Abstract
With the application of advanced molecular cytogenetic techniques, the number of patients identified as having abnormal chromosome 8p has increased progressively. Individuals with terminal 8p deletion have been extensively described in previous studies. The manifestations usually include cardiac anomalies, developmental delay/mental retardation, craniofacial abnormalities, and multiple other minor anomalies. However, some patients with proximal deletion also presented with similar phenotypic features. Here we describe a female child with an 18.5-Mb deletion at 8p11.23–p22 that include the cardiac-associated loci NKX2-6 and NRG1. Further mutation screening of these two candidate genes in 143 atrial septal defect patients, two heterozygous mutations NKX2-6 (c.1A > T) and NRG1 (c.1652G > A) were identified. The mutations were described for the first time in patients with congenital heart disease (CHD). The c.1A > T NKX2-6 generated a protein truncated by 45 amino acids with a decreased level of mRNA expression, whereas the NRG1 mutation had no significant effect on protein functions. Our findings suggest that 8p21-8p12 may be another critical region for 8p-associated CHD, and some cardiac malformations might be due to NKX2-6 haploinsufficiency. This study also links the NKX2-6 mutation to ASD for the first time, providing novel insight into the molecular underpinning of this common form of CHD.
Congenital heart disease (CHD) is the most common developmental defect. The prevalence of CHD at birth is estimated to be between 75 and 90 per 10000 live births1, and it is caused predominantly by genetic factors, including single-gene mutations and chromosomal aberrations. At present, the genetic mechanism underlying CHD is incompletely understood, and much attention is paid to the association between the disease and certain chromosomal aberrations. For example, the majority of individuals with trisomy 18 (Edwards syndrome) have ventricular septal defects (VSD) and patent ductus arteriosus (PDA)2. 22q11 deletions are a relatively common cause of CHD, such as interrupted aortic arch type B and tetralogy of Fallot (TOF) with absent pulmonary valve3. Abnormal dosage of one or more genes within these aberrant chromosomal fragments occurs frequently and at a higher frequency in CHD patients, and these genes are often also associated with extracardiac abnormalities.
Deletion of a segment of the short arm of chromosome 8, which is prone to rearrangements due to nonallelic homologous recombination4, has been described numerous times, and distal deletions of 8p are associated with CHD5. The CHD spectrum includes, but is not limited to, pulmonary stenosis6, secundum ASD, tetralogy of Fallot7, complete atrioventricular canal, double outlet right ventricle8. In addition to CHD, extracardiac manifestations usually present as low birth weight, growth deficiency, mental retardation, dolichocephaly, ears that are low-set and malformed, high-arched palate, thin lips and micrognathia9. The critical region associated with CHD, specifically ASD, is 8p23.110. However, individuals carrying more proximal deletions have also been reported to have CHD and similar associated extracardiac characteristics, suggesting that critical loci for heart defects exist more proximally as well.
A cluster of genes affecting cardiac differentiation is located on the distal 8p region11. Haploinsufficiency of GATA4, the cardiac transcription factor gene that maps to this locus, is considered to be the primary cause of CHD in patients with monosomy of distal 8p4,7. Additional studies also suggest that SOX7 haploinsufficiency may exacerbate the cardiac phenotype of individuals with GATA4 deletions8, and deletion of GATA4 alone or in conjunction with NEIL2 may result in cardiac defects in humans12. Nevertheless, it is interesting to note that some individuals without above genes on the deleted 8p also present with a wide spectrum of CHD. These observations caused us to wonder whether haploinsufficiency of any other genes in this interval may contribute to the heart defects observed in individuals with 8p deletion.
With this in mind, we present the case of a female child with an 18.5-Mb interstitial deletion of proximal 8p and a syndrome that include cardiac anomaly, developmental delay/mental retardation, and craniofacial abnormalities. By comparing our case and previously reported CHD cases with partially overlapping deletions, coupled with DNA sequence analysis and cytobiology experiments, we delineated another critical region of proximal 8p and identified candidate pathogenic genes for the CHD component of the comprehensive phenotype.
Results
Proband description
The proband is the first and only child of healthy, nonconsanguinous parents, who were both 25 years old at the time of her birth. Birth weight was 2800 g (<middle weight -1SD) and height was 49 cm; head circumference was not available. Delivery was by caesarean section after full-term gestation. The pregnancy and delivery were uneventful. At the age of 3 months the child was diagnosed with ostium secundum ASD, which was later surgically corrected. MRI of the brain at 9 months showed periventricular leukomalacia. At 11 months, weight was 4600 g (<middle weight -3SD); height was 62 cm (<average height -3SD); head circumference was 39.1 cm (<average head circumference -3SD). She could neither raise her head nor sit up, and physical examination revealed severe developmental delay and intellectual disability (Fig. 1). At 15 months, weight was 6800 g (<middle weight -3SD); height was 65.8 cm (<average height -3SD); head circumference was 41.2 cm (<average head circumference -3SD) and bone-strength test showed calcium deficiency. She was not able to walk or talk. Detailed clinical features of the patient are shown in Table 1.
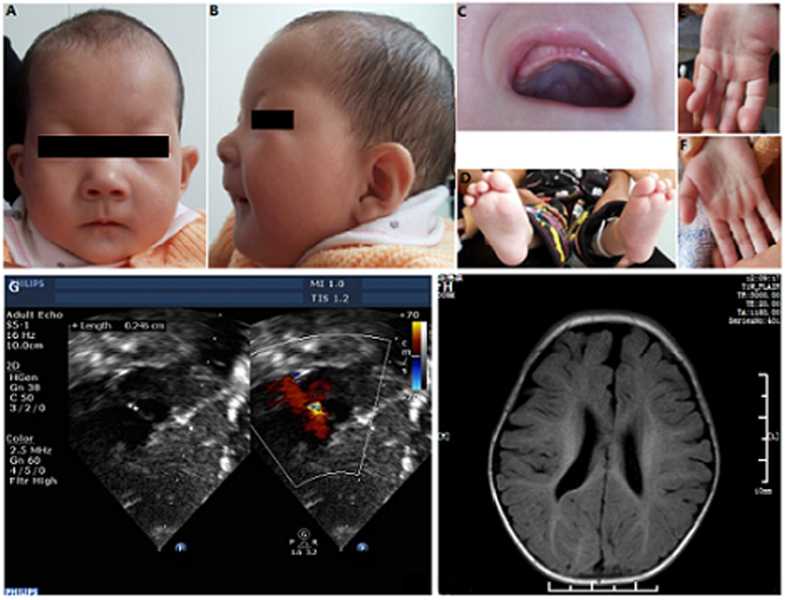
Figure 1. Proband at age 11 mouths.
(A) Frontal view of face. (B) lateral view. (C) The high arched palate is clearly visible. (D) Toe deformity. (E,F) The simian line. (G,H) Echocardiogram showing secundum ASD; MRI of the brain showing periventricular leukomalacia.
Table 1. Clinical features of the proband and previously reported CHD patients with partially overlapping chromosome 8p deletions.
| Reference | P1 | P 2 | P 3 | P 4 | P 5 | P6 | P7 | Present |
|---|---|---|---|---|---|---|---|---|
| 42 | 43 | 9 | 6 | 6 | 44 | 45 | — | |
| Gender | M | M | F | M | M | M | M | F |
| Deletion 8p | p21.lp23.1 | p11.23p21.1 | p11.23p21.3 | p21p23 | p21p23 | p12p21.3 | p22p12 | p11.23p22 |
| DD/MR | + | + | + | + | + | + | + | + |
| Microcephaly | + | − | + | + | + | + | − | + |
| Neuropathy | NA | + | NA | NA | NA | − | NA | + |
| Cerebral MRI abnormality | NA | − | NA | NA | NA | + | NA | + |
| Hypertelorism | − | + | + | − | + | NA | NA | + |
| Epicanthal folds | + | + | + | + | + | + | NA | + |
| Short nose | − | NA | + | NA | NA | NA | NA | + |
| Malformed ears | + | + | + | + | + | + | + | − |
| Micrognathia | + | + | + | NA | NA | NA | NA | − |
| Small mouth | + | + | + | + | − | NA | NA | − |
| Thin lips | NA | + | + | NA | NA | + | NA | + |
| Upslant palpebral fissures | − | − | − | NA | NA | + | NA | + |
| High arched palate | + | − | + | − | − | NA | NA | + |
| Heart Defects | CAVC, PS | ASD, PDA, MI | PS | AVC, PS, D, PLSVC, HRV | AVC, SAS, PLSVC | VSD | VSD, PDA, PA | ASD |
| Genitourinary anomalies | + | + | − | + | + | + | NA | − |
| Ocular abnormality | NA | + | NA | NA | NA | + | NA | + |
| Auditory problems | NA | NA | NA | NA | NA | + | NA | − |
| cleft palate | NA | + | − | NA | NA | NA | NA | − |
| Simian creases | NA | NA | − | NA | NA | NA | NA | + |
| Spherocytosis | NA | + | − | NA | NA | − | NA | − |
| malformedhands, feet | + | + | + | NA | NA | NA | NA | + |
| others | 1 | 2 | 3 | − | − | NA | 4 | − |
(since 1995) DD/MR: developmental delay/mental retardation; ASD: atrial septa1 defect; PDA: patent ductus arteriosus; MI: mitral insufficiency; PS: pulmonary stenosis.; VSD: ventricle septum defect; CAVC: complete atrioventricular canal; AVC: atrioventricular canal; SAS: subaortic stenosis; PLSVC: persistent left superior vena cava; HRV: hypoplastic right ventricle; D: dextrocardia; NA: not assessable. Others: 1. cryptorchidism and hypospadia; 2. bilateral undescended testis, 11 pairs of ribs; 3. bilateral inguinal hernias, anal atresia; 4. hypospadias, hirschrung disease.
Molecular and cytogenetic analysis
G-banding of the probannd’s chromosomes demonstrated a significant deletion of chromosome 8p and a paracentric inversion of chromosome 13q in all metaphases analyzed (Fig. 2A). To verify loss of genomic material from chromosome 8p, we further performed CytoScan HD array analysis and identified an 18.5-Mb region loss of genomic copy numbers at 8p22–p11.23, refining the karyotype to 46,XX,der(8)inv(13), arr 8p22–p11.23(18,634,986–37,160,315)x1 (Fig. 2B,C). Eighty-two genes documented in the OMIM database were located within the deleted region on chromosome 8p. The chromosome karyotype of the mother was normal and that of the father was 46,XY,22ps+, suggesting that the deletion of the proband was de novo.
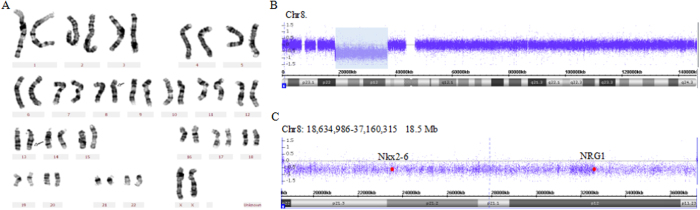
Figure 2. Results of molecular and cytogenetic analysis.
(A) G-banded karyotype showing a karyotype of 46,XX,del(8)(p22p11.23)inv(13). The arrows indicate the breakpoints on normal chromosomes. (B) CytoScan HD array present with an 18.5-Mb deletion at chromosome bands 8p22–p11.23(18,634,986–37,160,315)x1 (NCBI build 37/hg19). (C) zoomed-in view. Red spots indicate the sites of NKX2-6 and NRG1.
Mutation screening
Genes within the 18.5-M interval with reported expression in cardiac tissues or with roles in heart development, including NKX2-6 (NM_001136271) and NRG1 (NM_013956), were considered CHD candidates. To determine whether disruption of NKX2-6 or NRG1 could explain the CHD phenotype, we screened all coding exons and flanking introns in these genes in 143 sporadic ASD patients and 200 normal Chinese children. As a result, we identified two nonsynonymous mutations that result in amino acid changes in two unrelated ASD patients.
A heterozygous sequence variation of NKX2-6 was detected in 1/143 ASD patients, with a mutational prevalence of 0.6999% (Fig. 3A). This mutation site was not found in 200 normal Chinese children. Specifically, the NKX2-6 missense mutation (c.1A > T) was not located in the highly conserved DNA-binding homeodomain but disrupted the start codon of the gene (ATG to TTG). The mutation is not described in the dbSNP, HGMD, or 1000 GP database, and it has not been previously reported in the literature in association with any disease. The affected residue is highly conserved across vertebrates (Fig. 3B).
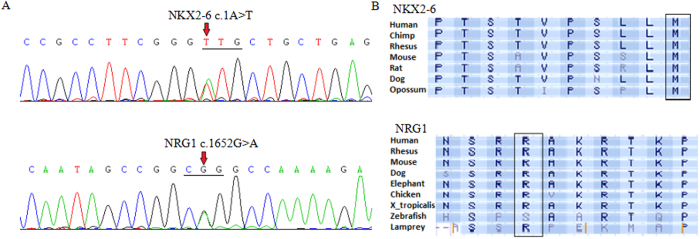
Figure 3. Sequence analysis of NKX2-6 and NRG1.
(A) Gene sequencing peak map showing the NKX2-6 and NRG1 mutations, the arrow indicates the altered heterozygous nucleotides of c.1A > T in NKX2-6 and c.1652G > A in NRG1. (B) Alignment of multiple NKX2-6 and NRG1 amino acid sequences among species. The NKX2-6 and NRG1 amino acid residues affected by these mutations are evolutionarily conserved across various species.
Sequence analysis of NRG1 identified a heterozygous mutation in 1/143 ASD patients with the same mutational prevalence as NKX2-6. The mutation consisted of a change from guanine to adenine at nucleotide position 1652 (c.1652G > A), corresponding to conversion of a basic arginine to a polar neutral glutamine at amino acid position 551 (p.R551Q), in the cytoplasmic tail domain (Fig. 3A). Mutation R551Q had been reported to the dbSNP database with ID rs141355195 and a frequency of 0.1% in the 1000 Genomes Project phase 1 population. The affected residue was evolutionarily well conserved in NRG1 orthologs in other species (Fig. 3B).
Impact of NKX2-6 mutation on mRNA and protein expression
To understand the effects of c.1A > T on NKX2-6 mRNA and protein levels, we constructed C-terminal FLAG-tagged NKX2-6 wild-type and c.1A > T mutant expression vector. COS7 cells were transfected with the NKX2-6 wild-type or mutant vector, or pcDNA3.1 vector as a control. We first compared the levels of mRNA in cultured cells by quantitative PCR (qPCR). NKX2-6 mRNA level in c.1A > T cells was dramatically reduced to ≈33% of the level in wild-type cell lines (Fig. 4A).
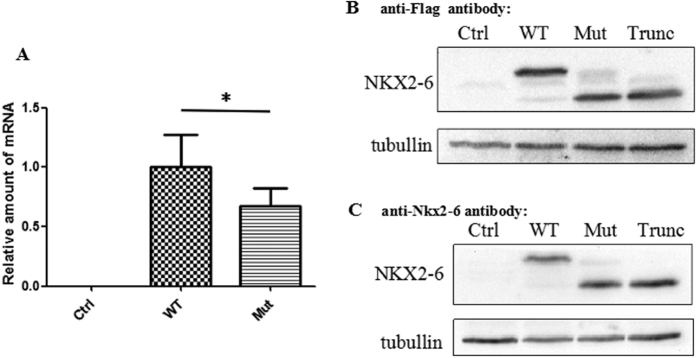
Figure 4. Expression analysis of NKX2-6 in COS7 cells.
(A) NKX2-6 mRNA expression of c.1A > T (Mut) compared with wild-type (WT). mRNA levels normalized to that of GAPDH and then to WT. (B,C) Western blots showing the proteins produced by cells transfected with wild-type, c.1A > T, and c.A1_A135del (Trunc) NKX2-6 vector. Tubullin was used as the loading control for normalization.
C.1A > T is a missense mutation predicted to start the NKX2-6 translation at the second AUG codon (amino acid M46), suggesting that the truncated protein is p.M1_R45del. M46 is in frame with the canonical start codon. We analysis the proteins by an immunoblotting using anti–Flag antibody as well as anti–NKX2-6 antibody. The results showed that the molecular weight of the mutant protein was 5 kDa less than wild-type (30kDa versus 35 kDa) (Fig. 4B,C). To test the hypothesis that c.1A > T cells produce p.M1_R45del NKX2-6 proteins, we next generated expression constructs encoding a C-terminal FLAG-tagged truncted NKX2-6 beginning at the M46 codon (c.A1_A135del). Overexpression of these proteins in COS7 cells followed by western analysis demonstrated that proteins encoded by c.1A > T mutant showed similar bands to c.A1_A135del NKX2-6 in the SDS-gel, (Fig. 4B,C), which was also further confirmed by mass spectrometer (Supplemental Fig. S1).
Mutation results in no change in NRG-1 protein and subcellular localization
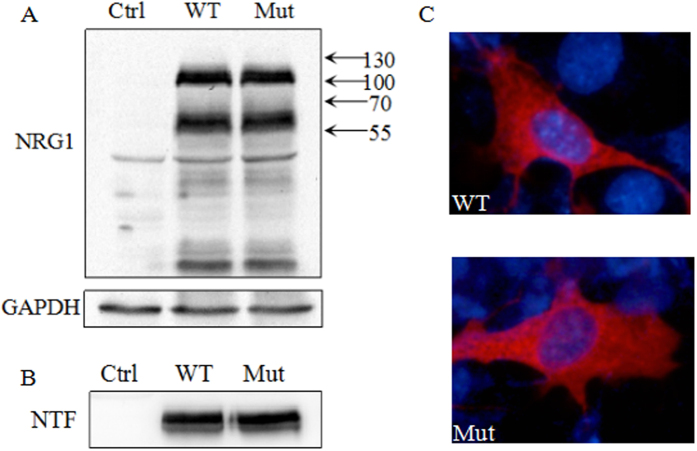
We evaluated the function of the NRG1 p.R551Q mutant protein by overexpressing the wild-type and mutant proteins in COS7 cell. Western analysis of lysates of cells expressing type I NRG1β1a using anti-NRG1α/β1/2 antibody against the cytoplasmic domain of the protein revealed prominent bands of 60 kDa and 110 kDa. It has been suggested that the 110-kD band represents the NRG1 pro-protein, and the 60-kD band the cleaved NRG1 intracellular fragments13. Based on amino acid sequences, the calculated molecular weight of type I NRG1β1a is 73 kDa. That the protein bands are larger than predicted is likely due to posttranslational modifications such as glycosylation. In the cultured medium, a 45-kDa FLAG-tagged peptide was observed, representing the N-terminal fragment of type I NRG1β1a. However, the R551Q mutation of NRG1 does not seem to grossly alter the protein expression (Fig. 5A) or the release of extracellular domains (Fig. 5B). Cellular localization of wild-type and mutant NRG1 was analyzed by immunofluorescence staining with anti–NRG1α/β1/2 antibody. Wild-type and mutant NRG1 localized to the cell membrane and the intracellular organelles (endoplasmic reticulum, Golgi apparatus, etc.) but not the nucleus. No apparent difference in localization patterns was observed between mutant and wild-type (Fig. 5C).
Figure 5. Functional analysis of NRG1 in COS7 cells.
(A) Intracellular levels of wild-type (WT) and R551Q mutant (Mut) NRG1 proteins. GAPDH was used as the loading control for normalization. (B) Western blot analysis of the release of the extracellular domain in the culture medium. (C) Cellular localization of wild-type and mutants NRG1 was analyzed by immunofluorescence staining.
Discussion
Proximal 8p deletions carried by CHD patients often occur in the 8p21–p12 region
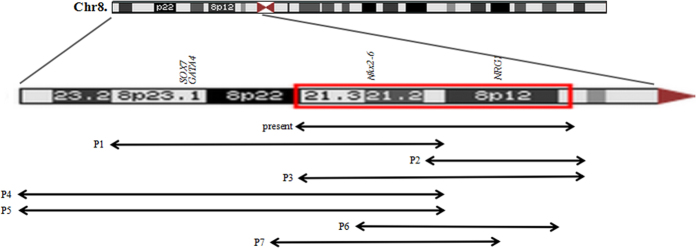
Individuals with a cytogenetically detectable 8p deletion display a series of congenital malformations that often includes CHD, congenital diaphragmatic hernia14, skeletal deformity15, autism16, hypospadias and seizures6. Of these, CHD has been well described in distal 8p deletion patients in the past years, and the phenotype-karyotype correlations have assigned the critical deletion region for heart defects to band 8p235,6. However, heart malformations are frequently described in patients with proximal 8p deletion in the literature6,9, which suggests that there is a need to broaden the critical deletion region on chromosome 8p that is associated with CHD. In clinical practice, we encountered a female patient with an aberrant proximal 8p region, who was diagnosed with ASD, delayed development, impaired intelligence, and abnormal facial appearance. Detailed examination using microarray chromosomal testing revealed the precise 18.5-Mb region of the deletion at 8p22–p11.23, which includes 82 OMIM genes. Subsequently, we summarized the clinical, cytogenetic, and molecular analyses of previously reported heart defects patients who carried overlapping deletions (Fig. 6 and Table 1). All of these patients had a similar phenotype including such features as CHD, delayed development, mental retardation, microcephaly, distinct craniofacial abnormalities. Importantly, all deleted 8p fragments in these patients encompassed 8p12, 8p21 or both, leading us to hypothesize that the 8p21–8p12 deletion is the another critical region accounting for CHD.
Figure 6. Summary overview of clinical characteristics in our patient and previously reported CHD patients with overlaping deletions.
NKX2-6 is the candidate gene for CHD on 8p21–p12
A particular feature of the comprehensive phenotype may result from haploinsufficiency of one or more genes in the chromosome copy number variation region. NKX2-6 is a member of the NK-2 family of transcription factors17 that maps to the deleted proximal 8p region. NKX2-6 is detected in embryos between embryonic day 8.0 (E8.0) and E11.5, but not in later-stage embryos. In the heart, NKX2-6 is expressed in mouse embryos from E8.0–8.5 in posterior myocardial progenitors, then in sinus venosus and dorsal pericardium, and from E9.5 in outflow tract myocardium18,19. NKX2-6 and NKX2-5 belong to the cardiac NK-2 family, which are the vertebrate relatives of drosophila tinman20. Spatiotemporal expression profiles and functions of NKX2-6 partially overlap with those of NKX2-5 during embryogenesis18, and loss of NKX2-6 could be compensated for by an increase in NKX2-5 expression21.
Missense or frameshift mutations in NKX2-6 have been found in different CHDs including persistent truncus arteriosus/common arterial trunk19,22, TFO, DORV, and VSD23,24. However, the functional role of NKX2-6 is far from clarified because there have been no detailed functional studies of NKX2-6 mutations. In this paper, a genetic analysis of NKX2-6 was performed in 135 patients with ASD, and only one missense mutation, c.1A > T, was identified. The NKX2-6 gene consists of two exons that encode the 301 amino acids of NKX2-6 and lacks 5′ or 3′ untranslated regions (UTRs). Subsequent cytobiology experiments of ≈33% reduction in NKX2-6 mRNA levels in c.1A > T cells compared with control cell lines suggests that the mutation at the transcription start site inhibits normal transcription. At the protein level, this mutation causes the loss of the translation start codon ATG for methionine. Further functional studies of the newly identified start codon mutation show that it shifts the initiation start codon to Met+46 (p.M1_R45del), resulting in a protein truncated by 45-amino acids, which contains a conserved transcription repressor tinman domain. Consequently, we propose the N-terminally truncated protein generated by NKX2-6 mutation may be an etiological factor in heart defects.
NRG1 might account for the intellectual disability in proximal 8p deleted patients
NRG1 belongs to the epidermal growth factor (EGF) gene family that is located in chromosomal region 8p12. NRG1 isoforms differ in their levels and patterns of expression in various tissues, including the brain25, heart26, breast27, and gut28. In the heart, NRG1 is expressed and released by the endocardial and microvasculature endothelium, and binds to ErbB receptors with its extracellular EGF-like domain, which further activates a large number of signaling cascades, including Erk 1/2, Akt, and FAK in cardiomyocytes29,30,31. However, no mutations within this gene have been previously identified in CHD, and the R551Q mutation of NRG1 identified in 143 sporadic ASD patients in this paper, does not alter protein functions. Additionally, extensively studies have shown that NRG1 and its receptor ErbB4 plays an important role in many aspects of nervous system, including neural development, myelination32, synaptic plasticity33, radial glia migration34, and neurotransmitter receptor expression35. NRG1 mutations have been reported to be associated with Alzheimers disease36 and schizophrenia37, and autism38. Therefore, we hypothesis that NRG1 haploinsufficiency might account for the mental retardation in most of patients with proximal 8p deletion.
Association of the syndromic phenotype with the deletion region
It should be noted that not all patients with proven proximal 8p deletion have cardiac anomalies. There are several possible explanations for this phenomenon. It may reflect the ample phenotypic variability of NKX2-6 mutations. Compensatory increases in NKX2-5 may mitigate the effects of NKX2-6 deletion, as seen in mice homozygous for a targeted mutation of NKX2-6. These animals are normal as a result of functional complementation between NKX2-5 and NKX2-618,21. Alternatively, other genetic modifier loci or stochastic events may also play an important role in determining the phenotype. Moreover, the size of the deletion had no obvious correlation with the severity of heart disease, which may be related to the presence of some other specific phenotypes. Extracardiac symptoms in these partients may be attributable to the neighboring genes within the deletion region. For example, polypeptides neurofilament medium (NEFM) and neurofilament light (NEFL) are subunits of neurofilaments, and NEFM localizes to axons and dendrites39. A mutation in NEFL associated with Charcot-Marie-Tooth disease (CMT2E) disrupts both neurofilament assembly and axonal transport of neurofilaments in neurons40. Therefore, NEFM and NEFL, to some extent, might impact the intellectual development in patients with proximal 8p deletion. More studies based on patients with similar phenotypes need to be conducted to test these hypotheses.
Conclusion
In our study, we describe a proximal 8p deletion in a female child and summarize the clinical and molecular cytogenetics findings for this patient, as well as review those for previously reported patients, and conclude that proximal 8p21–8p12 is another critical region for CHD. Mutations identified within NKX2-6 (p.M1_R45del) and NRG1 (p. R551Q), the genes for which map to the critical deletion interval, together with a series of functional experiments, expanded our understanding of the significance of the genes in the pathogenesis of CHD. We conclude that CHD associated with proximal 8p deletion is likely due to haploinsufficiency of the cardiac transcription factor NKX2-6. However, in the future, studies employing larger sample sizes, cell-based experiments, and animal models need to be performed to confirm this conclusion.
Materials and Methods
Ethics statement
This study was approved by local ethics committees of Xinhua Hospital and Shanghai Children’s Medical Center (SCMC). The written informed consent was obtained from participants or their guardians prior to commencement of investigation, the patients in the study have provided written informed consent to publishing these case details. All methods were performed in accordance with the relevant guidelines and regulations.
Proband and specimens
The proband consulted in the Department of Pediatric Cardiology at the Xinhua Hospital underwent a comprehensive evaluation including individual and familial histories, medical records, cerebrum MRI, intelligence test, oralo, audio, phthalmologic, neurologic, cardiopneumatic, genitourinary examinations. CHD diagnosis was based on 12-lead electrocardiogram, two-dimensional transthoracic echocardiography with color flow Doppler, and confirmed by echocardiography and cardiac catheterizations examinations. A total of 143 ASD sporadic patients and 200 normal Chinese children performed in the study had been recruited in the Department of Pediatric Cardiology at the Xinhua Hospital and Shanghai Children’s Medical Center from May 2013 to June 2014. The same procedure was followed for the sporadic samples. Individuals with known chromosomal abnormalities or syndromic cardiovascular defects, such as Down syndrome, DiGeorge syndrome, Alagille syndrome and Holt-Oram syndrome, were excluded from the study. Approximately 1–2 ml of peripheral venous blood sample was collected from every participant after acquiring informed consent.
G-banding chromosome analysis
G-banded chromosome analysis were performed using cultured peripheral blood lymphocytes from the proband and her parents. Two milliliters of peripheral blood samples were collected with heparin added and subjected to RPMI1640 culture medium with 20% calf serum (Invitrogen Gibco, USA) at 37 °C in 5% CO2. Preparation of metaphase and conventional cytogenetics followed standard cytogenetic protocols. A minimum of 20 metaphases with good chromosome separation were analyzed. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (International Standing Committee on Human Cytogenetic Nomenclature, 2009).
Affymetrix CytoScan HD array analysis
Genomic DNA of the proband was isolated using the QIAamp DNA Blood Midi Kit (Qiagen) from peripheral blood of the proband following the manufacturer’s instructions. The CytoScan HD Array platform (Affymetrix, USA), which is a high-density chip that contains 2.6 million copy number probes with a resolution of 25 kb, was used to detecte copy number variations (CNVs) at the DNA level. Duplications and deletions were analyzed using Chromosome Analysis Suite (ChAS) software and the annotations of the genome version GRCH37 (hg19). CNVs that affected a minimum of 50 markers in a 100 kb length were initially considered.
Mutation detection
Polymerase chain reaction amplification was used to screen for DNA variants in the entire coding regions and intron-exon boundaries of NKX2-6 (NM_001136271) in 143 ASD individuals. The primer sets are as follows: exon 1F 5′-AATGGGGGGCTACGGTCT-3′ and exon 1R 5′-CGTTAGGGGTGTGTGAAGC-3′; exon 2aF 5′-CCAGGGAGAGGAAAGTCTTG-3′ and exon 2aR 5′-CAGGACGGGCACAGCTACTC-3′; exon 2bF 5′-AGAACCGACGCTACAAATGC-3′ and exon 2bR 5′-GAGATCCCTCCGGAAAGAAG-3′24. The referential genomic DNA sequence of NKX2-6 was derived from GenBank (accession No. NG_030636). Both strands of each PCR product were sequenced with ABI 3730 genetic analyzer (Applied Biosystems).
All isoforms of NRG1 coding exons and flanking introns were amplified in the same 143 ASD specimens using the FastTargetTM technology (Genesky Biotechnologies Inc, Shanghai, China), which is based on improved multiplex PCR amplification and next-generation sequencing. Sequencing was performed on a Miseq bench-top sequencer (Illumina) with 2 × 300 bps methodology. Paired-end sequencing data from the Miseq reporter software was further analyzed off instrument and used to cross calibration the accuracy of sequences. Furthermore, 12–16 selected high-frequency single nucleotide polymorphisms (SNPs) in the target regions were genotyped to validate the target sequencing results. Candidate variants were confirmed by Sanger sequencing.
The identified NKX2-6 and NRG1 sequence variations were queried in the SNP database at NCBI (http://www.ncbi.nlm.nih.gov/), the human gene mutation (HGM) database (http://www.hgmd.org/) and the 1000 Genome Project (1000 GP) database (http://www.1000genomes.org/).
Plasmids and mutagenesis
A full-length human NKX2-6 cDNA (NKX2-6 NM_001136271) was inserted into the plasmid 3 × Flag-pcDNA3.1(+) expression vector at the site of HindIII and XhoI. We also generated C-terminally tagged NKX2-6 expression construct encoding a truncated coding sequence beginning with the M46 codon (c.A1_A145del) by PCR using the following primers: F 5′-GCCCAAGCTTGCCACC ATGGACGCAGAGCCGCGA-3′ and R 5′-CCGCTCGAGCCAGGCCCTGACACCCTGCA-3′. The NRG1 expression vector containing the cDNA of human neuregulin type I-1 isoform (NRG-1 NM_013956.3) was purchased from Origene (Sino, Biological, China). The NRG1 cDNA was digested and subcloned into a Flag-pCMV expression vector at the KpnI and XbaI sites. Specific mutations were introduced into NRG1 and NKX2-6 cDNAs using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, California, USA), and the wild-type plasmids were used as the templates. the primers are as follows: NKX2-6-F 5′-TTAAGCTTGCCACCTTGCTGCTGAGCCCC-3′ and NKX2-6-R 5′-GGGGCTCAGCAGCAAGGTGGCAAGCTTAA-3′; NRG1-F 5′-TGGTTCTTTTGGCCTGCCGGCTATTGGCG-3′ and NRG1-R 5′-CGCCAATAGCCGGCAGGCCAAAAGAACCA-3′. Mutant DNAs have been verified by sequencing.
Cell culture and transfection
COS7 cells were maintained in DMEM high glucose medium (HyClone logan, utah, USA) containing 10% fetal bovine serum (Invitrogen, California, USA) and 100 U/ml penicillin/streptomycin in humidified air at 37 °C with 5% CO2. 2ug of the wild-type or mutant constructs were transfected into COS7 cells in 6-well plate. For Western blotting of NRG1, 20ug of plasmids was transfected into cells in 100 mm dishes. FuGENE HD transfection reagent (Promega, Madison, Wisconsin, USA) was used for transfection according to the manufacturer’s protocol.
Antibodies
The primary antibodies used were anti-NKX2-6 (1:1000, Sigma-AldrichS, AB4500670), anti-NRG1α/β1/2 (C-20) (1:100, Santa Cruz, sc348), anti-FLAG (1:1000, Sigma-Aldrich, F7425), anti-GAPDH (1:10000, Sigma-Aldrich, G9545), anti-tubullin (1:1000, Sigma-Aldrich, T5201). The secondary antibodies used were anti-rabbit (1:10000, Jackson, 111035003), anti-mouse (1:10000, Jackson, 115035003), and Cy3-conjugated affinipure goat anti-rabbit lgG (H + L) (1:100, Jackson ImmunoResearch, 111165003).
Western blotting
At 40 hr after transfection, whole cell lysates were made using cell lysis buffer [20 mM Tris PH7.5, 150 mM NaCl, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM EDTA, 1% Na3VO4, 0.5 μg/ml leupeptin, 1 mM phenylmethanesulfonyl fluoride (PMSF)]. For NRG1 conditioned medium, it was collected and filtered through a 0.22 μm pore size sterile filter unit (Millipore) and then concentrated up to 50-fold by centrifugation through a Amicon-10 K (Millipore) concentrator. Protein concentration was determined with Bicinchoninic Acid assay (Beyotime biotechnology, China). 30μg of total protein was separated on 12% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked at room temperature for 1 h, incubated at 4 °C overnight with primary antibody, washed and incubated with HRP-conjugated secondary antibodies for 1 h, all in 0.1% Tween/TBS with 5% non-fat dry milk. Detection was performed by ECL western blotting detection reagents according to the manufacturer’s instructions.
qRT-PCR
Total RNA was isolated using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. Total RNA (1 μg) was retrotranscribed with PrimeScriptTM RT reagent (TaKaRa). qPCR was performed according to the SYBR® Premix Ex TaqTM II protocol (Applied TaKaRa). Expression levels were normalized to the GAPDH house-keeping gene. Primer sequences are as follows: NKX2-6-F 5′-TCCAGAACCGACGCTACAAAT-3′ and NKX2-6-R 5′-TAGCAAGAGTAGGGCGACACT-3′ (PrimerBank ID:343183349c2); GAPDH-F 5′-ACAACTTTGGTATCGTGGAAGG-3′ and GAPDH-R 5′-GCCATCACGCCACAGTTTC-3′.
Mass spectrum analysis
The lysate was separated with 12% SDS-PAGE and the related band was cut for in-gel digestion as described previously41. The tryptic peptides were analyzed on a nano-HPLC coupled Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, Waltham, MA). The raw spectra data was processed by protein discovery to extract MS/MS spectra data, which was searched against the Uniprot human database (88,817 sequences) by Mascot (v.2.4, Matrix Science, London, UK). Proteins identification was further filtered by perculator with 1% FDR.
Immunofluorescence and subcellular localization
COS7 cells were seeded onto glass coverslips at 5 × 105 cells/ml. Thirty-six hours after transfection, the cells were washed twice with phosphate buffered saline (PBS), fixed with 4% paraformaldehyde for 20 min at room temperature, washed three times in PBS, permeabilized in 0.1% Triton X/PBS at 4 °C for 10 min, and blocked in 1% BSA/0.1% Triton X-100 in PBS at room temperature for 1 h. The cells were incubated with a 1:50 dilution of an anti-NRG1α/β1/2 (C-20) rabbit polyclonal antibody for 1 h at room temperature, after washing three times in PBS, immunosignals were then detected using the secondary antibody Cy3-conjugated affinipure goat anti-rabbit lgG (H + L) at a dilution of 1:100, washed three times in PBS then counterstained with DAPI to detect nuclei. Cells were photographed using an Olympus BX51 fluorescence microscopy.
Additional Information
How to cite this article: Li, T. et al. Identification of candidate genes for congenital heart defects on proximal chromosome 8p. Sci. Rep. 6, 36133; doi: 10.1038/srep36133 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are grateful to the patients and their families for their participation. The project was funded by the grants from the National Natural Science Foundation of China (81270233/H0204), the grant of the Science Committee of Shanghai (13JC1401705), the grant of Shanghai Municipal Commission of Health and Family Planning three-year action plan (GWIV-23). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions T.L., Y.X. and R.X. designed the experiments. T.L. and C.L. performed the experiments. T.L. and R.X. analyzed the data and prepared the figures. Q.G., S.C. and K.S. provide the clinical information of the patients. T.L. wrote the main manuscript text. All authors reviewed the manuscript.
References
- Kahr P. C. & Diller G. P. Almanac 2014: congenital heart disease. Heart. 101, 65–71, doi: 10.5543/tkda (2015). [DOI] [PubMed] [Google Scholar]
- Rosa R. F. et al. Trisomy 18: review of the clinical, etiologic, prognostic, and ethical aspects. Rev. Paul. Pediatr. 31, 111–120, doi: 10.1590/S0103-05822013000100018 (2013). [DOI] [PubMed] [Google Scholar]
- Lindsay E. A. & Baldini A. Congenital heart defects and 22q11 deletions: which genes count? Mol. Med. Today. 4, 350–357, doi: 10.1016/S1357-4310(98)01302-1 (1998). [DOI] [PubMed] [Google Scholar]
- Shimokawa O. et al. Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am. J. Med. Genet. A. 136, 49–51, doi: 10.1002/ajmg.a.30778 (2005). [DOI] [PubMed] [Google Scholar]
- Mei M. et al. Analysis of genomic copy number variations in two unrelated neonates with 8p deletion and duplication associated with congential heart disease. Zhonghua. Er. Ke. Za. Zhi. 52, 460–463, doi: 10.3760/cma.j.issn.0578-1310.2014.06.013 (2014). [DOI] [PubMed] [Google Scholar]
- Digilio M. C. et al. Deletion 8p syndrome. Am. J. Med. Genet. 75, 534–536, doi: 10.1002/(SICI)1096-8628(19980217)75:5< 534::AID-AJMG15 > 3.0.CO;2-L (1998 ). [DOI] [PubMed] [Google Scholar]
- Pehlivan T. et al. GATA4 Haploinsufficiency in Patients With Interstitial Deletion of Chromosome Region 8p23.1 and Congenital Heart Disease. Am. J. Med. Genet. 83, 201–206, doi: 10.1002/(SICI)1096-8628(19990319)83:3< 201::AID-AJMG11 > 3.0.CO;2-V (1999 ). [DOI] [PubMed] [Google Scholar]
- Wat M. J. et al. Chromosome 8p23.1 Deletions as a Cause of Complex Congenital Heart Defects and Diaphragmatic Hernia. Am. J. Med. Genet. A. 149, 1661–1677, doi: 10.1002/ajmg.a.32896 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara M. et al. Interstitial deletion of 8p: report of two patients and review of the literature. Clin. Genet. 48, 41–45, doi: 10.1111/j.1399-0004.1995.tb04052.x (2008). [DOI] [PubMed] [Google Scholar]
- Devriendt K. et al. Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am. J. Hum. Genet. 64, 1119–1126 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio S. et al. Deletion of a 5-cM region at chromosome 8p23 is associated with a spectrum of congenital heart defect. Circulation. 102, 432–437, doi: 10.1161/01.CIR.102.4.432 (2000). [DOI] [PubMed] [Google Scholar]
- Keitges E. A. et al. Prenatal Diagnosis of Two Fetuses With Deletions of 8p23.1, Critical Region for Congenital Diaphragmatic Hernia and Heart Defects. Am. J. Med. Genet. A. 161, 1755–1758, doi: 10.1002/ajmg.a.35965 (2013). [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Miller S. J. & Falls D. L. The N-terminal Region of Neuregulin Isoforms Determines the Accumulation of Cell Surface and Released Neuregulin Ectodomain. J. Biol. Chem. 276, 2841–2851, doi: 10.1074/jbc.M005700200 (2001). [DOI] [PubMed] [Google Scholar]
- Longoni M. et al. Congenital Diaphragmatic Hernia Interval on Chromosome 8p23.1 Characterized by Genetics and Protein Interaction Networks. Am. J. Med. Genet. A. 158, 3148–3158, doi: 10.1002/ajmg.a.35665 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope K. et al. Dextrocardia, Atrial Septal Defect, Severe Developmental Delay, Facial Anomalies, and Supernumerary Ribs in a Child With a Complex Unbalanced 8;22 Translocation Including Partial 8p Duplication. Am. J. Med. Genet. A. 158, 641–647, doI: 10.1002/ajmg.a.34431 (2012). [DOI] [PubMed] [Google Scholar]
- Glancy M. et al. Transmitted duplication of 8p23.1–8p23.2 associated with speech delay, autism and learning difficulties. Eur. J. Hum. Genet. 17, 37–43, doi: 10.1038/ejhg.2008.133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C., Hatzistavrou T. & Harvey R. P. Expression of NK-2 class homeobox gene Nkx2-6 in foregut endoderm and heart. Mech. Dev. 73, 125–127, doi: 10.1016/S0925-4773(98)00037-9 (1998). [DOI] [PubMed] [Google Scholar]
- Nikolova M., Chen X. & Lufkin T. NKX2-6 expression is transiently and specifically restricted to the branchial region of pharyngeal-stage mouse embryos. Mech. Dev. 69, 215–218 (1997). [DOI] [PubMed] [Google Scholar]
- Ta-Shma A. et al. Conotruncal malformations and absent thymus due to a deleterious NKX2-6 mutation. J. Med. Genet. 51, 268–270, doi: 10.1136/jmedgenet-2013-102100 (2014). [DOI] [PubMed] [Google Scholar]
- Bartlett H., Veenstra G. J. & Weeks D. L. Examining the cardiac NK-2 genes in early heart development. Pediatr. Cardiol. 31, 335–341, doi: 10.1007/s00246-009-9605-0 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M. et al. Nkx2.5 and NKX2-6, homologs of Drosophila tinman, are required for development of the pharynx. Mol. Cell. Biol. 21, 4391–4398, doi: 10.1128/MCB.21.13.4391-4398.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heathcote K. et al. Common arterial trunk associated with a homeodomain mutation of NKX2-6. Hum. Mol. Genet. 14, 585–593, doi: 10.1093/hmg/ddi055 (2005). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. A novel NKX2-6 mutation associated with congenital ventricular septal defect. Pediatr. Cardiol. 36, 646–656, doi: 10.1007/s00246-014-1060-x (2015). [DOI] [PubMed] [Google Scholar]
- Zhao L. et al. Prevalence and spectrum of NKX2-6 mutations in patients with congenital heart disease. Eur. J. Med. Genet. 57, 579–586, doi: 10.1016/j.ejmg.2014.08.005 (2014). [DOI] [PubMed] [Google Scholar]
- Meyer D. & Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 378, 386–390, doi: 10.1038/378386a0 (1995). [DOI] [PubMed] [Google Scholar]
- Odiete O., Hill M. F. & Sawyer D. B. Neuregulin in Cardiovascular Development and Disease. Circ. Res. 111, 1376–1385, doi: 10.1161/CIRCRESAHA.112.267286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krane I. M. & Leder P. NDF/heregulin induces persistence of terminal end buds and adenocarcinomas in the mammary glands of transgenic mice. Oncogene. 12, 1781–1788 (1996). [PubMed] [Google Scholar]
- Paratore C. et al. Survival and glial fate acquisition of neural crest cells are regulated by an interplay between the transcription factor Sox10 and extrinsic combinatorial signaling. Development. 128, 3949–3961 (2001). [DOI] [PubMed] [Google Scholar]
- Baliga R. R. et al. NRG-1-induced cardiomyocyte hypertrophy. Role of PI-3-kinase, p70(S6K), and MEK-MAPK-RSK. Am. J. Physiol. 277, H2026–H2037 (1999). [DOI] [PubMed] [Google Scholar]
- Timolati F. et al. Neuregulin-1 beta attenuates doxorubicin-induced alterations of excitation-contraction coupling and reduces oxidative stress in adult rat cardiomyocytes. J. Mol. Cell. Cardiol. 41, 845–854, doi: 10.1016/j.yjmcc.2006.08.002 (2006). [DOI] [PubMed] [Google Scholar]
- Kuramochi Y., Guo X. & Sawyer D. B. Neuregulin activates erbB2-dependent src/FAK signaling and cytoskeletal remodeling in isolated adult rat cardiac myocytes. J. Mol. Cell. Cardiol. 41, 228–235, doi: 10.1016/j.yjmcc.2006.04.007 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velanac V. et al. Bace1 processing of NRG1 type III produces a myelin-inducing signal but is not essential for the stimulation of myelination. Glia. 60, 203–217, doi: 10.1002/glia.21255(2012) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L. & Xiong W. C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat. Rev. Neurosci. 9, 437–452, doi: 10.1038/nrn2392 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluch S. & Juliano S. L. A normal radial glial scaffold is necessary for migration of interneurons during neocortical development. Glia. 55, 822–830, doi: 10.1002/glia.20488 (2007). [DOI] [PubMed] [Google Scholar]
- Corfas G., Roy K. & Buxbaum J. D. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat. Neurosci. 7, 575–580, doi: 10.1038/nn1258 (2004). [DOI] [PubMed] [Google Scholar]
- Go R. C. et al. Neuregulin-1 polymorphism in late onset Alzheimer’s disease families with psychoses. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 139B, 28–32, doi: 10.1002/ajmg.b.30219(2005) . [DOI] [PubMed] [Google Scholar]
- Walss-Bass C. et al. A novel missense mutation in the transmembrane domain of neuregulin 1 is associated with schizophrenia. Biol. Psychiatry. 60, 548–553, doi: 10.1016/j.biopsych (2006). [DOI] [PubMed] [Google Scholar]
- McInnes L. A. et al. The NRG1 exon 11 missense variant is not associated with autism in the Central Valley of Costa Rica. BMC. Psychiatry. 7, 21, doi: 10.1186/1471-244X-7-21 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M. W. et al. The human mid-size neurofilament subunit: a repeated protein sequence and the relationship of its gene to the intermediate filament gene family. EMBO. J. 6, 1617–1626 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlees J. et al. Charcot-Marie-Tooth disease neurofilament mutations disrupt neurofilament assembly and axonal transport. Hum. Mol. Genet. 11, 2837–2844, doi: 10.1093/hmg/11.23.2837 (2002). [DOI] [PubMed] [Google Scholar]
- Wu Z. et al. Quantitative chemical proteomics reveals new potential drug targets in head and neck cancer. Mol. Cell. Proteomics. 10, M111.011635, doi: 10.1074/mcp.M111.011635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino B. et al. Nonrandom Association of Atrioventricular Canal and del(8p) Syndrome. Am. J. Med. Genet. 42, 424–427, doi: 10.1002/ajmg.1320420404 (2005). [DOI] [PubMed] [Google Scholar]
- Okamoto N. et al. Hereditary Spherocytic Anemia With Deletion of the Short Arm of Chromosome 8. Am. J. Med. Genet. 58, 225–229, doi: 10.1002/ajmg.1320580306 (1995). [DOI] [PubMed] [Google Scholar]
- Willemsen M. H. et al. Clinical and molecular characterization of two patients with a 6.75 Mb overlapping deletion in 8p12p21 with two candidate loci for congenital heart defects. Eur. J. Med. Genet. 52, 134–139, doi: 10.1016/j.ejmg.2009.03.003 (2009). [DOI] [PubMed] [Google Scholar]
- Yu S. et al. Genomic profile of copy number variants on the short arm of human chromosome 8. Eur. J. Hum. Genet. 18, 1114–1120, doi: 10.1038/ejhg.2010.66 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.