Abstract
The genus Aphanomyces (Saprolegniales, Oomycetes) includes species with a variety of ecologies from saprotrophs to plant and animal parasites. Two important species in this genus are A. astaci, the cause of crayfish plague and its close relative, A. invadans, which causes the epizootic ulcerative syndrome on fish. In this study, we have assembled and annotated the mitochondrial (mt) genomes of A. astaci and A. invadans from the whole genome shotgun sequence reads (PRJNA187372; PRJNA258292, respectively). The assembly was generated from A. astaci Pc-genotype strain APO3 and A. invadans strain NJM9701. The sizes of the mtDNAs were 49,489 bp and 49,061 bp for A. astaci and A. invadans, respectively. The species shared similar genetic content and organization encoding 35 proteins, two ribosomal RNAs, three putative open reading frames and 33 transfer RNAs of 19 amino acids for peptide synthesis. Both species also had a large inverted repeat region (LIR) of approximately 12 kb, the LIR contained large and small ribosomal RNAs and eight protein coding genes. These annotated mt genomes serve as a valuable genetic backbone for further development of diagnostic methods and phylogenetic and migration studies of the animal parasitic species of Aphanomyces.
Oomycetes (Stramenopiles) include a variety of species, both saprotrophic and pathogenic, having plant and animal hosts in terrestrial and aquatic ecosystems1. The plant pathogenic Oomycetes (Peronosporoales) have been reported to cause economically and culturally significant global epidemics, such as potato late blight (Phytophthora infestans) and sudden oak death (Phytophthora ramorum), thus having impacted both human society and natural ecosystems2. Aquatic clades of Oomycetes, like Saprolegniales, are a diverse group adapted to several hosts and different life styles in aquatic environment3. Some species of this aquatic clade impose notable ecological and economic impacts on aquaculture and fisheries4,5. The genus Aphanomyces (Saprolegniales) includes a variety of species belonging to plant pathogens, animal parasites and saprotrophs5, two important animal parasitic species are A. astaci6, the causative agent of crayfish plague, and A. invadans7, which causes epizootic ulcerative syndrome (EUS) in both wild and farmed fish8.
Aphanomyces astaci is a highly specialized parasite of freshwater crayfish species originating from the North American continent9,10. When it spread into European aquatic ecosystems via Italy after the 1860’s, A. astaci became a devastating pathogen for native European crayfish species6,11. The disease has since then dramatically altered the balance of European aquatic ecosystems12. To date, five A. astaci genotypes have been detected in Europe, each of them being introduced with a different North American crayfish species5,13,14,15. However, the true spectrum of A. astaci host species may be much wider, since a limited amount of information is available about genotypes that may infect other North American crayfish species16. The presumably rapid co-evolution of A. astaci and its native European crayfish hosts17, combined with its fairly simple life-history, makes it especially interesting model organism for host parasite co-evolution studies.
Aphanomyces invadans, formerly known as Aphanomyces piscisida, causes epizootic ulcerative syndrome (EUS) in both wild and farmed fish8. More than 100 fish species have been reported to be affected by EUS18. The disease was first detected in Japan19, but has been subsequently reported from continental Asia, Australia, North America and Africa, with different names being given20. Based on several molecular studies, a single clone of A. invadans has been broadly distributed worldwide21,22. A. astaci and A. invadans seem to be closely related on the genetic level5, and they also share several common life style characteristics; i.e. lack of a sexual reproduction cycle, repeated zoospore emergence, and highly specialized parasitic nature, (an alternative host for either species from different taxonomic class is currently unknown)5,21,23.
In Oomycetes, mitochondrial DNA (mtDNA) is maternally inherited24 and has been widely used for tracking the origin and geographic migration of a broad variety of species, perhaps most interestingly studying the historic migration of Phytophthora infestans using herbarium specimens25,26,27. Mitochondrial (Mt) genomes of several genera of Oomycetes have recently been characterized and their size and organization have been found to be highly variable, from approximately 37 kb for Phytophthora infestans28 to 60 kb for Pythium ultimum29. Currently, there are 15 Oomycete mt genomes published30,31,32,33,34,35,36,37, or available in sequence databases (KT946598), but with one exception these represent plant parasitic and saprophytic species while the animal pathogenic group has thus far been underrepresented.
The rapid development of sequencing techniques has offered new possibilities for detailed and comprehensive analyses of different organisms. Recently, A. astaci and A. invadans genomes were sequenced38. The present study describes the assembled mt genomes of these two closely related, animal pathogenic oomycetes, and are the first mt genomes representing the genus Aphanomyces. The presented results will form a solid basis that enables further studies of evolution, diversification and migration of A. astaci and A. invadans, and in a wider scale, the evolution of the animal parasitic lineages of Oomycetes.
Results
Genome size and organization
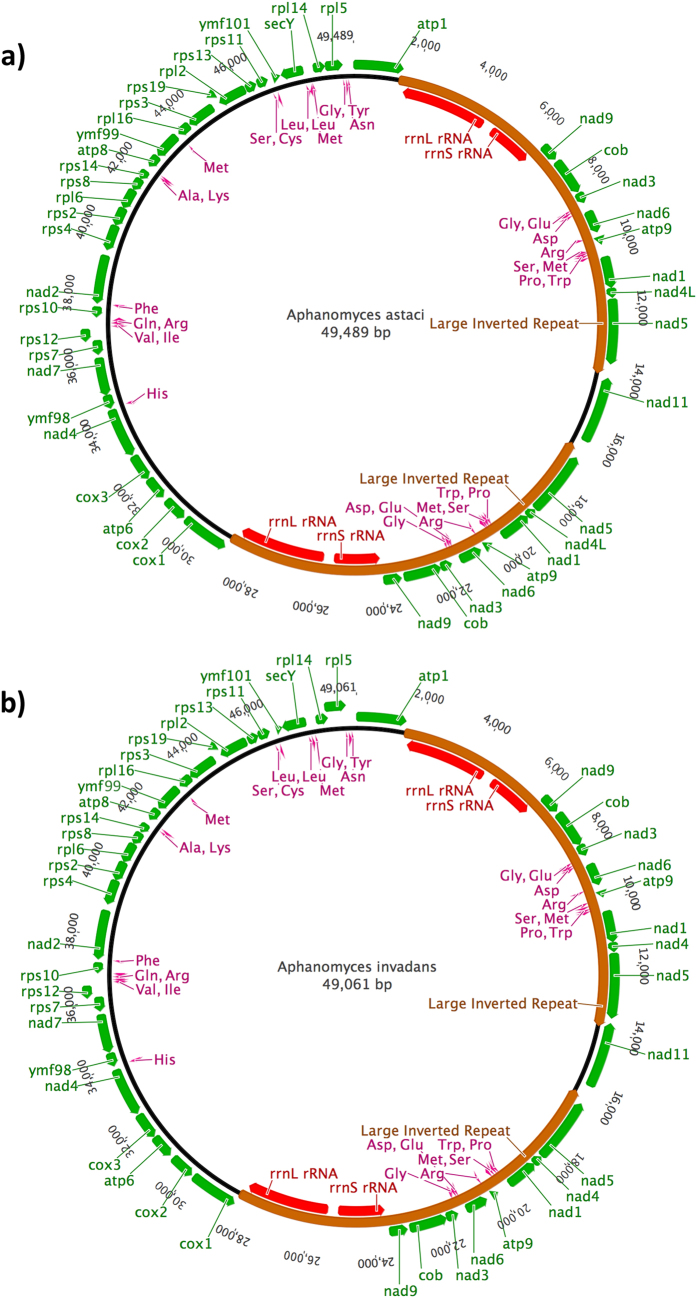
The mt genome size of A. astaci was 49, 489 bp (mean coverage depth in the assembly of 525 reads) (Fig. 1A) while the A. invadans mt genome was 49,061 bp (mean coverage depth in the assembly of 520 reads) (Fig. 1B). The mtDNA content and orientation was similar in both species. There were large inverted repeats (LIRs) in the genomes of these species. In A. astaci there was a 12,570 bp LIR while in A. invadans there was a 12,366 bp LIR. The LIRs in these species represent approximately 50% of the mt genome size. Between the LIRs there were single copy regions of 2,342 bp in A. astaci and 2,133 bp in A. invadans, which coded for the nad11 gene. There was also a 22,008 bp single copy region in A. astaci and a 22,213 bp region in A. invadans that encoded 26 genes and three putative open reading frames (ORFs). A unique feature observed in both Aphanomyces mtDNAs was that the large (rrnL) and small (rrnS) ribosomal RNA subunits, located in the LIR, were concatenated and having a tail-to-tail orientation.
Figure 1. The organization of the mitochondrial genome including protein coding genes (green), large inverted repeat region (brown), ribosomal RNAs (red) and tRNAs (pink).
(A) Aphanomyces astaci mtDNA (49,489 bp) and (B) Aphanomyces invadans mtDNA (49,061 bp).
Genome composition
The A. astaci mt genome was comprised of tightly packed coding regions (92.5%) with 60% of the intergenic spacer regions less than 20 bp. In contrast 32.0% of the intergenic regions in A. invadans were larger than 100 bp, while only 30% was less than 20 bp. The overall GC content of A. astaci mt genome was 22.6% with 23.9% GC content in coding regions and only 7.9% in intergenic regions. The mt genome of A. invadans was composed of 89.6% coding regions and 10.4% intergenic spacer regions. Overall GC content was 21.8% and the GC-content of coding regions was 22.2%, while for non-coding regions it was 15.8%. The largest intergenic spacer in both A. astaci (575 bp) and A. invadans (570 bp) was between nad5 in the second LIR-region and nad11 in the single copy region (Fig. 1A,B). The second largest intergenic spacer region of A. astaci (444 bp) was between nad5 in the first LIR-region and nad11, but for A invadans this spacer was only 129 bp and was completely located in the first LIR-region (Fig. 1B). In general, intergenic spacer regions were larger in the LIR-regions and the genes in the 22,213 bp single copy region were more tightly packed. In both species, the protein encoding genes had an ATG start codon and TAA stop codon. In both species, there were three exceptions for the stop codon for rpl2, rpl5 and ymf98 which were TAG not TAA. No introns were detected in the mt genome of either species.
Protein coding genes and tRNAs
Altogether, the mtDNA of the studied Aphanomyces species encoded 35 proteins and two ribosomal RNAs. The gene set contained 18 respiratory chain proteins, 16 ribosomal proteins and an import protein secY (Table 1). Moreover, three putative ORFs for ymf98 (orf147), ymf99 (orf217) and ymf101 (orf64) were observed. These ORFs have also been observed in other oomycetes. The LIR-region of both species encoded eight genes (atp9, cob, nad1, nad3, nad4L, nad5, nad6 and nad9) and the ribosomal RNAs rrnL and rrnS. The single copy -region of 22,008 bp on the opposite side of the LIRs encoded 29 genes, containing all of the ribosomal proteins, a part of complex I and all of the complex IV and V respiratory chain proteins, and secY import protein (Table 1, Fig. 1).
Table 1. Genetic content of Aphanomyces astaci and A. invadans mtDNA.
| Respiratory chain proteins (ATP synthesis) |
|---|
| Complex I (n = 10) |
| nad1*, nad2, nad3*, nad4, nad4L*, nad5*, nad6*, nad7, nad9*, nad11 |
| Complex III (n = 1) |
| cob* |
| Complex IV (n = 3) |
| cox1, cox2, cox3 |
| Complex V (n = 4) |
| atp1, atp6, atp8, atp9* |
| Ribosomal proteins |
| Small subunit (n = 11) |
| rps2, rps3, rps4, rps7, rps8, rps10, rps11, rps12, rps13, rps14, rps19 |
| Large subunit (n = 5) |
| rpl2, rpl5, rpl6, rpl14, rpl16 |
| Import proteins (n = 1) |
| secY |
| Translational RNAs |
| Ribosomal RNAs (n = 2) |
| rnnS*(SSU), rnnL*(LSU) |
| tRNAs (n = 33) |
| tRNA-GlyTCC*, tRNA-GluTTC*, tRNA-AspGTC*, tRNA-ArgTCT*, tRNA-SerGCT*, tRNA-MetCAT*, tRNA- |
| ProTGG*, tRNA-TrpCCA*, tRNA-TrpCCA*, tRNA-ProTGG*, tRNA-MetCAT*, tRNA-SerGCT*, tRNA- |
| ArgTCT*, tRNA-AspGTC*, tRNA-GluTTC*, tRNA-GlyTCC*, tRNA-HisGTG, tRNA-ValTAC, tRNA-IleGAT, |
| tRNA-GlnTTG, tRNA-ArgGCG, tRNA-PheGAA, tRNA-AlaTGC, tRNA-LysTTT, tRNA-MetCAT, tRNA-SerTGA, |
| tRNA-CysGCA, tRNA-LeuTAA, tRNA-LeuTAG, tRNA-MetCAT, tRNA-GlyGCC, tRNA-TyrGTA, tRNA-AsnGTT |
| Unassigned open reading frames (n = 3) |
| ymf98 (orf143), ymf99 (orf217), ymf101 (orf64) |
The genes and tRNAs marked with an asterisk located in inverted repeat region.
In the mt genome of both species, there was an identical set of 33 tRNAs for 19 different amino acids. No tRNA encoding amino acid threonine were detected. The sizes of the tRNAs varied between 71 and 89 bp and the GC content between 27.6 and 50.0%. Eight tRNAs were located in LIR-region (Table 1, Fig. 1).
Phylogenetic comparisons
The comparison of A. astaci and A. invandas mtDNAs indicated that these two closely related species had an identical mitochondrial gene order and orientation. The similarity percentage of the complete mt genome was 91.6%, being highest (93.4%) in the coding regions, and the lowest (66.7%) in the intergenic spacer regions. The pairwise identity between the LIR of each species was 91.6%, identical to comparisons of the complete mt genome. The individual gene showing the greatest level of similarity was rrnS rRNA, where the similarity reached 97.8%. The highest divergence was observed in the 444 bp intergenic spacer region between the genes nad11 and nad5, locating in the end of the LIR, where the similarity was only 23.4%.
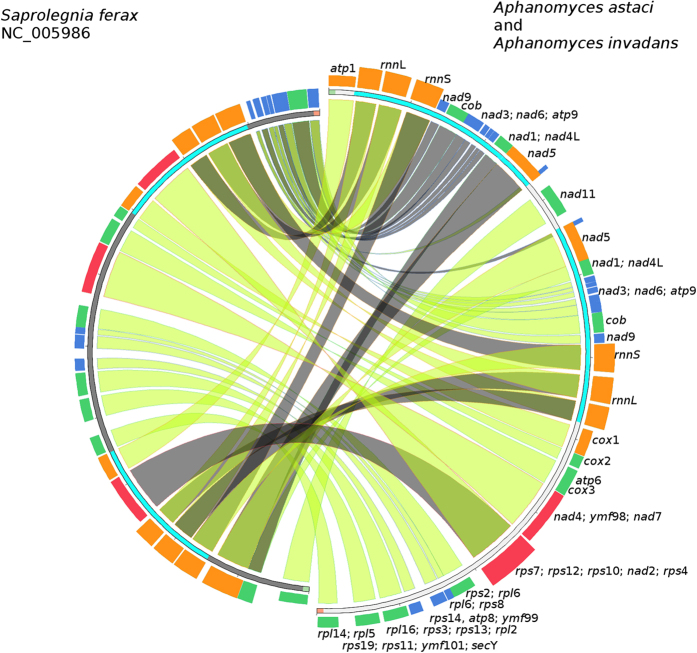
A phylogenetic comparison of a concatenated gene set (n = 37) of 17 oomycete mt genomes placed the Aphanomyces species into the Saprolegnian clade (Fig. 2). Furthermore, the genetic structure of A. astaci and A. invadans mt genomes were compared against S. ferax (Fig. 3), which is their most closely related species in the Saprolegniaceae with a mt genome currently available. The structural comparisons showed that in the approximately 22 kb single copy region, there was a conserved 25 gene order with the exception that in Aphanomyces the nad2-rps10-rps12-rps7 genes were inserted between rps4 and nad7. While the rnnS and rnnL were encoded in the LIR in both genera, in Aphanomyces spp. they had a tail-to-tail-positioning. The remaining list of the LIR encoded genes, containing 8 and 4 additional genes for Aphanomyces spp. and S. ferax, respectively, was completely different. The greatest sequence similarity between the genera were the two approximately 4 kb blocks, containing rps7, -12, -10, nad2 and rps4 genes, and nad4, ymf98 and nad7 -genes (Fig. 3). Overall, the sequence identities of the gene blocks varied between 83 and 95%.
Figure 2. Maximum likelihood (ML) tree of Aphanomyces astaci, A. invandans and 15 other oomycetes based on concatenated mitochondrial gene set of 37 genes.
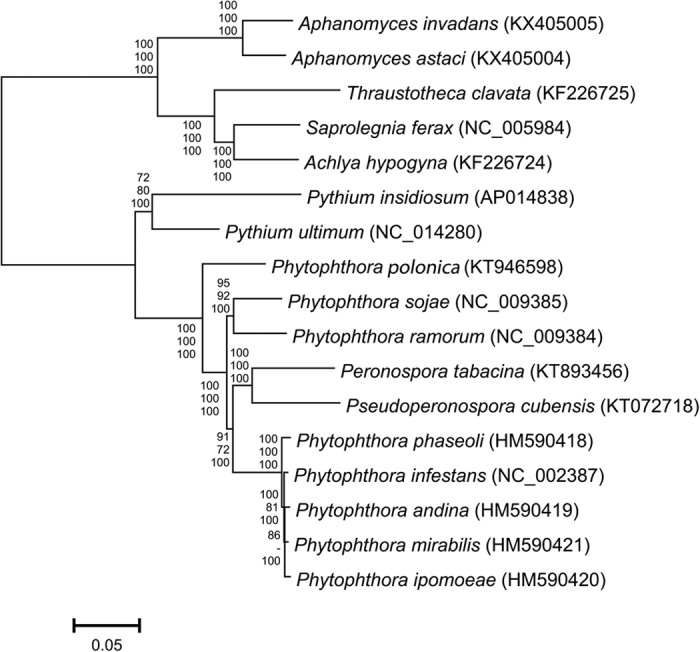
Values at the nodes represent bootstrap support for maximum likelihood (top), maximum parsimony (middle) and posterior probabilities for Bayesian (bottom).
Figure 3. Structural comparison of Aphanomyces astaci and A. invadans mtDNA against Saprolegnia ferax.
Green and red block in the beginning and end of the sequences indicate the sequence orientation. The inverted repeats are marked with turquoise color. The colored blocks outside the sequence describe the blast hit scoring, best quartile being red, then orange, green and blue, respectively. The connective lines are green, when the sequences have inverted orientation and grey, when coded on same strand.
Discussion
Here we assembled and annotated the mt genomes of A. astaci and A. invadans, which both represent a parasitic lineage of the genus Aphanomyces in the Saprolegniales lineage of the oomycete family. The species shared similar genetic content and organization and both species also had a large inverted repeat region (LIR) of approximately 12 kb, which contained large and small ribosomal subunits and eight protein coding genes.
The sizes of the A. astaci and A. invadans mt genomes (49,489 bp and 49,061 bp, respectively), are among the larger range of mtDNA sizes that have been observed in the oomycetes. The largest mt genome so far has been reported for two Pythium species, P. ultimum29 and P. insidiosum36, having a mt genome size of 59.7 and 55 kb, respectively (Table 2). A more closely related species to Aphanomyces, Saprolegnia ferax, had a mtDNA genome of approximately 47 kb30, being similar to the genome size found for Achlya hypogyna and Thraustotheca clavata32. In contrast, Phytophthora species and Peronospora tabacina do not have large inverted repeats, resulting in generally smaller mt genomes ranging from approximately 37 to 43 kb31,33,34. Comparison of the mt genomes of 17 oomycete taxa with and without inverted repeats indicates that genome sizes are fairly consistent when the second LIR copy is excluded (ranging from approximately 36 to 43 kb) with subsequent differences in genome size influenced by the numbers of putative ORFs that are present (Table 2). Also, the GC% of oomycete mtDNAs were uniform and generally low, from 19.0% of P. ramorum to 23.4% of T. clavata, with both Aphanomyces species coming between these.
Table 2. Mitochondrial genome size and number of putative open reading frames for a range of oomycete taxa with and without an inverted repeat.
| Taxa | GenBank accession | Mt genome size (bp) | Inverted repeat size (bp) | Genome size, less 1 arm of inverted repeat | Number putative open reading frames |
|---|---|---|---|---|---|
| Aphanomyces astaci | KX405004 | 49,489 | 12,570 | 36,919 | 3 |
| Aphanomyces invadans | KX405005 | 49,061 | 12,367 | 36,694 | 3 |
| Saprolegnia ferax | AY534144 | 46,930 | 8,133 | 38,797 | 4 |
| Thraustotheca clavata | NC022179 | 47,382 | 9,525 | 37,857 | 4 |
| Achyla hypogyna | KF226724 | 46,840 | 7,973 | 38,867 | 5 |
| Pythium ultimum | GU138662 | 59,689 | 21,950 | 37,737 | 7 |
| Pythium insidiosum | AP014838 | 54,989 | 18,266 | 36,723 | 4 |
| Phytophthora ramorum | DQ832718 | 39,314 | 1,150 | 38,164 | 9 |
| Phytophthora sojae | DQ832717 | 42,977 | — | 42,977 | 13 |
| Phytophthora polonica | KT946598 | 40,467 | — | 40,467 | 6 |
| Phytophthora andina | HM590419 | 37,874 | — | 37,874 | 5 |
| Phytophthora ipomoeae | HM590420 | 37,872 | — | 37,872 | 5 |
| Phytophthora mirabilis | HM590421 | 37,779 | — | 37,779 | 5 |
| Phytophthora infestans | AY898627 | 39,870 | — | 39,870 | 11 |
| Phytophthora phaseoli | HM590418 | 37,914 | — | 37,914 | 5 |
| Peronospora tabacina | NC028331 | 43,225 | — | 43,225 | 13 |
| Pseudoperonospora cubensis | KT072718 | 38,553 | — | 38,553 | 6 |
Number of unassigned putative reading frames, counted only once when present in an inverted repeat.
The large inverted repeat (LIR) regions, observed in mt genomes of A. astaci and A. invadans, ranged in sizes from 12,570 bp to 12,366 bp, respectively. The smallest inverted repeats have been detected in Phytophthora ramorum (~1.1 kb)31, and the largest from Pythium species (~20 kb) (Table 2)39. LIR’s have been considered as the major contributor to the mt genome size variation40 as exemplified among the 17 oomycete taxa in Table 2. The role of these repeat regions have been thought to be the stabilization of the mtDNA structure31. No signs of the flip-flop isomerization, previously observed in oomycetes Achlya ambisexualis41, A. klebsiena42, S. ferax30 and P. ramorum43, were detected in the above noted Aphanomyces species.
The mt genomes of A. astaci and A. invadans encoded 35 protein coding genes and three putative ORFs. In addition, two ribosomal RNAs and 33 tRNAs for 19 amino acids, were detected. In comparison to other sequenced aquatic oomycete species32, the genetic composition, number of genes, and GC% of the mtDNA observed was similar. The largest structural difference was that in both Aphanomyces species, a large part of nad-genes (Table 1, Fig. 3) were located in LIR-region. In addition, the set of putative ORFs was different; ymf98 (orf147), ymf99 (orf217) and ymf101 (orf64) were found in both Aphanomyces spp. with ymf98 and ymf101 shared with S. ferax while ymf99 was not. Furthermore, S. ferax also had unique putative orf’s −273 and −312, which were not present in the studied Aphanomyces mt genomes. Similarities were also observed in the single copy region between arms of the LIR with the same gene order between both genera with the exception that the Aphanomyces spp. had 4 genes and 5 tRNAs inserted between rps4 and nad7. The gene order between secY and rps4 (15 genes, 5 tRNAs) is highly conserved and also observed in the Peronosporales taxa29,31,33.
Based on the phylogenetic analyses, A. astaci and A. invadans group within the Saprolegnian lineage of Oomycetes (Fig. 2), the results are in agreement with previous studies utilizing nuclear molecular markers2,5,44. Furthermore, our unpublished results show a high intraspecific variation in the mt genomes of different A. astaci genotypes, the significance of which awaits additional analysis. Surprisingly the two downy mildews included in this analysis (Pseudoperonospora cubensis and Peronospora tabacina) were grouped within the Phytophthora clade, indicating additional analysis with a larger number of species is needed to clarify this phylogenetic relationship. Mitochondrial barcoding genes have been shown to be a valuable addition to the taxonomic resources of the oomycetes45. Especially in Phytophthora, mitochondrial variation, exhibiting both inter- and intraspecific variation46, has previously been used to trace the spread of the different mitochondrial haplotypes47,48, and as a marker for examining the population structure of historical samples of P. infestans25,26,27.
The two Aphanomyces species examined herein are the first oomycete aquatic animal pathogens with fully assembled mtDNA genomes. The present results provide a useful genetic resource to further explore the population structure, evolution and origins of these devastating animal pathogenic oomycetes. The assembled mtDNA’s will serve as a future resource basis and reference sequence. When working with clonally reproducing organisms such as A. astaci and A. invadans, the mt genome likely provides a valuable marker for population studies.
Material and Methods
The strains
Aphanomyces astaci strain AP0349 was isolated from indigenous European crayfish, Austropotamobius pallipes, suffering of crayfish plague in La Garrotxa National Park in North-Eastern Pyrenees, Cataluña, Spain. The disease outbreak was caused by the Pc-(D)-genotype of A. astaci, which was transmitted to native A. pallipes population from the neighboring invasive red swamp crayfish (Procambarus clarkii) population49. The strain was isolated and maintained in Real Jardín Botánico, Consejo Superior de Investigaciones Científicas (CSIC), Spain.
Aphanomyces invadans strain NJM9701, originally isolated from Ayu (Plecoglossus altivelis) in Japan in 199750 was the subject for the mt genome assembly and annotation in this study. The strain has been used as a starting material for the whole genome shotgun sequencing of A. invadans (PRJNA188085) and our study utilized the sequence data (SRS473776) produced in this sequencing project.
DNA extraction
For genomic DNA isolation, the selected A. astaci strain was grown for three days at 24 °C in peptone glucose broth51. Briefly, fresh mycelia (1 g), were ground to a fine powder under liquid nitrogen, mixed with 10 mL of extraction buffer (0.2 M Tris HCl pH 8.5, 0.25 M NaCl, 25 mM EDTA, 0.5% SDS), 7 mL Tris-equilibrated phenol and 3 mL of chloroform:isoamyl alcohol (24:1), incubated at room temperature for 1 h and centrifuged at 6 000× g for 30 min. The aqueous phase was extracted with equal volume of chloroform:isoamyl alcohol (24:1) and centrifuged at 10 000 × g for 15 min. A total of 50 μl of 10 mg/mL RNase A was added to the aqueous phase and the tubes were incubated at 37 °C for 30 min. Then, isopropanol (0.6 volumes) was added, the samples were mixed gently, and the DNA was precipitated on ice for 30 min. DNA was collected by centrifugation at 10,000 g for 20 min, washed with 70% ethanol, dried and resuspended in DNase/RNase-free water. DNA was checked for quality and RNA contamination by gel electrophoresis using an 0.8% agarose gel.
Library preparation and genome sequencing
A. astaci (PRJNA187372) whole genome shotgun sequencing was generated by BROAD Institute Illumina HiSeq2000TM and the original raw reads (SRS473784 and SRS473776, respectively) were stored in the NCBI sequence read archive database. The whole genome shotgun sequencing of genomic DNA was generated by paired-end library via random selection, 3–5 kb jumping library and Fossil jumping library project (SRP018895). For A. invadans, the sequence reads were directly downloaded from the NCBI sequence read archive (PRJNA188085).
Assembly and annotation
The generated reads were used to assemble the complete mt genome. One assembly of the WGS-reads of A. astaci was made using the RAY-assembler52. A scaffold representing a partial genome was generated and identified by BLASTn-search to match to the mt genome of Saprolegnia ferax (AY534144). To obtain a complete genome a de novo assembly was done with SeqMan NGen, version 4.1.2 (DNASTAR, Madison, WI, USA) with mitochondrial contigs identified by BLAST analysis with the S. ferax mt genome. The mitochondrial contigs were used in a templated assembly with all the sequencing reads with the resulting assembly evaluated for uniformity and depth of coverage. The contig was broken into smaller contigs when gaps/low coverage or inconsistences were observed and reassembled using the small templated assembly option of SeqMan NGen to extend the ends of the contigs and to close the gaps. The borders of the inverted repeat were identified when reviewing the assembly and noting a higher percentage of truncated reads at specific locations; additional sequence comparisons confirmed the border locations. Annotation of coding regions and prediction of ORFs was done with DS Gene v1.5 (Accelrys, San Diego, CA) using the universal genetic code with confirmation of gene identities done by BLAST analysis against published mt genome sequences from Saprolegnia, Pythium, and Phytophthora spp. Annotation of the tRNAs were conducted using tRNA Scan53. Assembly of the A. invadans mt genome followed the same procedure with SeqMan NGen. The sequences were submitted to NCBI GenBank with access numbers KX405004 (A. astaci) and KX405005 (A. invadans).
Comparative genomics
A concatenated mt gene set (n = 37) of 17 oomycetes (Table 2) were included to phylogenetic comparisons. The genes included atp1, atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad11, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl14, rpl16, rpl2, rpl5, rpl6, rps10, rps11, rps12, rps13, rps14, rps19, rps2, rps3, rps4, rps7, rps8, secY, rnnL, and rnnS and the concatenation was made in Geneious 8.054. The optimal substitution model, GTR + G, was selected with JModel Test in Topali v1.555 and three types of phylogenetic trees were generated. The maximum likelihood tree was generated with PhyML56 with 100 bootstrap replicates. The Bayesian tree (MrBayes57) was conducted in two runs with 1 000 000 generations with sampling frequency of 20 and burn in 30%. The maximum parsimony (MP) analysis was done using PAUP version 4.0b10 (Sinaur Associates, Sunderland, MA) with a heuristic tree search with MULPARS on, steepest decent option off, random addition of sequences (10 replicates) and TBR branch swapping. To determine support for the various clades of the trees, the analysis was bootstrapped with 100 replicates. No outgroups were used as the sequence divergence (over 36% in several cases) and the amount of length mutations was too high.
The structural comparisons of aquatic oomycetes (S. ferax, A. astaci and A. invadans) mtDNAs were generated with Circoletto58, combining Blast search59 with Circos output60.
Additional Information
How to cite this article: Makkonen, J. et al. Mitochondrial genomes and comparative genomics of Aphanomyces astaci and Aphanomyces invadans. Sci. Rep. 6, 36089; doi: 10.1038/srep36089 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This research has been supported by the strategic funding of the University of Eastern Finland (Innovative Research Initiatives) (HK, AV, JM), LIFE+ CrayMate (JJ) (LIFE12 INF/FI/233), Finnish Cultural Foundation (RK) and Maj and Tor Nessling Foundation (JJ, JM). JD-U was supported by project of the Ministerio de Economía y Competitividad, Spain (CGL2012-39357).
Footnotes
Author Contributions J.M. wrote the main manuscript text and prepared Figures 1–2 and Table 1. F.M. assembled the A. invadans and finalized the assembly of the A. astaci mt genomes, prepared Table 2, ran the phylogenetic analysis contributed text and corrected the language. A.V. and H.K. contributed the methodology. H.K., F.M., J.J. and R.K. were responsible for the planning and execution of the study. J.D.-U. was responsible for the A. astaci genome project and the collection and handling of the strain used in this study. All authors revised the manuscript.
References
- Kamoun S. Molecular genetics of pathogenic oomycetes. Eukaryot. Cell. 2(2), 191–199 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. N., Abad G. Z., Balci Y. & Ivors K. Identification and detection of Phytophthora: reviewing our progress, identifying our needs. Plant Dis. 96, 1080–1103 (2012). [DOI] [PubMed] [Google Scholar]
- Beakes G. W., Glockling S. L. & Sekimoto S. The evolutionary phylogeny of the oomycete “fungi”. Protoplasma. 249(1), 3–19, doi: 10.1007/s00709-011-0269-2 (2012). [DOI] [PubMed] [Google Scholar]
- Cerenius L., Andersson G. & Söderhäll K. Aphanomyces astaci and Crustaceans in Oomycete Genetics and Genomics - Diversity, Interactions and Research Tools (eds Lamour K. & Kamoun S.) 425–434 (Wiley-Blackwell, 2009). [Google Scholar]
- Diéguez-Uribeondo J. et al. Phylogenetic relationships among plant and animal parasites, and saprotrophs in Aphanomyces (Oomycetes). Fungal Gen. Biol. 46, 365–376 (2009). [DOI] [PubMed] [Google Scholar]
- Schikora F. Die Krebspest. Fisch. Zeit. 9, 529–532, in German (1906). [Google Scholar]
- Willoughby L. G., Roberts R. J. & Chinabut S. Aphanomyces invaderis sp. nov., the fungal pathogen of freshwater tropical fish affected by epizootic ulcerative syndrome. J. Fish Dis. 18(3), 273–275 (1995). [Google Scholar]
- Oidtmann B. Review of biological factors relevant to import risk assessments for Epizootic Ulcerative Syndrome (Aphanomyces invadans). Transbound. Emerg. Dis. 59, 26–39 (2012). [DOI] [PubMed] [Google Scholar]
- Unestam T. & Weiss D. W. The host-parasite relationship between freshwater crayfish and the crayfish disease fungus Aphanomyces astaci: responses to infection by a susceptible and a resistant species. J. Gen. Microbiol. 60(1), 77–90 (1970). [DOI] [PubMed] [Google Scholar]
- Unestam T. Defense reactions in and susceptibility of Australian and New Guinean freshwater crayfish to European crayfish plague fungus. Aust. J. Exp. Biol. Med. Sci. 53(5), 349–359 (1976). [DOI] [PubMed] [Google Scholar]
- Alderman D. J. Geographical spread of bacterial and fungal diseases of crustaceans. Rev. Off. Int. Epizoot. 15(2), 603–632 (1996). [DOI] [PubMed] [Google Scholar]
- Souty-Grosset C., Holdich D. M., Noël P. Y., Reynolds J. D. & Haffner P. (eds), Atlas of Crayfish in Europe (Muséum national d’Histoire naturelle, 2006). [Google Scholar]
- Huang T. S., Cerenius L. & Söderhäll K. Analysis of genetic diversity in the crayfish plague fungus, Aphanomyces astaci, by random amplification of polymorphic DNA. Aquaculture 126, 1–9 (1994). [Google Scholar]
- Diéguez-Uribeondo J., Huang T.-S., Cerenius L. & Söderhäll K. Physiological adaptation of an Aphanomyces astaci strain isolated from the freshwater crayfish Procambarus clarkii. Mycol. Res. 99(5), 574–578 (1995). [Google Scholar]
- Kozubíková E., Viljamaa-Dirks S., Heinikainen S. & Petrusek A. Spiny-cheek crayfish Orconectes limosus carry a novel genotype of the crayfish plague pathogen Aphanomyces astaci. J. Invert. Pathol. 108, 214–216 (2011). [DOI] [PubMed] [Google Scholar]
- Makkonen J., Jussila J. & Kokko H. The diversity of the pathogenic oomycete (Aphanomyces astaci) chitinase genes within the genotypes indicate adaptation to its hosts. Fungal Gen. Biol. 49(8), 635–642 (2012). [DOI] [PubMed] [Google Scholar]
- Jussila J., Vrezec A., Makkonen J., Kortet R. & Kokko H. Invasive crayfish and their invasive diseases in Europe with the focus on the virulence evolution of the crayfish plague in Biological invasions in changing ecosystems. Vectors, ecological impacts, management and predictions (ed. Canning-Clode J.) Ch. 8, 183–211 (De Gruyter Open Ltd, 2015). [Google Scholar]
- Kamilya D. & Baruah A. Epizootic ulcerative syndrome (EUS) in fish: history and current status of understanding. Rev. Fish Biol. Fisher. 24, 369–380 (2014). [Google Scholar]
- Egusa S. & Masuda N. A new fungal disease of Plecoglossus altivelis. Fish Pathol. 6, 41–46 (1971). [Google Scholar]
- Food and agriculture organization of the United Nations (FAO). Report of the international emergency disease investigation task force on a serious finfish disease in Southern Africa. Available at: http://www.fao.org/3/a-i0778e.pdf. (Accessed: 30th January 2016) (2009).
- Lilley J. H. et al. Molecular characterization of the fish-pathogenic fungus Aphanomyces invadans. J. Fish Dis. 26, 263–275 (2003). [DOI] [PubMed] [Google Scholar]
- Boys C. A. et al. Emergence of epizootic ulcerative syndrome in native fish of the Murray–Darling river system, Australia: hosts, distribution and possible vectors. PLoS One 7, e35568 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderhäll K. & Cerenius L. The crayfish plague fungus: history and recent advances. Freshw. Crayfish 12, 11–35 (1999). [Google Scholar]
- Förster H. & Coffey M. D. Mating behavior of Phytophthora parasitica: evidence for sexual recombination in oospores using DNA restriction fragment length polymorphisms as genetic markers. Exp. Mycol. 14, 351–359 (1990). [Google Scholar]
- Yoshida K. et al. The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2, e00731, doi: 10.7554/eLife.00731 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. D. et al. Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 4, 2172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. D., Ho S. Y. H., Wales N., Ristaino J. B. & Gilbert T. P. Persistence of the mitochondrial lineage responsible for the Irish potato famine in extant new world Phytophthora infestans. Mol. Biol. Evol. 31, 1414–1420 (2014). [DOI] [PubMed] [Google Scholar]
- Paquin B. et al. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet. 31, 380–395 (1997). [DOI] [PubMed] [Google Scholar]
- Lévesque C. A. et al. Genome sequence of the necrotrophic plant pathogen, Pythium ultimum, reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 11, R73, doi: 10.1186/gb-2010-11-7-r73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayburn W. S., Hudspeth D. S. S., Gane M. K. & Hudspeth M. E. S. The mitochondrial genome of Saprolegnia ferax: organization, gene content, and nucleotide sequence. Mycologia 96, 980–987 (2004). [PubMed] [Google Scholar]
- Martin F. N., Bensasson D., Tyler B. M. & Boore J. L. Mitochondrial genome sequences and comparative genomics of Phytophthora ramorum and P. sojae. Curr. Genet. 51, 285–296 (2007). [DOI] [PubMed] [Google Scholar]
- O’Brien M. A., Misner I. & Lane C. E. Mitochondrial genome sequences and comparative genomics of Achlya hypogyna and Thraustotheca clavata. J. Eukaryot. Microbiol. 61(2), 1550–7408 (2014). [DOI] [PubMed] [Google Scholar]
- Derevnina L. et al. Genome sequence and architecture of the tobacco downy mildew pathogen, Peronospora tabacina. Molec. Plant Microbe Interact. 28, 1198–2015 (2015). [DOI] [PubMed] [Google Scholar]
- Avila-Adame C. et al. Mitochondrial genome sequences and molecular evolution of the Irish potato famine pathogen, Phytophthora infestans. Curr. Genet. 49(1), 39–46 (2006). [DOI] [PubMed] [Google Scholar]
- Lassiter E. S. et al. Mitochondrial genome sequences reveal evolutionary relationships of the Phytophthora 1c clade species. Curr. Genet. 61, 567–577 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangphatsornruang S. et al. Comparative mitochondrial genome analysis of Pythium insidiosum and related oomycete species provides new insights into genetic variation and phylogenetic relationships. Gene 575(1), 34–41 (2016). [DOI] [PubMed] [Google Scholar]
- Lu W. J., Hu W. G. & Wang G. P. Complete mitochondrial genome of Pseudoperonospora cubensis. Mitochondr. DNA 27, 3487–3488 (2015). [DOI] [PubMed] [Google Scholar]
- Russ C., Tyler B., Diéguez-Uribeondo J. & Van West P. Aphanomyces WGS initiative. Available at: https://olive.broadinstitute.org/projects/aphanomyces_pathogens. (Accessed: 23th May 2016) (2014).
- McNabb S. A., Boyd D. A., Belkhiri A., Dick M. W. & Klassen G. R. An inverted repeat comprises more than three-quarters of the mitochondrial genome in two species of Pythium. Curr. Genet. 12(3), 205–208 (1987). [Google Scholar]
- McNabb S. A. & Klassen G. R. Uniformity of mitochondrial DNA complexity in oomycetes and the evolution of the inverted repeat. Exp. Mycol. 12(3), 233–242 (1988). [Google Scholar]
- Hudspeth M. E., Shumard D. S., Bradford C. J. & Grossman L. I. Organization of Achlya mtDNA: A population with two orientations and a large inverted repeat containing the rRNA genes. P. Natl. Acad. Sci. USA 80(1), 142–146 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D. A., Hobman T. C., Gruenke S. A. & Klassen G. R. Evolutionary stability of mitochondrial DNA organization in Achlya. Can. J. Biochem. Cell B. 62(7), 571–576 (1984). [Google Scholar]
- Martin F. N. Mitochondrial haplotype determination in the oomycete plant pathogen Phytophthora ramorum. Curr. Genet. 54, 23–34 (2008). [DOI] [PubMed] [Google Scholar]
- Riethmüller A., Michael Weiβ M. & Oberwinkler F. Phylogenetic studies of Saprolegniomycetidae and related groups based on nuclear large subunit ribosomal DNA sequences. Can. J. Bot. 77, 1790–1800 (1999). [Google Scholar]
- Robideau G. P. et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Res. 11(6), 1002–1011, doi: 10.1111/j.1755-0998.2011.03041.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattier R. A. M. et al. Sequence variation of intergenic mitochondrial DNA spacers (mtDNA-IGS) of Phytophthora infestans (Oomycetes) and related species. Mol. Ecol. Notes 3(1), 136–138 (2003). [Google Scholar]
- Martin F. N. & Coffey M. D. Mitochondrial haplotype analysis for differentiation of isolates of Phytophthora cinnamomi. Phytopathology 102(2), 229–239 (2012). [DOI] [PubMed] [Google Scholar]
- Yang Z.-H. et al. Mitochondrial DNA polymorphisms in Phytophthora infestans: New haplotypes are identified and re-defined by PCR. J. Microbiol. Meth. 95(2), 117–121 (2013). [DOI] [PubMed] [Google Scholar]
- Rezinciuc S., Galindo J. & Montserrat J. & Diéguez-Uribeondo J. AFLP-PCR and RAPD-PCR evidences of the transmission of the pathogen Aphanomyces astaci (Oomycetes) to wild populations of European crayfish from the invasive crayfish species, Procambarus clarkii. Fungal Biol. 118(7), 612–620 (2014). [DOI] [PubMed] [Google Scholar]
- Phadree P., Kurata O., Hatai K., Hirono I. & Aoki T. Detection and identification of fish-pathogenic Aphanomyces piscicida using polymerase chain reaction (PCR) with species-specific primers. J. Aquat. Anim. Health 16(4), 220–230 (2004). [Google Scholar]
- Söderhäll K., Svensson E. & Unestam T. Chinase and protease activities in germinating zoospore cysts of the parasitic fungus, Aphanomyces astaci, Oomycetes. Mycopathologia 64, 9–11 (1978). [Google Scholar]
- Boisvert S., Laviolette F. & Corbeil J. Ray: Simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J. Comput. Biol. 17(11), 1401–1415 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucl. Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12), 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I. et al. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics 20, 1806–1807 (2004). [DOI] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P. & Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8), 754–755 (2001). [DOI] [PubMed] [Google Scholar]
- Darzentas N. Circoletto: Visualizing sequence similarity with circos. Bioinformatics 26(20), 2620–2621 (2010). [DOI] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 19(9), 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]