, E2F1, miR-21 | Induce apoptosis, drug resistance | [74] |
| GAS6-AS1 | Down | Suppress metastasis | [75] | |
| PANDAR | Down | p53, NF-YA, Bcl-2 | Repress cell proliferation | [76] |
| HMlincRNA717 | Down | Associate with lymph node metastasis | [77] | |
| MEG3 | Down | P53 | Suppress cell proliferation, Induce apoptosis | [78] |
| TUG1 | Down | P53, PRC2 | Suppress cell proliferation | [79] |
| SPRY4-IT1 | Down | PRC2 | Induce apoptosis, Suppress cell proliferation | [80] |
| BANCR | Down | Suppress cell proliferation, Induce apoptosis | [81] | |
| AK126698 | Down | Wnt pathway | Mediate cisplatin resistance | [82] |
Note: AFAP1-AS1, actin filament associated protein 1 antisense RNA 1; ANRIL, antisense noncoding RNA in the INK4 locus; BANCR, BRAF-activated non-coding RNA; BCYRN1, brain cytoplasmic RNA 1; CAR10, chromatin associated RNA intergenic 10; CARLo-5, also known as colon cancer associated transcript 1 (CCAT1); CCAT2, colon cancer associated transcript 2; DLX6, distal-less homeobox 6; DLX6-AS1, distal-less homeobox 6 antisense RNA 1; DNMT1, DNA methyltransferase 1; EZH2, enhancer of Zeste homolog 2; GAS5, growth arrest-specific transcript 5; GAS6-AS1, growth arrest-specific transcript 6 antisense RNA 1; GHSROS, growth hormone secretagogue receptor opposite strand; HNF1A-AS1, HNF1 homeobox A antisense RNA 1; hnRNP C, heterogeneous nuclear ribonucleoprotein C; HOTAIR: Hox antisense intergenic RNA; lncRNA, long non-coding RNA; LSD1, lysine-specific demethylase 1; LUADT1, lung adenocarcinoma associated transcript 1; MALAT1, metastasis associated lung adenocarcinoma transcript 1; MEG3, maternally expressed gene 3; MVIH, microvascular invasion in HCC; NF-YA, A subunit of nuclear factor-Y; NKX2-AS1, NK2 homeobox-1 antisense RNA 1; NPM1, nucleophosmin 1; Nrf-2, NF-E2-related factor 2; NRG1, nickel-related gene 1; NSCLC, non-small-cell lung cancer; PANDAR, promoter of CDKN1A antisense DNA damage activated RNA; PARP1, poly (ADP-ribose) polymerase 1; PC2, subtilisin-related proprotein convertases 2; PRC2, polycomb repressive complex 2; PVT1, plasmacytoma variant translocation 1; RGMB, repulsive guidance molecule b; RGMBAS1, repulsive guidance molecule b antisense RNA1; SCAL1, smoke and cancer-associated lncRNA-1; SOX2-OT, SRY-box 2 overlapping transcript; SPRY4-IT1, SPRY4 intronic transcript 1; SR, serine/arginine RNA splicing protein; SUZ12, suppressor of Zeste 12; TUG1, taurine-upregulated gene 1; UTX, lysine demethylase 6A; YB-1, Y-box-binding protein 1; ZXF1, as known as ACTA2 antisense RNA 1 (ACTA2-AS1).
lncRNAs as biomarkers of NSCLCs
To improve overall survivals of patients, it is important to exploit new biomarkers for diagnosing, subtyping, and prognosing of NSCLCs. More and more studies have been focused on ncRNAs, particularly miRNAs in the past few years [83]. Likewise, studies have indicated that aberrant expression of lncRNAs is also a hallmark of carcinomas and some lncRNAs show tissue- or cell-specific expression pattern [84], suggesting their potentials as biomarkers. Several lncRNAs have been reported as candidate biomarkers, e.g., highly up-regulated in liver cancer (HULC) in human hepatocellular carcinoma [85] and prostate cancer gene 3 (PCA3) in prostate cancer [86]. Notably, many dysregulated lncRNAs have been identified in patients with NSCLCs (Table 1), suggesting that lncRNAs could be used for screening effective and specific biomarkers of NSCLCs.
To screen for lncRNAs as biomarkers for LADs at early-stage, Li et al summarized microarray data of 181 patients with early-stage LADs to examine their lncRNA expression profiles. As a result, they found that LINC00313 was highly expressed in patients with T2- and N1-stage LADs [87]. Therefore, LINC00313 could be used as a diagnostic biomarker of early-stage LADs. lncRNAs can be detected in serum, which makes it easier for clinical applications. Hence, researchers put more emphasis on circulating lncRNAs. As a results, MALAT1 [88], XIST, and HIF1A-AS1 [89] were found overexpressed in NSCLC patients’ serum when compared with controls. These lncRNAs may act as diagnosis biomarkers for screening NSCLCs via peripheral blood detection.
Subtyping of NSCLC cases is important for the selection of clinical treatment options. For instance, patients with LAD and LSCC differ in clinical outcomes. Zhao et al identified 72 differentially-expressed (23 upregulated and 49 downregulated) lncRNAs in patients with LADs and LSCCs by using human Affymetrix microarrays (HGU133plus2.0) [90]. Likewise, White et al identified 27 lung cancer-associated lncRNAs, which could be used as novel biomarkers for stratifying LADs and LSCCs [91]. Zhang et al found that expression of a novel lncRNA, LINC01133, was upregulated in LSCC but not in LAD samples [92]. All these findings indicate that some lncRNAs could serve as potential biomarkers for distinguishing subgroup of NSCLCs.
lncRNAs could also be used as prognostic biomarkers in patients with NSCLCs. For instance, expression levels of lncRNAs RP11-21L23.2, GPR158-AS1, RP11-701P16.5, and RP11-379F4.4 were negatively correlated with NSCLC patients’ overall survival. Conversely, expression levels of lncRNAs CTD-2358C21.4, RP11-94L15.2, KCNK15-AS1, and AC104134.2 were positively associated with the overall survival of NSCLC patients [93].
The observations above indicate that despite their obscure roles in lung tumorigenesis, these lncRNAs may be valuable for diagnosis of NSCLCs, selecting treatment protocols, and predicting the prognosis of patients with NSCLCs.
lncRNAs in the therapeutic resistance of NSCLCs
At present, surgical excision, chemotherapy, chest radiotherapy and targeted therapy are used alone or in combination to treat patients with NSCLC [94]. However, drug therapies fail in most NSCLCs due to development of drug resistance [95]. Studies have suggested an important role of dysregulated miRNAs in the development of drug resistance [96]. Additional studies also have demonstrated the association between the expression of certain lncRNAs and chemotherapeutic sensitivity of cancer cells. For instance, H19 induced P-glycoprotein- and MDR1-associated drug resistance in liver cancer cells [97].
Resistance to cisplatin, carboplatin, and EGFR-TKIs is inevitable in treating NSCLCs [98]. In an effort to explore the molecular mechanisms of cisplatin resistance, Yang et al found 1380 lncRNAs differentially expressed between regular A549 and cisplatin-resistant A549 cells, indicating the possible involvement of lncRNAs in cisplatin resistance. The authors identified a novel lncRNA, AK126698, which confers cisplatin resistance by targeting the Wnt pathway [82]. Likewise, other research groups showed that HOTAIR contributed to cisplatin resistance of NSCLC cells by downregulating p21WAF1/CIP1 expression [99] and that MEG3 mediated cisplatin resistance of NSCLC cells by regulating the expression of p53 and Bcl-xL [78]. Patients with low MEG3 expression showed poor response to cisplatin-based chemotherapy [78]. Notably, the effectiveness of cisplatin against LSCCs varied between individuals due to different gene expression profiles [100]. For instance, cisplatin-based chemotherapy was beneficial for LSCC patients with excision repair cross-complementation group 1 (ERCC1)-negative tumors after surgical operation, but not for LSCC patients with ERCC1-positive tumors [101]. Hou et al identified 1702 lncRNAs that were differentially expressed between cisplatin-sensitive and cisplatin-resistant LSCC patients. In particular, the expression of AC006050.3-003 was significantly downregulated in patients showing sensitivity to cisplatin compared with those with resistance to cisplatin, suggesting that AC006050.3-003 may be a biomarker for cisplatin treatment in patients with LSCCs [102]. In addition, Dong et al found that GAS5 enhanced the sensitivity of cells expressing wild-type EGFR to gefitinib treatment [103]. These studies demonstrate the correlations between lncRNAs and drug resistance, providing additional opportunities to overcome drug resistance by targeting lncRNAs and related signaling pathways.
Conclusions and perspectives
With the development of technological approaches, such as lncRNA microarray and RNA sequencing, more and more lncRNAs have been found to be dysregulated in NSCLCs, which function as oncogenes or tumor suppressors. Some of these lncRNAs are associated with different stages of NSCLCs, some are specifically overexpressed in one of the lung cancer subtypes, and some are involved in drug resistance. These findings suggest the important roles of lncRNAs in the pathogenesis and treatment of NSCLCs. However, only a small number of lncRNAs have been well characterized, whereas functions of most lncRNAs remain to be elucidated.
Many key questions still need to be addressed. For example, how lncRNAs regulate downstream pathways? Can we use lncRNAs as predictive markers for lung cancer risk or as early diagnostic or prognostic markers? How do lncRNAs mediate drug resistance? Can we use lncRNAs as appropriate therapeutic targets, how to target them if yes? How do we deliver the therapeutic lncRNAs into target tissues and evaluate their safety? Answers to these and other questions will provide new insights into the pathogenesis of lung cancers and help optimize therapeutic strategies to improve the clinical outcome of this deadly disease, which causes 1.59 million deaths each year worldwide [104].
Competing interests
The authors have declared that there are no competing interests.
Acknowledgments
This work was supported by the National Natural Science Funds for Distinguished Young Scholar (Grant No. 81425025) and the National Basic Research Program of China (Grant No. 2012CB910800). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Handled by William C.S. Cho
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Devesa S.S., Bray F., Vizcaino A.P., Parkin D.M. International lung cancer trends by histologic type: male: female differences diminishing and adenocarcinoma rates rising. Int J Cancer. 2005;117:294–299. doi: 10.1002/ijc.21183. [DOI] [PubMed] [Google Scholar]
- 2.Morgensztern D., Ng S.H., Gao F., Govindan R. Trends in stage distribution for patients with non-small cell lung cancer a national cancer database survey. J Thorac Oncol. 2010;5:29–33. doi: 10.1097/JTO.0b013e3181c5920c. [DOI] [PubMed] [Google Scholar]
- 3.Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X., Yang K., Kuang K. The efficacy and safety of EGFR inhibitor monotherapy in non-small cell lung cancer: a systematic review. Curr Oncol Rep. 2014;16:390. doi: 10.1007/s11912-014-0390-4. [DOI] [PubMed] [Google Scholar]
- 5.Anagnostou V.K., Brahmer J.R. Cancer immunotherapy: a future paradigm shift in the treatment of non-small cell lung cancer. Clin Cancer Res. 2015;21:976–984. doi: 10.1158/1078-0432.CCR-14-1187. [DOI] [PubMed] [Google Scholar]
- 6.Ettinger D.S., Akerley W., Borghaei H., Chang A.C., Cheney R.T., Chirieac L.R. Non-small cell lung cancer, version 2. J Natl Compr Canc Netw. 2013;11:645–653. doi: 10.6004/jnccn.2013.0084. [DOI] [PubMed] [Google Scholar]
- 7.Hainer S.J., Gu W., Carone B.R., Landry B.D., Rando O.J., Mello C.C. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes Dev. 2015;29:362–378. doi: 10.1101/gad.253534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spornraft M., Kirchner B., Pfaffl M.W., Riedmaier I. Comparison of the miRNome and piRNome of bovine blood and plasma by small RNA sequencing. Biotechnol Lett. 2015;37:1165–1176. doi: 10.1007/s10529-015-1788-2. [DOI] [PubMed] [Google Scholar]
- 9.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comings D.E. The structure and function of chromatin. In: Harris H., Hirschhorn K., editors. Advances in human genetics. Springer, US; New York: 1972. pp. 237–431. [DOI] [PubMed] [Google Scholar]
- 12.Thomas C.A., Jr. The genetic organization of chromosomes. Annu Rev Genet. 1971;5:237–256. doi: 10.1146/annurev.ge.05.120171.001321. [DOI] [PubMed] [Google Scholar]
- 13.Gall J.G. Chromosome structure and the C-value paradox. J Cell Biol. 1981;91:13s–14s. doi: 10.1083/jcb.91.3.3s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 15.Holmes D.S., Mayfield J.E., Sander G., Bonner J. Chromosomal RNA: its properties. Science. 1972;177:72–74. doi: 10.1126/science.177.4043.72. [DOI] [PubMed] [Google Scholar]
- 16.Pierpont M.E., Yunis J.J. Localization of chromosomal RNA in human G-banded metaphase chromosomes. Exp Cell Res. 1977;106:303–308. doi: 10.1016/0014-4827(77)90176-8. [DOI] [PubMed] [Google Scholar]
- 17.Berget S.M., Moore C., Sharp P.A. Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busch H., Reddy R., Rothblum L., Choi Y. SnRNAs, snRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- 19.Brannan C.I., Dees E.C., Ingram R.S., Tilghman S.M. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 21.Brown C.J., Hendrich B.D., Rupert J.L., Lafrenière R.G., Xing Y., Lawrence J. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 22.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 23.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 24.Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 25.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 26.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cancer. 2011;10:38. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Fu Z., Ji C., Gu P., Xu P., Yu N. Systematic gene microarray analysis of the lncRNA expression profiles in human uterine cervix carcinoma. Biomed Pharmacother. 2015;72:83–90. doi: 10.1016/j.biopha.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Chen W., Böcker W., Brosius J., Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183:345–351. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weikard R., Hadlich F., Kuehn C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genomics. 2013;14:789. doi: 10.1186/1471-2164-14-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z., Liu X., Liu L., Deng H., Zhang J., Xu Q. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19:561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moretti F., Thermann R., Hentze M.W. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16:2493–2502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forman J.J., Legesse-Miller A., Coller H.A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 39.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu X., Feng Y., Zhang D., Zhao S.D., Hu Z., Greshock J. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015;16:504–516. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steck E., Boeuf S., Gabler J., Werth N., Schnatzer P., Diederichs S. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med. 2012;90:1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 44.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berghoff E.G., Clark M.F., Chen S., Cajigas I., Leib D.E., Kohtz J.D. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu G., Chen J., Pan Q., Huang K., Pan J., Zhang W. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Lin J., Liu T., Chen T., Pan S., Huang W. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85:110–115. doi: 10.1016/j.lungcan.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 50.Gao L., Mai A., Li X., Lai Y., Zheng J., Yang Q. LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol Lett. 2013;223:205–210. doi: 10.1016/j.toxlet.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Hu G., Yang T., Zheng J., Dai J., Nan A., Lai Y. Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Mol Carcinog. 2015;54:E192–E204. doi: 10.1002/mc.22314. [DOI] [PubMed] [Google Scholar]
- 52.Thai P., Statt S., Chen C.H., Liang E., Campbell C., Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49:204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei M.M., Zhou Y.C., Wen Z.S., Zhou B., Huang Y.C., Wang G.Z. Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget. 2016 doi: 10.18632/oncotarget.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen L., Chen L., Wang Y., Jiang X., Xia H., Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 55.Loewen G., Jayawickramarajah J., Zhuo Y., Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li P., Li J., Yang R., Zhang F., Wang H., Chu H. Study on expression of lncRNA RGMB-AS1 and repulsive guidance molecule b in non-small cell lung cancer. Diagn Pathol. 2015;10:63. doi: 10.1186/s13000-015-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Y.R., Zang S.Z., Zhong C.L., Li Y.X., Zhao S.S., Feng X.J. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 59.Whiteside E.J., Seim I., Pauli J.P., O’Keeffe A.J., Thomas P.B., Carter S.L. Identification of a long non-coding RNA gene, growth hormone secretagogue receptor opposite strand, which stimulates cell migration in non-small cell lung cancer cell lines. Int J Oncol. 2013;43:566–574. doi: 10.3892/ijo.2013.1969. [DOI] [PubMed] [Google Scholar]
- 60.Cao Y., Gao Q., Lakshminarayanan M., Huang J., Ren M., Ramirez M.I. Role of a human long non-coding RNA antisense to Nk2–1 in lung tumorigenesis. Am J Respir Crit Care Med. 2013;A4750 [Google Scholar]
- 61.Hu T., Lu Y.R. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J., Li P., Zhao W., Yang R., Chen S., Bai Y. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48. doi: 10.1186/s12935-015-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng Z., Bo H., Gong Z., Lian Y., Li X., Li X. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–737. doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 64.Hou Z., Zhao W., Zhou J., Shen L., Zhan P., Xu C. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 65.Luo J., Tang L., Zhang J., Ni J., Zhang H.P., Zhang L. Long non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non-small cell lung cancer. Tumour Biol. 2014;35:11541–11549. doi: 10.1007/s13277-014-2442-7. [DOI] [PubMed] [Google Scholar]
- 66.Luo H., Sun Y., Wei G., Luo J., Yang X., Liu W. Functional characterization of long noncoding RNA lnc_bc060912 in human lung carcinoma cells. Biochemistry. 2015;54:2895–2902. doi: 10.1021/acs.biochem.5b00259. [DOI] [PubMed] [Google Scholar]
- 67.FANTOM Consortium and the RIKEN PMI and CLST (DGT), Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y., Liu H., Shi F., Yao Y., Yang W., Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2014;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu M., Xu Y., Yang X., Wang J., Hu J., Xu L. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–5380. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 70.Qiu M., Xu Y., Wang J., Zhang E., Sun M., Zheng Y. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:e1858. doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang L., Zhou X.F., Pan G.F., Zhao J.P. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2014;68:401–407. doi: 10.1016/j.biopha.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 72.Nie F.Q., Sun M., Yang J.S., Xie M., Xu T.P., Xia R. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 73.Sang H., Liu H., Xiong P., Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. 2015;36:4027–4037. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 74.Shi X., Sun M., Liu H., Yao Y., Kong R., Chen F. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54:E1–E12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 75.Han L., Kong R., Yin D.D., Zhang E.B., Xu T.P., De W. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol. 2013;30:694. doi: 10.1007/s12032-013-0694-5. [DOI] [PubMed] [Google Scholar]
- 76.Han L., Zhang E.B., Yin D.D., Kong R., Xu T.P., Chen W.M. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie X., Liu H.T., Mei J., Ding F.B., Xiao H.B., Hu F.Q. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2014;7:8881–8886. [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J., Wan L., Lu K., Sun M., Pan X., Zhang P. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One. 2015;10:e0114586. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang E.B., Yin D.D., Sun M., Kong R., Liu X.H., You L.H. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun M., Liu X.H., Lu K.H., Nie F.Q., Xia R., Kong R. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun M., Liu X.H., Wang K.M., Nie F.Q., Kong R., Yang J.S. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang Y., Li H., Hou S., Hu B., Liu J., Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeringer E.M., Rai A.J., DeCastro J., Qu L., Gonzalez M., Chapman L. A complete workflow for high throughput isolation of serum microRNAs and downstream analysis by qRT-PCR: application to cancer biomarker discovery. Cancer Res. 2015;75:3387. [Google Scholar]
- 84.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xie H., Ma H., Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leyten G.H., Hessels D., Jannink S.A., Smit F.P., de Jong H., Cornel E.B. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2014;65:534–542. doi: 10.1016/j.eururo.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 87.Li M., Qiu M., Xu Y., Mao Q., Wang J., Dong G. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol. 2015;36:9969–9978. doi: 10.1007/s13277-015-3714-6. [DOI] [PubMed] [Google Scholar]
- 88.Weber D.G., Johnen G., Casjens S., Bryk O., Pesch B., Jöckel K.H. Evaluation of long noncoding RNA MALAT1 as a biomarker for lung cancer in blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tantai J., Hu D., Yang Y., Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao W., Luo J., Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci Rep. 2014;4:6591. doi: 10.1038/srep06591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.White N.M., Cabanski C.R., Silva-Fisher J.M., Dang H.X., Govindan R., Maher C.A. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J., Zhu N., Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol. 2015;36:7465–7471. doi: 10.1007/s13277-015-3460-9. [DOI] [PubMed] [Google Scholar]
- 93.Zhou M., Guo M., He D., Wang X., Cui Y., Yang H. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Albain K.S., Swann R.S., Rusch V.W., Turrisi A.T., 3rd, Shepherd F.A., Smith C. Radiotherapy plus chemotherapy with or without surgical resection for stage III non-small-cell lung cancer: a phase III randomised controlled trial. Lancet. 2009;374:379–386. doi: 10.1016/S0140-6736(09)60737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Massarelli E., Varella-Garcia M., Tang X., Xavier A.C., Ozburn N.C., Liu D.D. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 96.Ma J., Dong C., Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 97.Tsang W.P., Kwok T.T. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–4881. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 98.Schneider-Merck T., Pohnke Y., Kempf R., Christian M., Brosens J.J., Gellersen B. Physical interaction and mutual transrepression between CCAAT/enhancer-binding protein beta and the p53 tumor suppressor. J Biol Chem. 2006;281:269–278. doi: 10.1074/jbc.M503459200. [DOI] [PubMed] [Google Scholar]
- 99.Liu Z., Sun M., Lu K., Liu J., Zhang M., Wu W. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schiller J.H., Harrington D., Belani C.P., Langer C., Sandler A., Krook J. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 101.Olaussen K.A., Dunant A., Fouret P., Brambilla E., André F., Haddad V. DNA repair by ERCC1 in non–small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 102.Hou Z., Xu C., Xie H., Xu H., Zhan P., Yu L. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS One. 2014;9:e108133. doi: 10.1371/journal.pone.0108133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dong S., Qu X., Li W., Zhong X., Li P., Yang S. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol. 2015;8:43. doi: 10.1186/s13045-015-0140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]