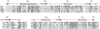Abstract
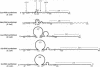
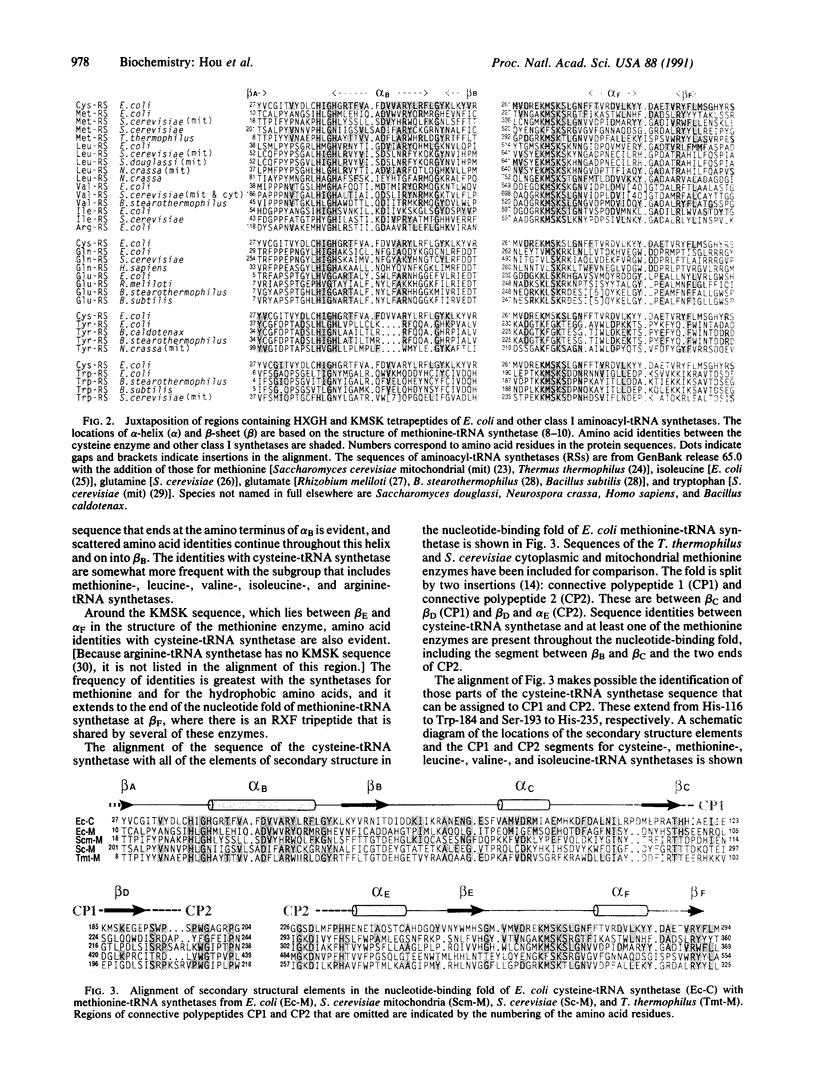
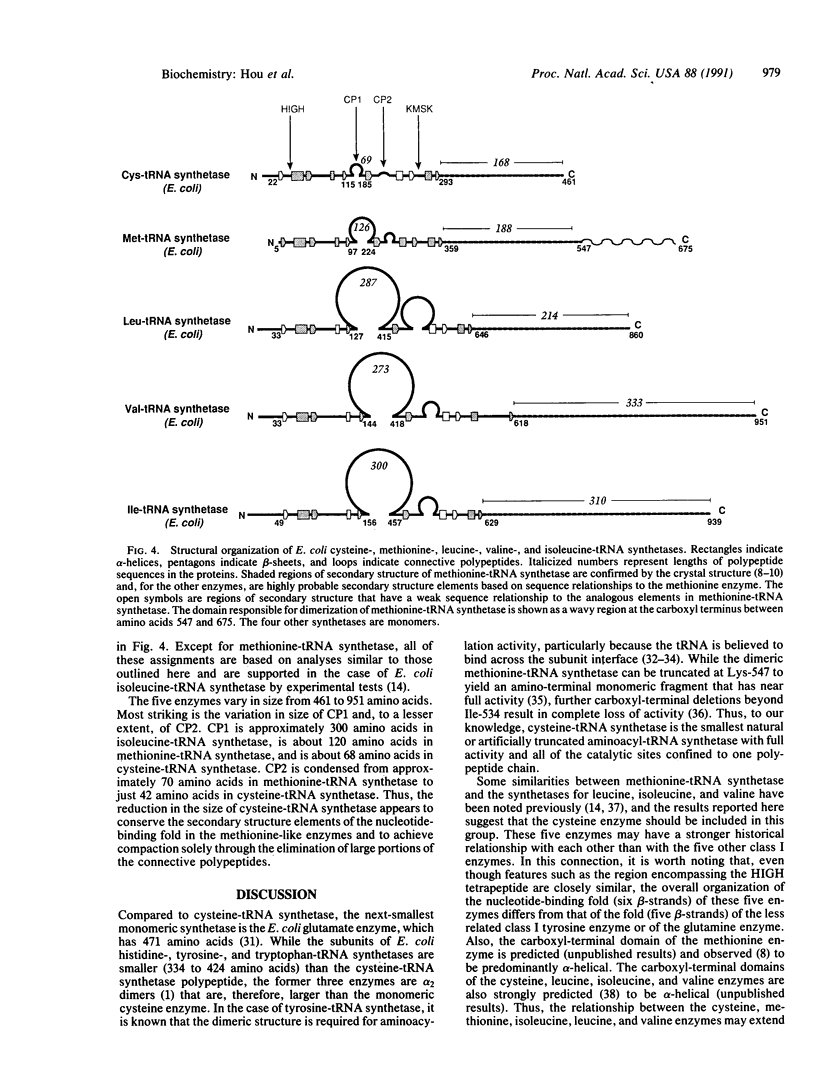
Polypeptide chains of 19 previously studied Escherichia coli aminoacyl-tRNA synthetases are as large as 951 amino acids and, depending on the enzyme, have quaternary structures of alpha, alpha 2, alpha 2 beta 2, and alpha 4. These enzymes have been organized into two classes which are defined by sequence motifs that are associated with specific three-dimensional structures. We isolated, cloned, and sequenced the previously uncharacterized gene for E. coli cysteine-tRNA synthetase (EC 6.1.1.16) and showed that it encodes a protein of 461 amino acids. Biochemical analysis established that the protein is a monomer, thus establishing this enzyme as the smallest known monomeric synthetase. The sequence shows that cysteine-tRNA synthetase is a class I enzyme that is most closely related to a subgroup that includes the much larger methionine-, isoleucine-, leucine-, and valine-tRNA synthetases, which range in size from 677 to 951 amino acids. The amino-terminal 293 amino acids of the cysteine enzyme can be modeled as a nucleotide-binding fold that is more compact than that of its closest relatives by virtue of truncations of two insertions that split the fold. This smaller nucleotide-binding fold accounts for much of the reduced size of the cysteine enzyme and establishes the limit to which the structure of this domain is contracted in the five members of this subgroup of class I enzymes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedouelle H., Winter G. A model of synthetase/transfer RNA interaction as deduced by protein engineering. 1986 Mar 27-Apr 2Nature. 320(6060):371–373. doi: 10.1038/320371a0. [DOI] [PubMed] [Google Scholar]
- Bhat T. N., Blow D. M., Brick P., Nyborg J. Tyrosyl-tRNA synthetase forms a mononucleotide-binding fold. J Mol Biol. 1982 Jul 15;158(4):699–709. doi: 10.1016/0022-2836(82)90255-8. [DOI] [PubMed] [Google Scholar]
- Bohman K., Isaksson L. A. Temperature-sensitive mutants in cysteinyl-tRNA ligase of E. coli K-12. Mol Gen Genet. 1979 Oct 2;176(1):53–55. doi: 10.1007/BF00334295. [DOI] [PubMed] [Google Scholar]
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Breton R., Watson D., Yaguchi M., Lapointe J. Glutamyl-tRNA synthetases of Bacillus subtilis 168T and of Bacillus stearothermophilus. Cloning and sequencing of the gltX genes and comparison with other aminoacyl-tRNA synthetases. J Biol Chem. 1990 Oct 25;265(30):18248–18255. [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Starzyk R. M., Schimmel P. Understanding structural relationships in proteins of unsolved three-dimensional structure. Proteins. 1990;7(2):99–111. doi: 10.1002/prot.340070202. [DOI] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Construction of heterodimer tyrosyl-tRNA synthetase shows tRNATyr interacts with both subunits. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1189–1192. doi: 10.1073/pnas.83.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassio D., Waller J. P. Modification of methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1971 May 28;20(2):283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Clarke N. D., Lien D. C., Schimmel P. Evidence from cassette mutagenesis for a structure-function motif in a protein of unknown structure. Science. 1988 Apr 22;240(4851):521–523. doi: 10.1126/science.3282306. [DOI] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Isolation and characterization of the gene coding for Escherichia coli arginyl-tRNA synthetase. Nucleic Acids Res. 1989 Jul 25;17(14):5725–5736. doi: 10.1093/nar/17.14.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J. D., Hatfield G. W. Valyl-tRNA synthetase gene of Escherichia coli K12. Primary structure and homology within a family of aminoacyl-TRNA synthetases. J Biol Chem. 1988 Jan 15;263(2):868–877. [PubMed] [Google Scholar]
- Hill K., Schimmel P. Evidence that the 3' end of a tRNA binds to a site in the adenylate synthesis domain of an aminoacyl-tRNA synthetase. Biochemistry. 1989 Mar 21;28(6):2577–2586. doi: 10.1021/bi00432a035. [DOI] [PubMed] [Google Scholar]
- Laberge S., Gagnon Y., Bordeleau L. M., Lapointe J. Cloning and sequencing of the gltX gene, encoding the glutamyl-tRNA synthetase of Rhizobium meliloti A2. J Bacteriol. 1989 Jul;171(7):3926–3932. doi: 10.1128/jb.171.7.3926-3932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludmerer S. W., Schimmel P. Gene for yeast glutamine tRNA synthetase encodes a large amino-terminal extension and provides a strong confirmation of the signature sequence for a group of the aminoacyl-tRNA synthetases. J Biol Chem. 1987 Aug 5;262(22):10801–10806. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Mellot P., Mechulam Y., Le Corre D., Blanquet S., Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989 Aug 5;208(3):429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A. MSW, a yeast gene coding for mitochondrial tryptophanyl-tRNA synthetase. J Biol Chem. 1985 Dec 5;260(28):15371–15377. [PubMed] [Google Scholar]
- Ralph W. W., Webster T., Smith T. F. A modified Chou and Fasman protein structure algorithm. Comput Appl Biosci. 1987 Sep;3(3):211–216. doi: 10.1093/bioinformatics/3.3.211. [DOI] [PubMed] [Google Scholar]
- Risler J. L., Zelwer C., Brunie S. Methionyl-tRNA synthetase shows the nucleotide binding fold observed in dehydrogenases. Nature. 1981 Jul 23;292(5821):384–386. doi: 10.1038/292384a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. R., Söll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schreier A. A., Schimmel P. R. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972 Apr 25;11(9):1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Burbaum J. J., Schimmel P. Insertion of new sequences into the catalytic domain of an enzyme. Biochemistry. 1989 Oct 17;28(21):8479–8484. doi: 10.1021/bi00447a031. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Webster T. A., Schimmel P. Evidence for dispensable sequences inserted into a nucleotide fold. Science. 1987 Sep 25;237(4822):1614–1618. doi: 10.1126/science.3306924. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A., Vambutas A., Akai A. Characterization of MSM1, the structural gene for yeast mitochondrial methionyl-tRNA synthetase. Eur J Biochem. 1989 Feb 1;179(2):365–371. doi: 10.1111/j.1432-1033.1989.tb14562.x. [DOI] [PubMed] [Google Scholar]
- Waye M. M., Winter G., Wilkinson A. J., Fersht A. R. Deletion mutagenesis using an 'M13 splint': the N-terminal structural domain of tyrosyl-tRNA synthetase (B. stearothermophilus) catalyses the formation of tyrosyl adenylate. EMBO J. 1983;2(10):1827–1829. doi: 10.1002/j.1460-2075.1983.tb01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- Zelwer C., Risler J. L., Brunie S. Crystal structure of Escherichia coli methionyl-tRNA synthetase at 2.5 A resolution. J Mol Biol. 1982 Feb 15;155(1):63–81. doi: 10.1016/0022-2836(82)90492-2. [DOI] [PubMed] [Google Scholar]