Abstract
In parasitic plants, the reduction in plastid genome (plastome) size and content is driven predominantly by the loss of photosynthetic genes. The first completed mitochondrial genomes (mitogenomes) from parasitic mistletoes also exhibit significant degradation, but the generality of this observation for other parasitic plants is unclear. We sequenced the complete mitogenome and plastome of the hemiparasite Castilleja paramensis (Orobanchaceae) and compared them with additional holoparasitic, hemiparasitic and nonparasitic species from Orobanchaceae. Comparative mitogenomic analysis revealed minimal gene loss among the seven Orobanchaceae species, indicating the retention of typical mitochondrial function among Orobanchaceae species. Phylogenetic analysis demonstrated that the mobile cox1 intron was acquired vertically from a nonparasitic ancestor, arguing against a role for Orobanchaceae parasites in the horizontal acquisition or distribution of this intron. The C. paramensis plastome has retained nearly all genes except for the recent pseudogenization of four subunits of the NAD(P)H dehydrogenase complex, indicating a very early stage of plastome degradation. These results lend support to the notion that loss of ndh gene function is the first step of plastome degradation in the transition to a parasitic lifestyle.
One of the defining characteristics of plants is the presence of a plastid, which enables the fixation of carbon to produce organic molecules via photosynthesis. Parasitic plants represent a dramatic departure from the typical autotrophic lifestyle of plants because they obtain organic carbon sources heterotrophically, using specialized organs called haustoria to make direct connections with the vascular tissue in the roots or shoots of a host plant. Parasitic plants, which comprise approximately 1% of all angiosperms1, can be subdivided based on the extent of their reliance on heterotrophy: hemiparasites retain the ability to photosynthesize and obtain only some of their nutrients from their hosts, while holoparasites have lost photosynthetic ability and must obtain all of their nutrition from hosts.
The transition from an autotrophic to a heterotrophic lifestyle has had a dramatic impact on the plastid genome (plastome) of parasitic plants. Studies of parasitic plant plastomes have established a wide range of genomic degradation, defined primarily by the presence or absence of photosynthetic activity and degree of dependence of the host. For example, in Orobanchaceae, the facultative hemiparasite Triphysaria versicolor has not lost any plastid genes, while the obligate hemiparasites Schwalbea americana and Striga hermonthica (also in Orobanchaceae) exhibit minimal pseudogenization/loss of only a few ndh genes, which encode subunits of the plastid NAD(P)H dehydrogenase complex2,3. Plastomes of hemiparasitic mistletoes (Viscaceae) are slightly more degraded, exhibiting both a reduction in size (down to 126–147 kb) and the loss of all 11 ndh genes plus a small number (1–6) of non-photosynthetic genes4. Within Cuscuta (Convolvulaceae), the four sequenced plastomes range from 85 to 125 kb in size and have experienced more extensive gene loss, yet they still retain all (or all but one) photosynthetic genes5,6, which is consistent with at least low levels of photosynthetic activity7. Other Cuscuta species are clearly non-photosynthetic and their plastomes have lost numerous photosynthetic and non-photosynthetic genes7,8. Plastomes in the holoparasitic species of Orobanchaceae are also heavily degraded2,3,9,10,11, most extensively in Conopholis americana whose plastome is only 46 kb in size with just 21 intact protein-coding genes. Similar levels of degradation were found in the plastomes of other holoparasites in Cynomoriaceae and Hydnoraceae12,13. Even greater genomic reduction was reported in Pilostyles (Apodanthaceae), whose plastomes are reduced to just 11–15 kb and may contain only five or six functional genes14. In some holoparasites, such as Rafflesia lagascae (Rafflesiaceae), the entire plastome may have been lost15.
Much less is known about the effects of a parasitic lifestyle on the mitochondrial genomes (mitogenomes) of plants. In fact, only a single genus of parasitic plants has a completely sequenced mitogenome, from the hemiparasitic mistletoes Viscum scurruloideum and Viscum album, along with draft genomes from two additional Viscum species16,17. Compared with other land plants, V. scurruloideum has the smallest mitogenome (66 kb) and all four Viscum sequences have lost functional copies of all nine nad genes encoding subunits of the mitochondrial NADH dehydrogenase complex I, the first reported loss of this complex from any multicellular eukaryote16. In contrast, the draft mitogenome from the holoparasite R. lagascae has a typical size (estimated at > 300 kb) for an angiosperm and contains a nearly complete set of protein-coding genes, including at least seven of nine nad genes15. The draft mitogenome of Cynomorium coccineum is even larger (>1 Mb) and also contains a nearly complete set of mitochondrial genes13. Thus, the effect of a plant parasitic lifestyle on the mitogenome is still unclear, requiring the analysis of mitogenomes from additional parasitic lineages.
Despite the limited mitogenomic information for parasitic plants, it is well established that their mitochondrial DNA undergoes frequent horizontal transfer, which is likely facilitated by the direct physical connection between parasitic and host plants18,19,20. Perhaps the best studied example of plant horizontal transfer involves the mobile group I intron of the cytochrome oxidase subunit 1 (cox1) gene. This intron was originally acquired from fungi and has been subsequently transferred many times during angiosperm evolution21,22,23,24. Intriguingly, this cox1 intron is highly overrepresented in the parasitic plants that have been examined to date, suggesting that parasitic plants may serve as mediators of horizontal intron transfer among angiosperms25. Although this hypothesis was not supported in an analysis with limited sampling of parasitic plants25, denser sampling of parasites and closely related nonparasitic taxa is needed before the hypothesis should be rejected.
The Orobanchaceae is an ideal family for studies on parasitic plant evolution because it contains the full range of trophic specialization, including a nonparasitic lineage (Lindenbergia), numerous hemiparasitic lineages with varying degrees of photosynthetic activity and host dependence (e.g., Bartsia, Castilleja, Schwalbea, Striga), and at least three transitions to holoparasitism (e.g., Lathraea, Orobanche, Hyobanche) resulting in a complete loss of photosynthesis26,27,28. Complete plastome sequences are available from more than a dozen species in Orobanchaceae, but only a few are from hemiparasites, while data from the mitogenome in this family is lacking. To improve our understanding of organellar genomic evolution in hemiparasitic plants, we sequenced the complete mitochondrial and plastid genomes from the facultative hemiparasite Castilleja paramensis. Furthermore, to assess mitogenomic diversity within the Orobanchaceae, we generated draft mitogenome sequences from six additional species representing the range of trophic diversity: the autotroph Lindenbergia philippensis, the hemiparasites Bartsia pedicularioides and S. americana, and the holoparasites Orobanche crenata, Orobanche gracilis, and Phelipanche ramosa. These sequences were compared to assess the degree of genomic degradation in response to a parasitic lifestyle.
Results
The mitochondrial genome of the hemiparasite Castilleja paramensis
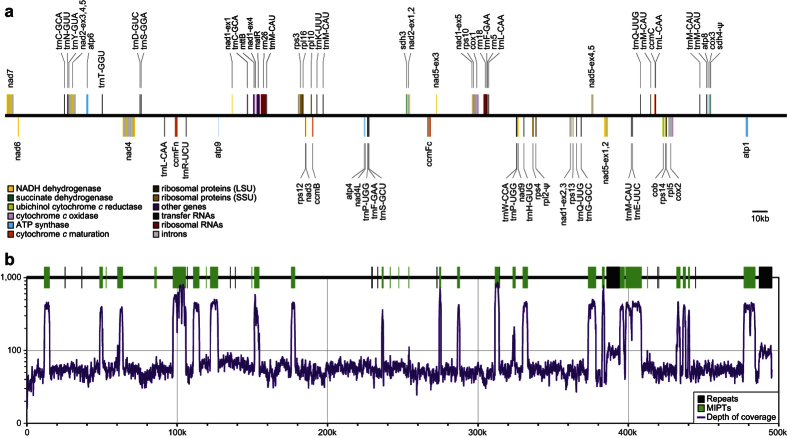
The complete mitogenome of C. paramensis maps as a single circular chromosome that is 495,499 bp in length (Fig. 1A). The genome includes a total of 67 genes (34 protein-coding, 3 rRNA, and 30 tRNA) and 23 introns (17 cis-spliced and 6 trans-spliced). In addition to these functional elements, repeats and MIPTs (mitochondrial DNA of plastid origin) comprise a substantial component of this genome (Fig. 1B). There is one large repeat of 8.7 kb, 15 intermediate repeats from 100 to 447 bp, and 32 small repeats between 50 and 100 bp. Together, these repeats cover 2.7% (13,525 bp) of the genome. A total of 43 MIPTs are also present. Ranging in size from 116 bp to 7.7 kb, these MIPTs cover 16.6% (82,133 bp) of the mitogenome, which is the highest MIPT percentage observed in any plant yet sequenced. The MIPTs contain 55 full-length or nearly full-length plastid genes, about half of which are pseudogenes based on the presence of frameshifting indels and/or premature stop codons. With the exception of the MIPTs and repeats, the depth of sequencing coverage is consistently at ~50x throughout most portions of the mitogenome. The greatly increased coverage depth at most MIPTs is likely due to mismapping of reads in the data set that were derived from the plastome. The two-fold increase in coverage depth of the 8.7 kb repeat relative to the rest of the mitogenome is an indication that this region is in fact present in two copies in the genome, consistent with its status as a repeat.
Figure 1. The Castilleja paramensis mitogenome.
(a) Gene and intron map. Top genes are transcribed in the forward direction; bottom genes are transcribed in the reverse direction. Colors correspond to the functional categories listed in the key. (b) Correlation of repeats and MIPTs with depth of sequencing coverage. The location of all repeats (black) and MIPTs (green) >100 bp in length are shown. Genome maps were drawn with OgDraw (http://ogdraw.mpimp-golm.mpg.de/).
Limited gene and intron loss from the parasitic Orobanchaceae mitogenomes
In contrast to the extensive mitochondrial gene and intron loss observed in mistletoes, there is only minor variation in gene and intron content in Orobanchaceae (Fig. 2), based on comparative mitogenomic analysis of a nonparasite (L. philippensis), three hemiparasites (B. pedicularioides, C. paramensis, S. americana), and three holoparasites (O. crenata, O. gracilis, P. ramosa). The mitogenomes of all seven Orobanchaceae species share 29 protein-coding genes. This conserved set encompasses 23 of the 24 core genes that are nearly universally present in angiosperm mitogenomes29, including nine subunits for the NADH dehydrogenase complex (nad1, 2, 3, 4, 4L, 5, 6, 7, 9), the apocytochrome b gene for the cytochrome bc1 complex (cob), three subunits for the cytochrome c oxidase complex (cox1, 2, 3), four of the five subunits for the ATP synthase complex (atp1, 4, 6, 8), the four cytochrome c maturation factors (ccmB, C, Fc, Fn), an intron maturase (matR), and a protein translocase (mttB/tatC). For the remaining ATP synthase subunit (atp9), the gene is present in all species except L. philippensis. However, the lack of detection of this very short gene (only 225 bp) should be interpreted with caution because it could be an artefact of an incomplete draft assembly.
Figure 2. Mitochondrial gene and intron content in Orobanchaceae and selected asterids.
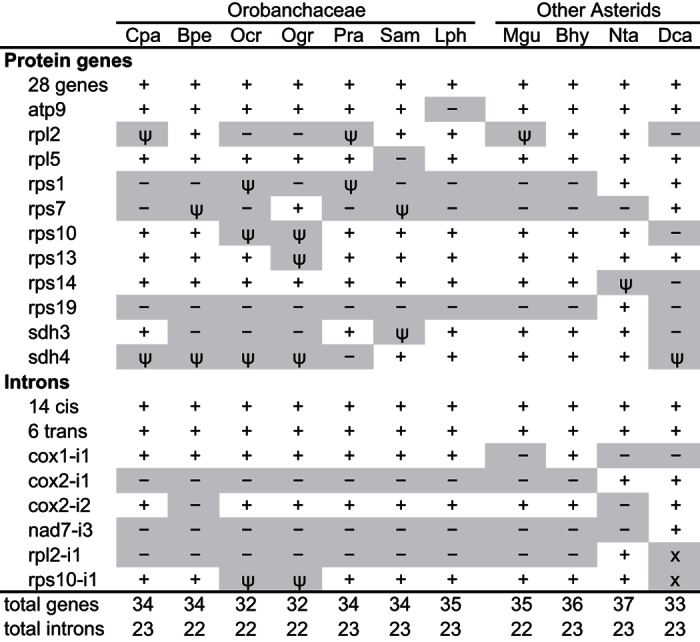
Genes and introns present in each genome are marked with a plus symbol (“+”). Lost genes and introns (“−”), pseudogenized genes and introns (“ψ”), and missing introns due to loss of the host gene (“x”) are shaded gray. The 28 genes include atp1, atp4, atp6, atp8, ccmB, ccmC, ccmFc, ccmFn, cob, cox1, cox2, cox3, matR, mttB, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl10, rpl16, rps3, rps4, and rps12. The 14 cis-arranged introns include ccmFc-i1, nad1-i2, nad2-i1, nad2-i3, nad2-i4, nad4-i1, nad4-i2, nad4-i3, nad5-i1, nad5-i4, nad7-i1, nad7-i2, nad7-i4, and rps3-i1. The six trans-arranged introns include nad1-i1, nad1-i3, nad1-i4, nad2-i2, nad5-i2, and nad5-i3. Cpa = Castilleja paramensis; Bpe = Bartsia pedicularioides; Ocr = Orobanche crenata; Ogr = Orobanche gracilis; Pra = Phelipanche ramosa; Sam = Schwalbea americana; Lph = Lindenbergia philippensis; Mgu = Mimulus guttatus; Bhy = Boea hygrometrica; Nta = Nicotiana tabacum; Dca = Daucus carota.
There is more variability in the presence of genes encoding subunits of the ribosomal protein and succinate dehydrogenase complexes among the Orobanchaceae species (Fig. 2). Six protein members of the large (rpl10, 16) and small (rps3, 4, 12, 14) ribosomal subunits are conserved in all seven Orobanchaceae mitogenomes, whereas the remaining seven ribosomal proteins and both succinate dehydrogenase genes have been lost or pseudogenized in at least one species. Also, several genes (O. gracilis rps7, O. crenata and P. ramosa rps13, C. paramensis and L. philippensis sdh3) have been tentatively scored as present and putatively functional in this study, although they are truncated by 20–30% and may be pseudogenes. Further analysis is required to assess whether they retain functionality. The other listed pseudogenes exhibit clearer loss-of-function mutations because they are heavily truncated (O. crenata rps1, O. gracilis rps10 and rps13, P. ramosa rpl2 and rps1, S. americana rps7) or they have one or more frameshifts (O. crenata rps10 and sdh4, O. gracilis sdh4, S. americana sdh3) that cannot be attributed to pyrosequencing errors at mononucleotide repeats.
In terms of intron content, all examined Orobanchaceae species contain either 22 or 23 introns (Fig. 2). In all seven Orobanchaceae species, there are 15 introns removed by cis splicing and 6 by trans splicing. All seven species lack cox2-i1, nad7-i3, and rpl2-i1, as do other sequenced Lamiales species (e.g., Boea, Mimulus), suggesting that the introns were lost early in Lamiales evolution prior to the radiation of Orobanchaceae. Within the Orobanchaceae, the cox2-i2 intron was uniquely lost from B. pedicularioides, while in the rps10 pseudogenes from O. crenata and O. gracilis, remnants of the rps10 intron are still retained.
The Orobanchaceae cox1 intron was acquired vertically from a non-parasitic ancestor
Previous studies have identified the mobile cox1 intron in a small fraction of angiosperms, between 4% and 25% of the hundreds of examined species in the two most extensive analyses22,23. In contrast to the general scarcity of this intron among angiosperms, it was previously observed that a large fraction of parasitic plants (15 out of 17 examined species, representing 12 distinct parasitic lineages) possess the intron, including Epifagus virginiana, the only Orobanchaceae parasite to be examined thus far25. In agreement with this observation, this intron is present in all six parasitic Orobanchaceae species examined in the current study, and also in the nonparasitic L. philippensis (Fig. 2).
The mobile nature of the cox1 intron, coupled with the overrepresentation of this intron in parasitic plants and the fact that parasitic plants are known to facilitate the horizontal transfer of mitochondrial DNA among species18,19,20, raises two possibilities: 1) parasitic plants may frequently transfer this intron to other angiosperms, explaining the abundant horizontal transmission of the intron among angiosperms, and 2) parasitic plants may frequently acquire this intron from other angiosperms, explaining the overrepresentation of the intron in parasitic plants. Both hypotheses can be tested phylogenetically. If parasitic plants are frequent donors of the intron to other angiosperms, then we would expect to find the introns of recipient angiosperms nested within the parasitic plant clade of introns. If parasitic plants are frequently receiving the intron from other angiosperms, then we would expect to see the horizontally acquired introns of parasitic plants cluster with the donating angiosperm clades rather than in the expected organismal position for Orobanchaceae species within Lamiales.
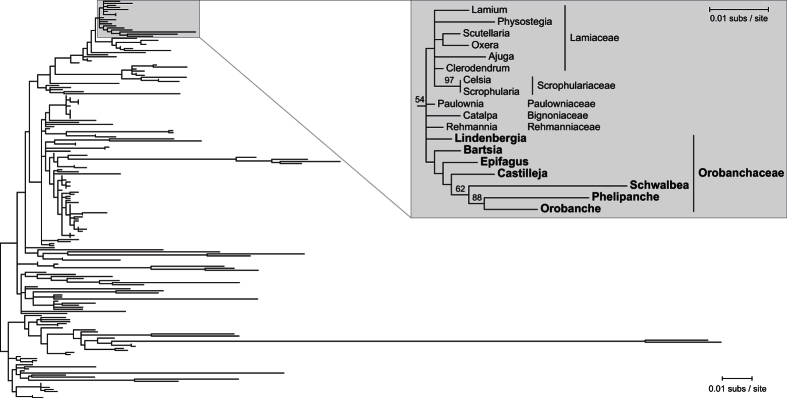
Phylogenetic analysis of the cox1 intron from Orobanchaceae sequences and a diverse collection of other angiosperms demonstrates that neither hypothesis is correct for the parasitic plants in this family (Fig. 3; Figure S1). Within the tree, there is a clade that comprises all Orobanchaceae parasites, which is nested within a larger clade of Lamiales that includes the nonparasitic Lindenbergia, also from Orobanchaceae, plus other species (Catalpa, Paulownia, Rehmannia) from families that are closely related to the Orobanchaceae. Support for most relationships within this larger Lamiales clade is generally weak (<50% bootstrap support for most branches). Nevertheless, the monophyletic clustering of Orobanchaceae species in the more-or-less expected position within Lamiales indicates that this cox1 intron was most likely acquired vertically in parasitic Orobanchaceae from a nonparasitic ancestor. Furthermore, the absence of any unexpected species nested within the parasitic Orobanchaceae clade indicates that the parasites did not donate the cox1 intron to any of the other angiosperm species sampled in the analysis.
Figure 3. Phylogenetic analysis of the mobile cox1 intron.
The tree results from maximum likelihood evaluation of 194 intron sequences from diverse angiosperms. The expanded section of the tree depicts a clade of Lamiales sequences from the seven Orobanchaceae species (large, bold text) and closely related families. Family names are labeled to the right of the subtree. Bootstrap values ≥ 50% from 500 replicates are shown on the branch. The subtree is drawn to a 2-fold expanded scale relative to the full tree; scale bars for the subtree and full tree are shown at top right and bottom right, respectively.
Minimal degeneration of the Castilleja paramensis plastid genome
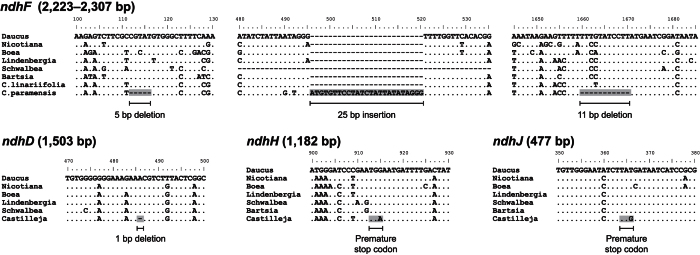
The C. paramensis plastome (Figure S2) is 152,926 bp in length, with a typical quadripartite structure that includes the large and small single-copy regions separated by two copies of an inverted repeat. Relative to the gene and intron content present in a typical asterid, the C. paramensis plastome contains nearly a full set of protein-coding genes, a full set of 4 rRNAs and 31 tRNAs, and a full set of 21 introns (Figure S2 and S3). The few exceptions involve the pseudogenization of ndhD and ndhF due to frameshifting indels and ndhH and ndhJ due to the presence of premature stop codons (Fig. 4). The pseudogenization of ndhF does not occur in all Castilleja species, as an intact gene was sequenced from Castilleja linariifolia in a previous study30. Like C. paramensis, the obligate hemiparasite S. americana has also lost functionality of several ndh genes (pseudogenization of ndhA, ndhD, ndhF, ndhG, ndhJ and loss of ndhI), whereas Orobanchaceae holoparasites including Cistanche, Conopholis, and Orobanche have lost ~70% of all of their genes, including nearly all of the photosynthesis-related genes and numerous tRNAs (Figure S3)3,10,11. Similar patterns of minor degeneration in hemiparasites and more extensive degeneration in holoparasites were observed in a recent broad analysis of Orobanchaceae species2.
Figure 4. Evidence for pseudogenization of Castilleja paramensis ndh genes.
Shown are sections of four ndh gene alignments with evidence of pseudogenization. The frameshifting indels and premature stop codons leading to loss of function are shaded in gray. The full length of functional versions of each gene is shown in parentheses next to each gene name. C. = Castilleja.
Discussion
Gene loss from Orobanchaceae mitogenomes is unrelated to parasitism
In this study, we generated one complete mitogenome from the hemiparasite C. paramensis and draft mitogenomes from six additional Orobanchaceae species, including two more hemiparasites (B. pedicularioides and S. americana), three holoparasites (O. crenata, O. gracilis and P. ramosa), and a nonparasite (L. philippensis). Despite the wide range of trophic strategies among the examined Orobanchaceae species, their mitogenomes display no evidence of functional degeneration that can be attributed to the adoption of a parasitic lifestyle. The relatively few mitochondrial genes that were lost or pseudogenized are limited to ribosomal proteins and succinate dehydrogenase subunits (Fig. 2). The loss of these genes is not attributable to the adoption of a parasitic lifestyle because these same genes have also been lost from the mitogenomes of many non-parasitic land plants16,29,31,32. Importantly, their loss is unlikely to have a detrimental effect on mitochondrial activity, as each loss event is usually preceded by the establishment of a homolog in the nucleus that maintains a functional product33,34,35,36. Like in these other examples, we suggest that the functions of the missing Orobanchaceae mitochondrial genes have been supplanted by nuclear-encoded homologs, although sequencing of the nuclear genome will be required to test this prediction.
In addition to the Orobanchaceae data reported here, large-scale mitogenomic data from a parasitic plant is available from four hemiparasitic mistletoes16,17, four holoparasitic members of Rafflesiaceae15,37, and a holoparasite in Cynomoriaceae13. As in the Orobanchaceae parasites described here, the Cynomoriaceae and Rafflesiaceae holoparasites contain a nearly complete set of the expected mitochondrial genes, although a substantial fraction were reported to have been acquired horizontally13,37. By contrast, in the hemiparasitic V. scurruloideum, the mitogenome has been greatly reduced in size, and in all four mistletoes the coding content has undergone extreme reduction, including the pseudogenization or loss of all nine nad genes encoding subunits of mitochondrial complex I, a NADH dehydrogenase16,17. The coordinated loss of functional copies of all nine nad genes, which has not been reported for any other multicellular eukaryote, argues against a nuclear transfer scenario and instead suggests that the entire complex I was lost16, with nuclear-encoded alternative dehydrogenases38 compensating for the loss of complex I activity.
Thus, while the reduced mitochondrial sequences from mistletoes suggested the possibility of general mitochondrial upheaval in parasitic plants, this does not appear to be the case, at least in the members of Orobanchaceae examined here or in the members of Rafflesiaceae and Cynomoriaceae examined previously. Overall, based on the available data from parasitic plants, there does not appear to be any clear correlation between mitogenomic degradation and the degree of host dependence. This is perhaps not surprising as the mitochondrion is essential for respiration and the production of ATP, and these processes are still required by parasitic plants to generate amino acids and other essential organic molecules. The putative loss of complex I from Viscum may reflect the first step in mitogenomic degradation in a parasitic plant, which may be tolerated due to the partially overlapping abilities of the alternative dehydrogenases16. Regardless, the unusual mitogenomic features observed for Viscum are clearly not representative of all parasitic plants. Whether this complex or any other seemingly essential mitochondrial genes have been lost in other parasitic lineages awaits further investigation.
No evidence that the cox1 intron was acquired or distributed horizontally by Orobanchaceae parasites
Studies have indicated that the angiosperm cox1 intron was acquired from a fungal donor and then horizontally transferred numerous times among species, evidenced primarily by the sporadic distribution of the intron among species and extensive phylogenetic incongruence in the intron tree21,22,23,24. Barkman et al. made the intriguing observation that nearly all examined parasitic plants possess this intron, but they found no evidence that the intron was acquired from their putative hosts25. Alternatively, parasitic plants, particularly those with nonspecific host preferences, may serve as key players in the horizontal spread of the intron.
Using the multiple Orobanchaceae cox1 introns assembled in this study, we demonstrate in this study that the Orobanchaceae introns were acquired vertically from a nonparasitic ancestor (Fig. 3; Figure S1), consistent with the initial results of Barkman et al. using a single Orobanchaceae intron sequence25. Furthermore, there is no evidence that the Orobanchaceae parasites facilitated the spread of the intron to any of the other intron-containing species included in the phylogeny. Overall, there are no indications that the parasitic lifestyle has had any influence on the presence of the cox1 intron in Orobanchaceae or its transfer from Orobanchaceae to any of the other species that were included in the analysis. Thus, it remains unclear why parasitic plants tend to have the cox1 intron, or whether the proclivity of parasitic plants for horizontal transfer plays any role in the intron’s spread. Broad sampling from additional parasitic plant lineages may help to shed light on any potential connections between the parasitic lifestyle and the distribution cox1 intron.
Plastid genome degeneration in parasitic plants
Unlike the mitogenome of C. paramensis, which exhibits few signs of functional degradation, the C. paramensis plastome has frameshift mutations or premature stop codons in four subunits of the plastid NAD(P)H dehydrogenase complex (Fig. 4). These mutations in the well-conserved ndh genes are likely to lead to a reduction or loss of gene function. This pattern of ndh-specific degradation in the C. paramensis plastome is similar to observations in other Orobanchaceae hemiparasites and some species of Cuscuta (Figure S3)2,3,5,6. The draft plastome from the hemiparasite Bartsia inaequalis also lacks intact, full-length copies of several ndh genes (ndhD, ndhE, ndhG, and ndhI), although it cannot be ruled out that these genes were missed due to the incomplete nature of the genome39. Compared with other sequenced hemiparasites, the C. paramensis plastome appears to be in the very earliest stages of degradation, as indicated by the small number of genes so-far affected, the limited number of deleterious mutations that have accumulated in each affected gene and the lack of any genes that were deleted completely. Furthermore, an intact ndhF gene is present in another Castilleja species30, indicating that the pseudogenization of the C. paramensis ndhF gene occurred recently within the genus, at some point after C. paramensis diverged from other members of the genus. The C. paramensis plastome thus provides strong support for the idea that loss of the NAD(P)H dehydrogenase complex is the first step of plastome degradation in the evolution of heterotrophy in plants3,40. By contrast, the plastomes from nonphotosynthetic holoparasites are generally much more degraded than those of hemiparasites, affecting not only the full spectrum of photosynthetic genes but also many genes not directly related to photosynthesis (Figure S3)2,3,8,9. Taken together, the collective evidence from available parasitic plastomes suggests a connection between the degree of plastomic degeneration and heterotrophic dependence.
Although it is possible that these genes have been functionally transferred to the nuclear genome in C. paramensis, there has been no demonstration of functional ndh gene transfer for any seed plants that have lost the plastid ndh genes. Fragments of some ndh genes were identified in the nucleus of several Orobanchaceae species11, but there is no indication that these fragments produce functional proteins. Instead, mounting evidence in multiple lineages—including the pine family, gnetophytes, several orchids, and several species of Erodium (Geraniaceae)—has shown that these lost plastid genes were not relocated to the nucleus, and furthermore, that many of the nuclear-encoded subunits of this complex have also been lost41. These results strongly suggest that the entire NAD(P)H complex has been eliminated from these species.
Materials and Methods
Sample collection and organellar genome sequencing
A C. paramensis individual was collected from a páramo in the department of Boyacá, Colombia on March 21, 2014 (voucher N. Pabón-Mora et al. 299, HUA). A B. pedicularioides individual was collected from a páramo in Cajas National Park, Ecuador on December 17, 2010 (voucher J. P. Mower et al. 2064, QCA). Total genomic DNA was extracted from silica-dried leaves using the Plant DNeasy Kit (Qiagen). DNA samples were sequenced on the Illumina HiSeq2000 platform at BGI (Shenzhen, China), which generated 6 Gb (for B. pedicularioides) or 8 Gb (for C. paramensis) of 100-bp paired-end reads from an 800-bp library.
Genome assembly and annotation
Draft organellar genomes of C. paramensis and B. pedicularioides were assembled from the Illumina sequence reads with Velvet version 1.2.0342 using multiple combinations of kmer (61, 71, 81, 91) and expected coverage (50, 100, 200, 500, 1000) values, as described previously43,44. Organellar contigs were identified in each assembly by using default blastn searches with known organellar gene sequences from related Lamiales species as queries. For each targeted genome, the best assembly that maximized total mitochondrial or plastid length in the fewest number of contigs was used for further scaffolding. Scaffolding was performed by mapping read pairs onto the contig sequences using blastn (e-value ≤ 1 × 10−10, hit length ≥ 90 bp, sequence identity ≥ 90%), and read pairs spanning two different contigs were used to infer contig joins and repeat regions. Using this strategy, circular-mapping plastid and mitochondrial genomes were assembled for C. paramensis, and a draft mitogenome was assembled for B. pedicularioides. The C. paramensis and B. pedicularioides mitogenome assemblies were annotated as described previously32,44,45. The C. paramensis plastid genome was annotated using DOGMA46 followed by manual adjustment as necessary.
To survey mitochondrial gene content in additional Orobanchaceae species, 454 pyrosequencing data from a previous study47 were downloaded from the NCBI sequence read archive (accession SRA047928) for one hemiparasite (S. americana), three holoparasites (O. crenata, O. gracilis, P. ramosa) and one nonparasite (L. philippensis). The downloaded 454 data were assembled with Velvet 1.2.03 as described above using various pairwise combinations of kmer (41, 51, 61, 71) and expected coverage (5, 10, 20, 50, 100) values. Lower kmer and expected coverage values were required for these data sets given the lower amount of data available (<1 Gb total genomic DNA for each species), resulting in assemblies with 5–10x depth of mitochondrial sequence coverage for each species. Scaffolding was not performed because the reads were unpaired. The presence of mitochondrial genes and introns was scored by using blastn searches with mitochondrial gene sequences from other Lamiales species as queries against the best 454 assemblies. Gene and intron sequences of interest were manually extracted from these 454 assemblies for further analysis.
Genes identified from each assembly were assessed for potential loss of function by searching for frameshifting indels and/or premature stop codons. Genes were scored as pseudogenes if the mutations disrupted >20% of their conserved domain structure, as defined by a search of the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), or if >30% of the gene was disrupted overall. For the genes assembled from 454 data, raw reads were mapped back against the assembled gene sequence using blastn to ensure that pseudogene calls were not the result of errors involving mononucleotide repeats or other errors due to the low-coverage nature of the data.
Phylogenetic evaluation of horizontal transfer of the cox1 intron
An angiosperm cox1 intron alignment containing sequences used in previous studies22,23, including the intron from the Orobanchaceae parasite E. virginiana, was provided by Dr. Virginia Sanchez-Puerta. Additional Orobanchaceae cox1 intron sequences were extracted from their best assemblies generated in this study and then manually aligned to the data set. Alignments were trimmed of poorly aligned regions with Gblocks 0.91b using relaxed parameters (b2 = half+1, b4 = 5, b5 = half). The final trimmed data set contained 958 aligned nucleotide positions and 194 intron sequences, representing 191 angiosperm species from 60 families (Table S1). The cox1 intron alignment was then used to construct a phylogenetic tree using maximum-likelihood in PhyML 3.048. A GTR+G+I model with four substitution rate categories was employed. Tree topologies, branch lengths, and rate parameters were optimized during the run. Branch support was calculated from 500 bootstrap replicates.
Additional Information
How to cite this article: Fan, W. et al. Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae. Sci. Rep. 6, 36285; doi: 10.1038/srep36285 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
The authors thank Nancy Hepburn and Danilo Minga for assistance with plant collection in Ecuador, Katya Romoleroux and Hugo Navarrete for helping to obtain exportation permits from Ecuador, Kanika Jain and Yizhong Zhang for preparation of DNA samples, Susann Wicke and Susanne Renner for providing access to 454 pyrosequencing data from several Orobanchaceae species prior to publication, and Virginia Sanchez-Puerta for providing an alignment of cox1 introns. This work was supported by the National Science Foundation (awards IOS 1027529 and MCB 1125386 to JPM) and by a China Scholarship Council award (to WF).
Footnotes
The authors declare no competing financial interests.
Author Contributions J.P.M. planned and designed the research. W.F., A.Z., M.K., N.S. and J.P.M. performed experiments and analyzed results. N.P.M. and F.G. conducted field work and analyzed results. W.F. and J.P.M. wrote the paper, with contributions from all other authors. All authors read and approved the final version of the text.
References
- Westwood J. H., Yoder J. I., Timko M. P. & dePamphilis C. W. The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235 (2010). [DOI] [PubMed] [Google Scholar]
- Wicke S. et al. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. USA 113, 9045–9050 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S. et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25, 3711–3725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G., Cuenca A. & Seberg O. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 7, 2520–2532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal J. R., Kuehl J. V., Boore J. L. & de Pamphilis C. W. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7, 57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk H. T., Berg S., Krupinska K., Maier U. G. & Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7, 45 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooij T. A., Krause K., Dorr I. & Krupinska K. Molecular, functional and ultrastructural characterisation of plastids from six species of the parasitic flowering plant genus Cuscuta. Planta 210, 701–707 (2000). [DOI] [PubMed] [Google Scholar]
- Braukmann T., Kuzmina M. & Stefanovic S. Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. J. Exp. Bot. 64, 977–989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Morden C. W. & Palmer J. D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89, 10648–10652 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One 8, e58747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusimano N. & Wicke S. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. 210, 680–693 (2016). [DOI] [PubMed] [Google Scholar]
- Naumann J. et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol. Evol. 8, 345–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot S. et al. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biol. Evol. 8, 2214–2230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot S. & Renner S. S. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol. Evol. 8, 189–201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina J. et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 31, 793–803 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E., Barkman T. J., Rice D. W. & Palmer J. D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 112, E3515–E3524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen G., Cuenca A., Moller I. M. & Seberg O. Massive gene loss in mistletoe (Viscum, Viscaceae) mitochondria. Sci. Rep. 5, 17588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. C. & Xi Z. Horizontal gene transfer in parasitic plants. Curr. Opin. Plant Biol. 26, 14–19 (2015). [DOI] [PubMed] [Google Scholar]
- Mower J. P., Jain K. & Hepburn N. J. The role of horizontal transfer in shaping the plant mitochondrial genome. Adv. Bot. Res. 63, 41–69 (2012). [Google Scholar]
- Sanchez-Puerta M. V. Involvement of plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal gene transfer. Acta Soc. Bot. Pol. 83, 317–323 (2014). [Google Scholar]
- Cho Y., Qiu Y.-L., Kuhlman P. & Palmer J. D. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl. Acad. Sci. USA 95, 14244–14249 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V. et al. Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended co-conversion of flanking exons. BMC Evol. Biol. 11, 277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta M. V., Cho Y., Mower J. P., Alverson A. J. & Palmer J. D. Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Mol. Biol. Evol. 25, 1762–1777 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. C., Mason M. T., Sper-Whitis G. L., Kuhlman P. & Palmer J. D. Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J. Mol. Evol. 41, 563–572 (1995). [DOI] [PubMed] [Google Scholar]
- Barkman T. J. et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol. Biol. 7, 248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett J. R. & Mathews S. Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 93, 1039–1051 (2006). [DOI] [PubMed] [Google Scholar]
- McNeal J. R., Bennett J. R., Wolfe A. D. & Mathews S. Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot. 100, 971–983 (2013). [DOI] [PubMed] [Google Scholar]
- Young N. D., Steiner K. E. & Depamphilis C. W. The evolution of parasitism in Scrophulariaceae/Orobanchaceae: plastid gene sequences refute an evolutionary transition series. Ann. MO Bot. Gard. 86, 876–893 (1999). [Google Scholar]
- Adams K. L., Qiu Y. L., Stoutemyer M. & Palmer J. D. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 99, 9905–9912 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refulio-Rodriguez N. F. & Olmstead R. G. Phylogeny of Lamiidae. Am. J. Bot. 101, 287–299 (2014). [DOI] [PubMed] [Google Scholar]
- Hecht J., Grewe F. & Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol. Evol. 3, 344–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. Ginkgo and Welwitschia mitogenomes reveal extreme contrasts in gymnosperm mitochondrial evolution. Mol. Biol. Evol. 33, 1448–1460 (2016). [DOI] [PubMed] [Google Scholar]
- Adams K. L., Ong H. C. & Palmer J. D. Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 18, 2289–2297 (2001). [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Knoop V., Fukuzawa H., Brennicke A. & Ohyama K. Interorganellar gene transfer in bryophytes: the functional nad7 gene is nuclear encoded in Marchantia polymorpha. Mol. Gen. Genet. 256, 589–592 (1997). [DOI] [PubMed] [Google Scholar]
- Mower J. P. & Bonen L. Ribosomal protein L10 is encoded in the mitochondrial genome of many land plants and green algae. BMC Evol. Biol. 9, 265 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent J. M. & Palmer J. D. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66, 473–481 (1991). [DOI] [PubMed] [Google Scholar]
- Xi Z. et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 9, e1003265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson A. G., Soole K. L. & Elthon T. E. Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu. Rev. Plant Biol. 55, 23–39 (2004). [DOI] [PubMed] [Google Scholar]
- Uribe-Convers S., Duke J. R., Moore M. J. & Tank D. C. A long PCR-based approach for DNA enrichment prior to next-generation sequencing for systematic studies. Appl. Plant Sci. 2, 1300063 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C. F. & Davis J. I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 99, 1513–1523 (2012). [DOI] [PubMed] [Google Scholar]
- Ruhlman T. A. et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15, 100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. et al. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol. Evol. 6, 580–590 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A., Guo W., Jain K. & Mower J. P. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol. Biol. Evol. 31, 1228–1236 (2014). [DOI] [PubMed] [Google Scholar]
- Mower J. P., Case A. L., Floro E. R. & Willis J. H. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 4, 670–686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K. & Boore J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255 (2004). [DOI] [PubMed] [Google Scholar]
- Piednoel M. et al. Next-generation sequencing reveals the impact of repetitive DNA across phylogenetically closely related genomes of Orobanchaceae. Mol. Biol. Evol. 29, 3601–3611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.