Abstract
A co-infection comprising to at least seven papillomavirus (PV) types was detected by next generation sequencing (NGS) of randomly primed rolling circle amplification (RCA) products of a bovine (Bos taurus) papilloma lesion from the Brazilian Amazon region. Six putative new PV types that could not be detected by commonly used PCR protocols were identified. Their overall L1 nucleotide identities were less than 90% compared to described PV species and types. L1 nucleotide BLAST sequence hits showed that each new type was related to Beta, Gamma, Dyokappa, Dyoeta, and Xipapillomavirus, as well as two likely new unclassified genera. Our results show that the employment of NGS is relevant to the detection and characterization of distantly related PV and is of major importance in co-infection studies. This knowledge will help us understand the biology and pathogenesis of PV, as well as contribute to disease control. Moreover, we can also conclude that there are many unknown circulating PVs.
Papillomaviruses (PVs) are circular double-stranded DNA viruses containing approximately 8,000 base pairs (bp)1,2. They belong to the Papillomaviridae family and these complex viruses can infect a wide range of amniotes1,2,3. This large family is composed of viruses phylogenetically assigned to 39 genera with several species, types, subtypes and variants1,2. The entire genome must be sequenced for considering new PV types and the L1 open reading frame (ORF) have to differ by more than 10% in comparison to the closest PV types. PV species share between 71% and 89% nucleotide identity within the complete L1 ORF. A PV genus is defined when the similarities are larger than 60% in the L1 ORF. When this difference is between 2% and 10% or less than 2% a subtype or variant occurs, respectively1,2.
Human PV (HPV) encompasses more than 200 types that are fully sequenced, characterized and cataloged, in contrast to the low number of Bos taurus papillomavirus (BPV), which comprises only 15 types (http://pave.niaid.nih.gov). PVs are usually characterized by PCR amplicon sequencing, which is performed using degenerate primer pairs that amplify a relatively conserved L1 gene region from all known PV types and species4. This technique has allowed the identification of divers putative new PVs types in humans and other animals, including BPV types in cattle herds from distinct continents worldwide5,6,7,8.
Although PV detection and characterization in animal species are still poorly studied3,9, primers directed to the PV genera have been commonly used and have shown an increase in the specificity of detection and characterization of PV DNA compared to general primers10,11,12. Nearly two hundred human PVs (HPVs) have been recognized and PV co-infections have been reported using numerous molecular techniques that detect many distinct HPVs genotypes13,14,15,16. Several generic PV primer systems have been developed for the detection of HPV types rather than BPV types17. Additionally, there is much less research of cattle in this field, reflected by the vague BPV types reported and the few cases of BPV co-infection reported18,19,20.
Multiply primed rolling circle amplification (RCA) of circular genomes followed by classic sequencing enabled the discovery of novel animal PVs21. Although this study did not show evidence of co-infections, the RCA followed by NGS has enabled the detection of HPV co-infections22. Moreover, the extraordinary diversity of PV types that infect the animal skin combined with the low numbers of PV types detected by degenerate primers23 indicate that these techniques allow the discovery of only closely related PVs from known genera. Furthermore, PVs could be cultivated only in a sophisticated and arduous raft cell culture system, thereby hampering whole genome analysis due to the lack of necessary adequate amount of purified genomic viral DNA24.
Therefore, an efficient method for amplification and sequencing is essential for improving the identification of PV species that are not detected by current methods mainly in animal researches. The knowledge of PV types is important for epidemiological studies of viral variants in different PV-affected species and to determine the full picture of genetic diversity. Such information will help clarify the biological relationship between distinct viruses with respect to both pathogenesis and treatment. Along these lines, randomly primed RCA followed by NGS was performed to investigate the complete genetic diversity of PVs present in a papilloma lesion.
Results
NGS from RCA products reveals distinct BPV genomes
NGS from RCA products of one papilloma lesion enabled the amplification of seven full-length PV genomes. The contigs associated with PV were assembled from 113,616 high-quality reads (Table 1). The seven contigs were named BPV13 BR/14RO and putative BPV BR/14RO-16 to BPV BR/14RO-21. Chimeric forms were not detected using the RDP4 software. Primer alignment with the Geneious software (version 9) (http://www.geneious.com)25 found mismatches in all sequences at the 3’ end of the FAP59 and MY09 primers (Supplementary Figure 1). A low number of reads probably corresponding to the sequence 14RO_12 (GenBank accession number KP701419) was also identified in the sample. Since the genome was not complete it was not further analyzed.
Table 1. Overall genome coverage and GC-content of the seven contig papillomavirus sequences recovered from a papillomatous lesion.
| GenBank acession no. | Read count* | Mean genome coverage# | GC-content (%) | |
|---|---|---|---|---|
| BPV13 BR/14RO | KU519390 | 1645 | 31 | 45.1 |
| BPV BR/14RO-16 | KU519391 | 650 | 12 | 44.5 |
| BPV BR/14RO-17 | KU519392 | 1366 | 27 | 42.4 |
| BPV BR/14RO-18 | KU519393 | 578 | 12 | 41.0 |
| BPV BR/14RO-19 | KU519394 | 774 | 16 | 42.7 |
| BPV BR/14RO-20 | KU519395 | 378 | 8 | 44.6 |
| BPV BR/14RO-21 | KU519396 | 630 | 13 | 47.7 |
*Number of reads mapped to the reference.
#Average number of times each base was sequenced.
The genomes are 7,149 to 7,961 Kb in length and display the archetypal organization of PVs (Fig. 1). The first nucleotide in E6 was assigned the number 1 in the sequences. All putative new viruses (BPV BR/14RO-16 to BPV BR/14RO-21) were predicted to contain six (BPV BR/14RO-18 and 19) to seven ORFs (BPV BR/14RO-16, BPV BR/14RO-17, BPV BR/14RO-20 and BPV BR/14RO-21), coding for early (E6, E7, E1, E2, and E4) and late (L1 and L2) proteins (Fig. 1). The BPV13 variant sequence showed the same characteristics as the reference genome17. The overall L1 nucleotide identities of the putative new types were less than 90% in comparison to other PV species and types1.
Figure 1. PV genomes found in this study and their archetypical organization.
The first nucleotide in the ORF6 was assigned a number 1 in the sequences. All putative new papillomavirus genomes (BPV BR/RO-16 to BPV BR/RO-21) were predicted to contain six to eight ORFs, coding for early (E6, E7, E1, E2, E4 and E5) and late (L1 and L2) proteins.
Phylogenetic inferences
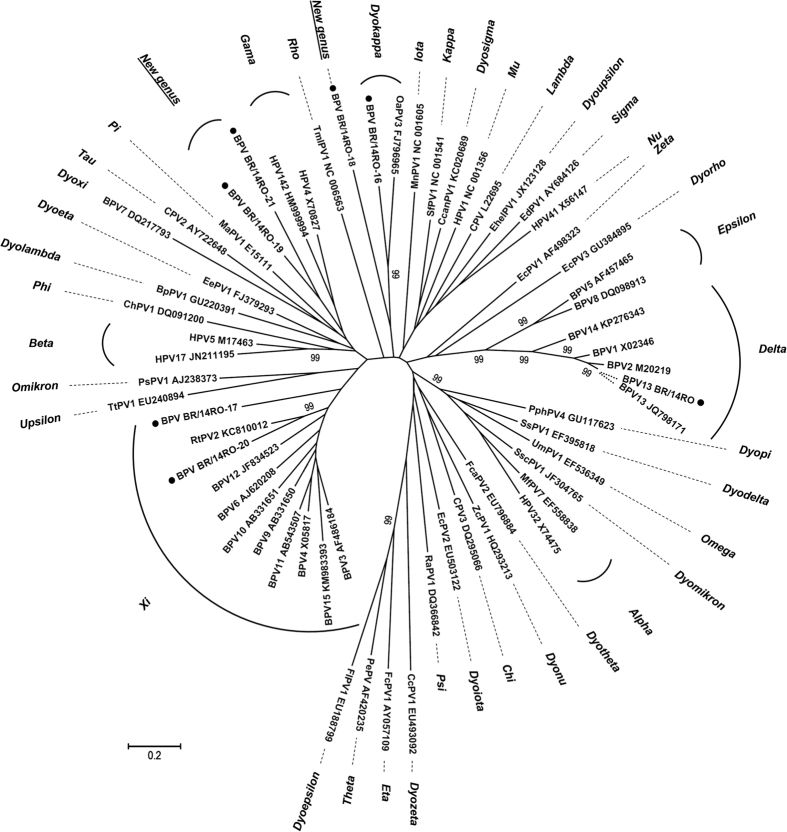
Phylogenetic inferences showed that the six new PV genomes clustered with three known and two unknown PV genera (Fig. 2). Their nucleotide and amino acid identities to the closest related PVs are summarized in Table 2. L1 identities to the most closely related PVs with corresponding GenBank accession numbers following phylogenetic analysis are summarized in Table 2. Putative BPV BR/14RO-16 clustered with members of Dyokappapapillomavirus and was most closely related to Ovis aries PV3 (OvPV3) (62% sequence identity). Putative BPV BR/14RO-17 was most closely related to BPV3 (62%), and putative BPV BR/14RO-20 was most closely related to RtPV2 (Rangifer tarandus PV2) (74% sequence identity), both members of the Xipapillomavirus genus. Putative BPV BR/14RO-18was most closely related to members of the Xi and Dyokappapapillomavirus genera and likely represents a new genus. Putative BPV BR/14RO-19 and BPV BR/14RO-21 constituted a distinct cluster and were most closely related to PVs belonging to Gamma, Xi and Dyoetapapillomavirus. Both are probable representatives of a new genus in the Papillomaviridae family. The BPV13 BR/14RO and the putative new BPV BR/14RO-16 to BPVBR/14RO-21 sequences were deposited in GenBank under accession numbers KU519390 to KU519396.
Figure 2. Phylogenetic tree of the papillomaviruses based on complete sequences of the L1 ORF.
Bootstrap repetitions (higher than 99%) are indicated above the main branches. Samples belonging of this study with representatives of other PV genera were included on the analysis. A total of 61 PV types of different species and genera were analyzed. Accession numbers for the sequences are included and abbreviations are used according to PaVE. Putative new types and genera are indicated with black dots.
Table 2. L1 identities to most closely related PVs with corresponding GenBank accession numbers following phylogenetic analysis.
| PNT# | Identity (%) | PV type | PV species | PV genera | GenBank acession |
|---|---|---|---|---|---|
| BPV BR/14RO-16 | 62 | OaPV3 | Dyokappapapillomavirus 1 | Dyokappa | FJ796965 |
| 55 | BPV3 | Xipapillomavirus 1 | Xi | AF486184 | |
| 55 | BPV6 | Xipapillomavirus 1 | Xi | AJ620208 | |
| 55 | BPV9 | Xipapillomavirus 1 | Xi | AB331650 | |
| 55 | BPV11 | Xipapillomavirus 1 | Xi | AB543507 | |
| BPV BR/14RO-17 | 62 | BPV3 | Xipapillomavirus 1 | Xi | AF486184 |
| 61 | BPV4 | Xipapillomavirus 1 | Xi | X05817 | |
| 61 | BPV6 | Xipapillomavirus 1 | Xi | AJ620208 | |
| 61 | RtPV2 | Xipapillomavirus 3 | Xi | KC810012 | |
| BPV BR/14RO-18 | 54 | RtPV2 | Xipapillomavirus 3 | Xi | KC810012 |
| 53 | OaPV3 | Dyokappapapillomavirus 1 | Dyokappa | FJ796965 | |
| 51 | BPV BR/14RO-16 | * | Dyokappa | KU519391 | |
| BPV BR/14RO-19 | 66 | BPV BR/14RO-21 | * | * | KU519396 |
| 60 | HPV4 | Gammapapillomavirus 10 | Gamma | X70827 | |
| 60 | EePV1 | Dyoetapapillomavirus 1 | Dyoeta | FJ379293 | |
| 59 | MaPV1 | Pipapillomavirus 1 | Pi | E15111 | |
| 59 | HPV142 | Gammapapillomavirus 10 | Gamma | HM999994 | |
| BPV BR/14RO-20 | 74 | RtPV2 | Xipapillomavirus 3 | Xi | KC810012 |
| 68 | BPV12 | Xipapillomavirus 2 | Xi | JF834523 | |
| 67 | BPV9 | Xipapillomavirus 1 | Xi | AB331650 | |
| BPV BR/14RO-21 | 66 | BPV BR/14RO-19 | * | * | KU519394 |
| 58 | HPV142 | Gammapapillomavirus 10 | Gamma | HM999994 | |
| 58 | BPV6 | Xipapillomavirus 1 | Xi | AJ620208 |
*Genomes not assigned to PV species or genera.
#Putative new types (PNT) are designated as BPV BR/14RO-16 to BPV BR/14RO-21.
Discussion
Remarkable efforts have been made to identify human PVs using numerous clinical arrays that can detect dozens of distinct HPV genotypes in the same sample13,14 using several generic PV primer systems17. These efforts reflect over than 200 HPV genomes that are fully sequenced, characterized and cataloged (PaVE). In comparison to HPV, only 15 BPV types have been detected and fully sequenced thus far (PaVE). This scenario is probably pictured because of the lower efforts in BPV studies when compared to HPV due clinical relevance and funding applied. Co-infections comprising HPV are commonly reported in young or immunodepressed women26,27,28. On the other hand, BPV co-infections comprising up to six known PV types based on multiplex BPV genotyping assay20 or specific primers18,29 are rarely reported. Although the majority of BPV types and putative new types have been characterized using generic or genus-specific primers8,12,17,30, such protocols have important limitations because they allow the discovery of only closely related PVs but rarely detect mixed infections. In the present study, the combination of RCA and NGS allowed the detection of at least seven BPVs co-infecting the same lesion, including six putative new BPV types.
An isothermal protocol that uses ϕ29 DNA polymerase to amplify complete PV genomes was previously developed31. Following the amplification of the genome, there is the need to analyze the DNA using restriction enzymes, cloning, and sequencing which is labor-intensive and time-consuming. Additionally, multiply primed RCA and primer-walking for entire genome sequencing already enabled the discovery of novel PVs21. Moreover, there is no evidence of co-infection. One possibility is that the specific degenerate primer pairs used in these studies selected a virus population with higher affinity to primer binding than unknown viruses that may be present in the same sample. This could lead to an underestimation of the detection of other viruses in mixed infections and even a failure to detect distant phylogenetic PVs.
A RCA followed by NGS approach was applied to analyze a sample from which have been previously found a putative new BPV type8. The method enabled the detection of seven PVs with six being uncharacterized so far. The randomly primed RCA followed by NGS offers the possibility to amplify and detect any circular DNA that is present in a sample without specificity, thus allowing a great overview of unknown PVs. This technique could magnify the sensitivity of all PVs present in one sample and allow the understanding of the natural history of infection by different PV types. This approach is meaningful once there is more evidence suggesting that cervical infections caused by some HPV types may also depend on the existence of other HPV types13, thereby suggesting a synergistic pattern. NGS present some restrictions as limiting capability to find mutations like single nucleotide polymorphisms (SNP), insertions and deletions in regions of lower coverage32,33. To minimize the possible base calling errors, in the present study, only high quality reads (Q ≥ 30) were used for de novo assembly. Also, although this Illumina platform displays some underestimation in AT-rich and CG- rich regions33 all putative new types and the BPV13 described in this study presented a GC content considered normal when compared to other PV family members.
The present method enables the detection of a large number of putative new types suggesting the existence of many other BPV types that may have been underestimated thus far. Such a massive PV co-infection indicates that these mammals can harbor genetically diverse PVs similar to humans. Additionally, the Amazon region ecosystem harbors one of the largest global biodiversities and it is quite propitious for the emergence of novel strains. However, there is a need for deeper investigations on this issue by applying this method in all PV affected animals, including other cattle herds worldwide and humans.
We have shown that the enrichment method together with the Illumina NGS platform works for a range of PV genera detection such as Dyokappa, Xi and Gammapapillomavirus. Moreover, the identity of three new types showed inter-genera localization, and these types probably compose two new genera in the Papillomaviridae family. These findings indicate a high number of undetected PVs ignored in the usual assays. Therefore, it is essential to use unbiased methods for the discovery of highly divergent novel viruses as has been done with numerous human and animal agents34.
Most of the xipapillomaviruses present E5 or E8 gene in the E6 genomic position, including BPV4, BPV9, BPV10, BPV11 and BPV15 (GenBank accession no. X05817, AB331650, AB331651, AB543507, and KM983393, respectively) that encode E5, and BPV3, BPV6 and BPV12 (GenBank accession no. AF486184, AJ620208, and JF834523, respectively) that encode E8. The novel xipapillomaviruses detected in the present study present E6 in the E6 genomic position as well as RtPV2 that clustered in the same terminal node that BR/14RO-20. BR/14RO-17 formed a separated branch within Xipapillomavirus and probably is a novel species.
In conclusion, the combination of two relatively simple and fast methods already developed to amplify and genotype PV genomes proved to be very effective in the detection of known and unknown PV viruses using small amounts of DNA derived from one PV lesion. Furthermore, viral genomes can be largely reconstituted by currently available de novo assembly algorithms. The presence of multiple PV types and variants in the same lesion will allow the development of new studies regarding the roles of these different viruses in the biology and pathogenesis of the diseases in which they are involved.
Material and Methods
Ethics Statement
Lesions were collected by veterinarians to prepare autogenous vaccines and all efforts were made to minimize animal suffering. All procedures were performed in compliance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes (European Treaty Series—No. 170 revised 2005) and the procedures of the Brazilian College of Animal Experimentation (COBEA). It must be highlighted that this project was approved by Universidade Federal do Rio Grande do Sul Animal Ethics Committee (number 28460) and we had the owner’s permission to access the animals for the purposes of this study.
Sample processing, rolling circle amplification, and Illumina sequencing
Biopsy material was obtained from a bovine of the Brazilian Amazonian region, diagnosed with epidermal papillomatosis. The lesion was removed using scalpels after local anesthesia (performed with 2% lidocaine, Bravet, Brazil). One putative new PV type was previously detected when a L1 fragment was sequenced using FAP primers8. To obtain the complete genome sequence of this putative new type, the tissue was processed and genomic DNA was extracted as described previously8. To amplify the full PV genomes, randomly-primed rolling circle amplification (RCA) was performed essentially as described by Rector et al.31 using 100 ng of purified DNA from the biopsy specimen. The amplicons were analyzed by agarose gel electrophoresis stained with Blue Green Loading Dye I (LGC, Brazil) and examined under UV light with the Molecular Imaging Software Gel Logic (Kodak, USA).
Following RCA, DNA was purified using a Genomic DNA Clean & Concentrator (Zymo Research). The quality and quantity of the DNA were assessed using a Nanodrop spectrophotometer (Thermo Scientific) and a Qubit fluorometer (Invitrogen). DNA fragment libraries were further prepared with one ng of purified RCA DNA using a Nextera XT DNA sample preparation kit and sequenced using an Illumina MiSeq instrument (2 × 150 paired-end reads with the Illumina v2 reagent kit).
Genome assemblies and sequence analyses
The paired-end sequence reads were assembled into contigs using SPAdes 3.535 and compared to sequences in the GenBank nucleotide and protein databases using BLASTn/BLASTx. Geneious software version 925 was used for open reading frame (ORF) and conserved domain predictions as well as genome annotation. Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motifscan) was used to confirm the conserved domain prediction pointed by Geneious software version 925.
Similarities searches were performed using local sequence alignments BLAST36. Global sequence alignments were accomplished to determine sequence identities with MUSCLE software37. Representative sequences within genera and sequences with the highest identities to the sequences from the present study that are available in GenBank were retrieved from the NCBI homepage (http://www.ncbi.nlm.nih.gov/) for phylogenetic analysis. Altogether, the dataset consisted of 45 sequences of the L1 gene. The multiple sequence alignments was performed through the MUSCLE software37.
The phylogeny was reconstructed with a maximum likelihood method using the MEGA6 software38. These analyses were performed using the GTR substitution model, and the algorithm was modeled with a gamma distribution (shape parameter = 5). The nucleotide substitution model was defined by the tool “find best DNA/Protein model (ML)” of MEGA6 software38. Statistical support was provided by 1,000 non-parametric bootstrap analyses.
RDP4 software39, using the RDP40, GENECONV41, BOOTSCAN42, MAXCHI43, CHIMAERA44, SISCAN45 and 3 SEQ46 methods using default settings were used to look for the presence of chimeric genomes that can arise during the building of contigs. Recombination was considered credible in sequences only if they were detected by more than three methods having significant P values coupled with strong phylogenetic support for recombination. To verify any mismatches that could make difficult the annealing of the viruses detected with the degenerate primer regions, all generated sequences were aligned with primers FAP59/644 and MY09/1147 using ClustalX software38.
Additional Information
How to cite this article: Daudt, C. et al. How many papillomavirus species can go undetected in papilloma lesions? Sci. Rep. 6, 36480; doi: 10.1038/srep36480 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado do Acre (FAPAC), Propesq/UFRGS and Financiadoras de Estudos e Projetos (FINEP) supported this study.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.D. conceived the study, performed the rolling circle amplification, analyzed the data and co-wrote the paper. F.R.C.S analyzed the data and co-wrote the paper. A.F.S. performed the phylogenetic analysis and contributed to the discussion of the paper. M.N.W. performed the RDP4 software analysis and contributed to discussion and paper writing. F.Q.M. contributed to discussion, interpretation of the data, and writing of the paper. S.P.C. conceived the data analysis, discussion, interpretation of the data and paper writing. C.W.C. contributed to the data analysis, discussion, interpretation of the data and paper writing. All authors approved the final version of the manuscript.
References
- Bernard H. U. et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401, 70–79 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers E.-M., Fauquet C., Broker T. R., Bernard H.-U. & zur Hausen H. Classification of papillomaviruses. Virology 324, 17–27 (2004). [DOI] [PubMed] [Google Scholar]
- Rector A. & Van Ranst M. Animal papillomaviruses. Virology 445, 213–223 (2013). [DOI] [PubMed] [Google Scholar]
- Forslund O., Antonsson A., Nordin P., Stenquist B. & Hansson B. G. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 80, 2437–2443 (1999). [DOI] [PubMed] [Google Scholar]
- Antonsson A. & Hansson B. G. Healthy skin of many animal species harbors papillomaviruses which are closely related to their human counterparts. J. Virol. 76, 12537–12542 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T. et al. Broad-spectrum detection of papillomaviruses in bovine teat papillomas and healthy teat skin. J. Gen. Virol. 85, 2191–2197 (2004). [DOI] [PubMed] [Google Scholar]
- Claus M. P. et al. Identification of unreported putative new bovine papillomavirus types in Brazilian cattle herds. Vet. Microbiol. doi: 10.1016/j.vetmic.2008.05.026 (2008). [DOI] [PubMed] [Google Scholar]
- da Silva F. R. C. et al. Genetic characterization of Amazonian bovine papillomavirus reveals the existence of four new putative types. Virus Genes 51, 77–84 (2015). [DOI] [PubMed] [Google Scholar]
- Munday J. S., Hanlon E. M., Howe L., Squires R. a. & French a F. Feline cutaneous viral papilloma associated with human papillomavirus type 9. Vet. Pathol. 44, 924–927 (2007). [DOI] [PubMed] [Google Scholar]
- Silva M. A. R. et al. Comparison of two PCR strategies for the detection of bovine papillomavirus. J. Virol. Methods 192, 55–58 (2013). [DOI] [PubMed] [Google Scholar]
- Karlsen F. et al. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J. Clin. Microbiol. 34, 2095–2100 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday J. S. et al. Genomic characterisation of the feline sarcoid-associated papillomavirus and proposed classification as Bos taurus papillomavirus type 14. Vet. Microbiol. doi: 10.1016/j.vetmic.2015.03.019 (2015). [DOI] [PubMed] [Google Scholar]
- Mejlhede N., Pedersen B. V., Frisch M. & Fomsgaard A. Multiple human papilloma virus types in cervical infections: Competition or synergy? Apmis 118, 346–352 (2010). [DOI] [PubMed] [Google Scholar]
- Goldman B. et al. Patterns of cervical coinfection with multiple human papilloma virus types in a screening population in Denmark. Vaccine 31, 1604–1609 (2013). [DOI] [PubMed] [Google Scholar]
- Poljak M. & Kocjan B. Poljak et al. Commer. available assays Mult. Detect. alpha Hum. papillomaviruses 1139–62. at http://go.galegroup.com/ps/i.do?id=GALE|A263223820&v=2.1&u=capes&it=r&p=AONE&sw=w&asid=f6f964b899426405057147175a0248e7 (2010). [DOI] [PubMed]
- Arroyo L. S. et al. Next generation sequencing for human papillomavirus genotyping. J. Clin. Virol. 58, 437–442 (2013). [DOI] [PubMed] [Google Scholar]
- Lunardi M. et al. Genetic characterization of a novel bovine papillomavirus member of the Deltapapillomavirus genus. Vet. Microbiol. 162, 207–213 (2013). [DOI] [PubMed] [Google Scholar]
- Araldi R. P. et al. Bovine papillomavirus clastogenic effect analyzed in comet assay. Biomed Res. Int. doi: 10.1155/2013/630683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey C. J. et al. Bovine papillomavirus DNA in milk, blood, urine, semen, and spermatozoa of bovine papillomavirus-infected animals. Genet. Mol. Res. 8, 310–318 (2009). [DOI] [PubMed] [Google Scholar]
- Schmitt M., Fiedler V. & Müller M. Prevalence of BPV genotypes in a German cowshed determined by a novel multiplex BPV genotyping assay. J. Virol. Methods 170, 67–72 (2010). [DOI] [PubMed] [Google Scholar]
- Rector A. et al. Characterization of a novel close-to-root papillomavirus from a Florida manatee by using multiply primed rolling-circle amplification: Trichechus manatus latirostris papillomavirus type 1. J. Virol. 78, 12698–12702 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiring T. L. et al. Next-generation sequencing of cervical DNA detects human papillomavirus types not detected by commercial kits. Vor. J. 9, 1–10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas A. C., Mariz F. C., Silva M. A. R. & Jesus A. L. S. Human papillomavirus vertical transmission: review of current data. Clin. Infect. Dis. an Off. Publ. Infect. Dis. Soc. Am. 56, 1451–1456 (2013). [DOI] [PubMed] [Google Scholar]
- Johne R., Müller H., Rector A., van Ranst M. & Stevens H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 17, 205–211 (2009). [DOI] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A. K. et al. Human papillomavirus infection with multiple types: Pattern of coinfection and risk of cervical disease. J. Infect. Dis. 203, 910–920 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarella S., Franceschi S., Snijders P. J. F., Herrero R. & Meijer C. J. L. M. Concurrent Infection with Multiple Human Papillomavirus Types: Pooled Analysis of the IARC HPV Prevalence Surveys Concurrent Infection with Multiple Human Papillomavirus Types: Pooled Analysis of the IARC HPV Prevalence Surveys. 19, 503–510 (2010). [DOI] [PubMed]
- Chaturvedi A. K. Prevalence and Clustering Patterns of Human Papillomavirus Genotypes in Multiple Infections. Cancer Epidemiol. Biomarkers Prev. 14, 2439–2445 (2005). [DOI] [PubMed] [Google Scholar]
- Carvalho C. C. R., Batista M. V. A., Silva M. A. R., Balbino V. Q. & Freitas A. C. Detection of bovine papillomavirus types, co-infection and a putative new BPV11 subtype in cattle. Transbound. Emerg. Dis. 59, 441–447 (2012). [DOI] [PubMed] [Google Scholar]
- Zhu W. et al. Characterization of novel bovine papillomavirus type 12 (BPV-12) causing epithelial papilloma. Arch. Virol. 157, 85–91 (2012). [DOI] [PubMed] [Google Scholar]
- Rector A., Tachezy R. & Ranst M. Van. A Sequence-Independent Strategy for Detection and Cloning of Circular DNA Virus Genomes by Using Multiply Primed Rolling-Circle Amplification A Sequence-Independent Strategy for Detection and Cloning of Circular DNA Virus Genomes by Using Multiply Primed. 78, 4993–4998 (2004). [DOI] [PMC free article] [PubMed]
- Basho R. K. & Eterovic A. K. Clinical Applications and Limitations of Next-Generation Sequencing. Am. J. Hematol. Oncol. 11, 17–22 (2015). [Google Scholar]
- Goodwin S., Mcpherson J. D. & Mccombie W. R. Coming of age: ten years of next- generation sequencing technologies. Nat. Publ. Gr. 17, 333–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. Y. Viral pathogen discovery. Curr. Opin. Microbiol. 16, 468–478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 19, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P., Murrell B., Golden M., Khoosal a. & Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, vev003–vev003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. & Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16, 562–563 (2000). [DOI] [PubMed] [Google Scholar]
- Padidam M., Sawyer S. & Fauquet C. M. Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225 (1999). [DOI] [PubMed] [Google Scholar]
- Martin D. P., Posada D., Crandall K. a. & Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 21, 98–102 (2005). [DOI] [PubMed] [Google Scholar]
- Smith J. M. Analyzing the mosaic structure of genes. J. Mol. Evol. 34, 126–129 (1992). [DOI] [PubMed] [Google Scholar]
- Posada D. & Crandall K. A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98, 13757–13762 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M. J., Armstrong J. S. & Gibbs A. J. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16, 573–582 (2000). [DOI] [PubMed] [Google Scholar]
- Boni M. F., Posada D. & Feldman M. W. An Exact Nonparametric Method for Inferring Mosaic Structure in Sequence Triplets. Genetics 176, 1035–1047 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manos M. M. et al. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cell 7, 209–214 (1989). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.