Abstract
This was a first-in-human study of the novel phosphodiesterase-2A (PDE2A) PET ligand 18F-PF-05270430. The primary goals were to determine the appropriate tracer kinetic model to quantify brain uptake and to examine the within-subject test–retest variability. Methods: In advance of human studies, radiation dosimetry was determined in nonhuman primates. Six healthy male subjects participated in a test–retest protocol with dynamic scans and metabolite-corrected input functions. Nine brain regions of interest were studied, including the striatum, white matter, neocortical regions, and cerebellum. Multiple modeling methods were applied to calculate volume of distribution (VT) and binding potentials relative to the nondisplaceable tracer in tissue (BPND), concentration of tracer in plasma (BPP), and free tracer in tissue (BPF). The cerebellum was selected as a reference region to calculate binding potentials. Results: The dosimetry study provided an effective dose of less than 0.30 mSv/MBq, with the gallbladder as the critical organ; the human target dose was 185 MBq. There were no adverse events or clinically detectable pharmacologic effects reported. Tracer uptake was highest in the striatum, followed by neocortical regions and white matter, and lowest in the cerebellum. Regional time–activity curves were well fit by multilinear analysis-1, and a 70-min scan duration was sufficient to quantify VT and the binding potentials. BPND, with mean values ranging from 0.3 to 0.8, showed the best intrasubject and intersubject variability and reliability. Test–retest variability in the whole brain (excluding the cerebellum) of VT, BPND, and BPP were 8%, 16%, and 17%, respectively. Conclusion: 18F-PF-05270430 shows promise as a PDE2A PET ligand, albeit with low binding potential values.
Keywords: phosphodiesterase 2A, positron emission tomography, test-retest reproducibility, dosimetry, brain imaging
Phosphodiesterase enzymes (PDEs) hydrolyze cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP) into their corresponding monophosphates. By this hydrolysis, the signals generated by these molecules are terminated. The phosphodiesterase gene superfamily consists of 11 gene families that express PDEs (1). PDE2 (phosphodiesterase-2A), which possesses protein motifs (GAF domains) that regulate dimerization of the enzyme, and may also constitute an allosteric site that is activated by cGMP, is characterized by a high Michaelis constant Km for both cGMP and cAMP (>5 μM) (2). Agents that stimulate cGMP production, such as nitric oxide synthase activators or natriuretic peptides, activate PDE2A and cause hydrolysis of cAMP.
In the central nervous system, PDE2A is highly expressed in the striatum, hippocampus, frontal cortex, olfactory neurons, and medial habenula (3) and plays a role in regulating neurotransmission. In non–central nervous system tissues, PDE2A is highly expressed in adrenal glands and vascular endothelial cells in which it may regulate vascular permeability (4). The development of a PDE2A radioligand would allow the evaluation of disease-specific changes (e.g., in schizophrenia) and potential inhibitor drugs.
We recently reported selection criteria for successful site-specific PET ligands, including PDE2A radiotracers (5). The companion paper reports that the PET ligand 18F-PF-05270430 is a potent inhibitor with high binding affinity for PDE2A (6). It shows excellent selectivity over other PDE subtypes and has good properties for PET imaging, as evaluated in nonhuman primates.
In this study, we present nonhuman primate dosimetry results and first-in-human PET data. Along with evaluating the safety and tolerability of 18F-PF-05270430 in healthy human participants, we evaluated appropriate analytic strategies for tracer binding to PDE2A sites and assessed subject test–retest variability.
MATERIALS AND METHODS
Radiotracer Synthesis
18F-PF-05270430 was prepared as described in the companion paper (6) and in the supplemental materials (available at http://jnm.snmjournals.org). Radiochemical purity of the final product solution was greater than 98%. Specific activity at the end of synthesis was 115 ± 53 GBq/μmol (n = 12).
Monkey Dosimetry Studies
Four rhesus monkeys (2 male and 2 female; mean weight ± SD, 8.95 ± 3.42 kg) underwent whole-body dynamic PET scans on a Biograph mCT (Siemens Medical Systems) after intravenous injection of 18F-PF-05270430. Animals were scanned for approximately 3 h in a sequence of 14–17 passes with either 4 or 5 bed positions from the top of the head to mid thigh. Scans were visually inspected for organ activity exceeding background level. Included organs were the liver, kidneys, urinary bladder, heart, spleen, gallbladder, brain, red marrow (femur), upper large intestines, small intestines, stomach wall, pancreas, ovaries, and cortical bone. Using early and later summed images, we delineated manually regions of interest (ROIs), and mean organ activity values were computed to form time–activity curves.
Decay correction was removed to reflect actual activity in each organ, and integrated activity (Bq⋅h/cm3) was computed using isotope decay to extrapolate to infinity after the scan period. These values were multiplied by the organ volumes of a standard 70-kg (male) and 55-kg (female) reference mathematic phantom (7), scaled by animal mass/body weight and then normalized to injected activity to obtain organ residence times (N, hour), which were entered into the OLINDA software (8) to obtain absorbed doses in all organs. Doses to the stomach were adjusted to reflect the presence of activity in the stomach wall rather than the contents.
Human Subjects
Healthy male volunteers (age, 24–54 y; weight, 87.5 ± 14.1 kg) with no clinically significant diseases completed a test–retest study. The institutional review board–approved study was also approved by the Yale-New Haven Hospital Radiation Safety Committee and performed in accordance with federal guidelines and regulations of the United States for the protection of human research subjects contained in Title 45 part 46 of the Code of Federal Regulations. All subjects signed a written informed consent form.
Human Brain PET Studies
PET Imaging
Each subject underwent 2 PET scans with 18F-PF-05270430. In advance of each scan, an intravenous line was placed for tracer injection, and an arterial line was placed for measurement of the input function. The test and retest scans were 4.7 ± 0.8 d apart. Each subject was limited to a maximum dose of 185 MBq per scan, based on dosimetry study results. The mass dose in clinical PET studies was limited to 10 μg or less per administration. The choice of dose was based on rat safety toxicity studies, which showed that doses were well tolerated up to the maximum dose tested (3 μg/kg). In the clinic, safety was assessed by measurement of vital signs (blood pressure, heart rate, oxygen saturation, and hematocrit) and recording of adverse symptoms after tracer administration.
18F-PF-05270430 was administered intravenously over 1 min by an automatic pump (Harvard PHD 22/2000; Harvard Apparatus). PET scans were then obtained with an ECAT EXACT HR+ scanner (Siemens Medical Systems) in 3-dimensional mode. For the first 3 subjects, the acquisition duration was 240 min, consisting of a 120-min scan, 30-min break, and a 90-min scan. The scan duration was shortened to 120 min for subjects 4–6 based on analysis of the initial data. Dynamic scan data were reconstructed with corrections for attenuation (transmission scan), normalization, scatter, randoms, and dead time using the ordered subset-expectation maximization algorithm (4 iterations, 16 subsets) in 33 frames (6 × 0.5, 3 × 1, 2 × 2, and 22 × 5 min) for the 120-min scan plus 18 frames (18 × 5 min) for the scan from 150 to 240 min.
MRI
Before the PET scans, each subject underwent an MR scan for PET image registration. MR imaging was performed using a 3-dimensional magnetization-prepared rapid gradient-echo pulse sequence with an echo time of 2.78 ms, repetition time of 2,500 ms, inversion time of 1,100 ms, and flip angle of 7° on a 3-T whole-body scanner (Trio; Siemens Medical Systems) with a circularly polarized head coil.
Arterial Input Function Measurement
Continuous and discrete blood samples were taken for determination of whole-blood activity and the metabolite-corrected plasma input function. For the first 7 min, arterial blood was continuously counted using an automated blood counter (PBS-101; Veenstra Instruments). Discrete blood samples were manually collected at 3, 5, 8, 10, 15, 20, 25, 30, 40, 50, 60, 75, 90, 105, and 120 min after dosing. Additional samples were obtained at 150, 180, 210, and 240 min for the 240-min scan.
Plasma analysis of radiotracer metabolism was performed from samples collected at 5, 15, 30, 60, 90, and 120 min after injection (plus 180 and 240 min for the 240-min scans). Metabolite analysis was performed using the column-switching high-performance liquid chromatography (HPLC) method (9) to determine the parent fraction. An ultrafiltration-based method (Centrifree; Millipore) was used for measuring the plasma free fraction (fP). Details are provided in the supplemental materials.
At the retest scan of 1 subject, no arterial line was available. This subject was excluded from calculation of test–retest variability. For another subject, the fP value of the test scan was excluded from calculation of mean and SD of fP because of contaminated samples.
Image Registration and ROIs
For each subject, motion correction of each dynamic frame was performed by registration to a summed PET image (0–10 min after injection) using a 6-parameter mutual information algorithm (FMRIB's Linear Image Registration Tool [FLIRT], FSL). The summed PET image was then coregistered to the subject’s T1-weighted MR image (6-parameter affine registration), which was subsequently coregistered to the automated anatomic labeling template (10) in Montreal Neurologic Institute (11) space using a nonlinear transformation (Bioimage Suite; Yale University) (12). Regional time–activity curves were generated for the putamen, caudate, nucleus accumbens, temporal cortex, occipital cortex, parietal cortex, frontal cortex, cerebellum, and centrum semiovale using the combined transformations from template-to-PET space.
Quantitative Analysis
Regional distribution volumes (VT) and binding potentials relative to the nondisplaceable tracer in tissue (BPND), concentration of tracer in plasma (BPP), and free tracer in tissue (BPF) were computed using 1-tissue (1TC) and 2-tissue (2TC)-compartment models, and multilinear analysis-1 (MA1) with the plasma input function. The simplified reference tissue model (SRTM) was also used to derive BPND directly. Blood volume in the brain was assumed to be 5% in all models. The equation for MA1 with blood volume correction (13) was:
where CPET(t), CP(t), and CWB(t) represent the tissue, plasma, and whole-blood time–activity curves, respectively, and FV is the fractional blood volume. To calculate binding potentials, the cerebellum was used as a reference region because no PDE2A signal was detected in the cerebellum in an in vitro immunoreactivity study of human brain extracts with an anti-PDE2A antibody (4). For parameter estimation, data points were weighted on the basis of noise equivalent counts. Percentage SE (%SE; [SE]/[estimated parameter]) was estimated from the theoretic covariance matrix.
Effect of Scan Time
The time stability of VT estimates was evaluated by fitting the MA1 model (t* = 40 min) to time–activity curves with shortened scan durations ranging from 50 to 120 min (n = 11). The ratio, r, of VT values from the shortened scan to those from the 120-min scan was computed for each ROI and duration. The minimum acceptable scan duration was the one that satisfied the following 2 criteria (14): the mean ratio (r) across subjects was between 0.95 and 1.05, and the interindividual SD of the ratio (r) was less than 0.1. We also compared 120-min VT estimates with those from longer scans (160–240 min; n = 6: 3 subjects × 2 scans each).
Statistical Analyses
The reproducibility of the outcome measures was assessed with test–retest variability (TRV) and absolute TRV (aTRV). TRV was calculated as:
where p(T) and p(R) are parameter estimates at test and retest conditions, respectively. The mean of TRV indicates the presence of a trend between the 2 scans, and the SD of TRV is an index of the variability of the percentage difference between 2 measurements. aTRV uses absolute values of TRV, and its mean indicates the coefficient of variation when the mean of TRV was 0 (15). Reliability was also assessed by the intraclass correlation coefficient (ICC), ranging from −1 (no reliability) to +1 (maximum reliability).
RESULTS
Nonhuman Primate Dosimetry Study
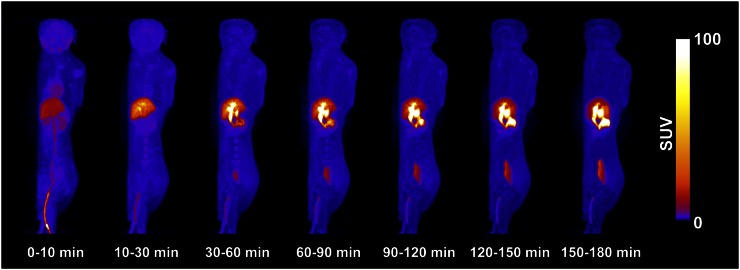
Scan parameters for these experiments are summarized in Table 1. Figure 1 shows an example of the whole-body distribution over time. The main route of elimination was hepatobiliary, with the gallbladder having the highest radioactivity uptake. Organ residence times are listed in Table 2, and accounted for 61% ± 23% of the theoretic value. Table 3 shows absorbed doses derived from the male and female phantoms (n = 4). Under CFR 361.1 (for single study, 50 mSv per organ or 30 mSv to selected organs undergoing rapid cell division, whichever is less), the gallbladder is the dose-limiting organ with a single-study dose limit of 306 and 249 MBq for the male and female phantoms, respectively. The dose to the liver was slightly less than that of the gallbladder. On the basis of these values, a target dose of 185 MBq (5 mCi) was chosen for human studies.
TABLE 1.
Subject Information and PET Scan Parameters
| Humans |
|||
| Parameter | Test (n = 6) | Retest (n = 6) | Monkeys, dosimetry (n = 4) |
| Injected dose (MBq) | 182 ± 3 | 174 ± 9 | 135 ± 71 |
| Specific activity at time of injection (MBq/nmol) | 83.9 ± 51.4 | 83.6 ± 32.4 | 72.9 ± 46.9 |
| Injected mass (μg/kg) | 0.024 ± 0.032 | 0.013 ± 0.010 | 0.11 ± 0.09 |
Data are mean ± SD. Age was 37 ± 12 and 8 ± 2 y for humans and monkeys, respectively. Body weight was 87.5 ± 14.1 and 9.0 ± 3.4 kg for humans and monkeys, respectively.
FIGURE 1.
Maximum-intensity projection (coronal view) over 3 h of 18F-PF-05270430 in male rhesus monkey.
TABLE 2.
Organ Residence Times
| Organ | N (h) |
| Marrow | 0.032 ± 0.007 |
| Brain | 0.033 ± 0.013 |
| Gallbladder | 0.095 ± 0.031 |
| Heart | 0.009 ± 0.002 |
| Intestine | 0.107 ± 0.052 |
| Kidneys | 0.069 ± 0.030 |
| Liver | 1.147 ± 0.306 |
| Lungs | 0.025 ± 0.005 |
| Stomach | 0.008 ± 0.006 |
| Urinary bladder | 0.063 ± 0.040 |
| Total | 1.588 ± 0.306 |
Values are mean ± SEM (n = 4).
TABLE 3.
Radiation Absorbed Dose for 18F-PF-05270430
| 70-kg man |
55-kg woman |
|||
| Organ | Mean (mSv/MBq) | SD (mSv/MBq) | Mean (mSv/MBq) | SD (mSv/MBq) |
| Adrenals | 1.55E–02 | 6.41E–03 | 1.84E–02 | 7.19E–03 |
| Brain | 4.76E–03 | 3.18E–03 | 6.80E–03 | 4.57E–03 |
| Breasts | 3.01E–03 | 1.43E–03 | 3.51E–03 | 1.65E–03 |
| Gallbladder wall | 1.63E–01 | 8.79E–02 | 2.01E–01 | 1.08E–01 |
| Lower large intestine wall | 6.61E–02 | 6.27E–02 | 7.01E–02 | 6.58E–02 |
| Small intestine | 7.32E–03 | 2.67E–03 | 9.04E–03 | 3.29E–03 |
| Stomach wall | 9.56E–03 | 6.66E–03 | 1.18E–02 | 7.80E–03 |
| Upper large intestine wall | 8.97E–03 | 3.42E–03 | 1.10E–02 | 4.14E–03 |
| Heart wall | 9.83E–03 | 4.17E–03 | 1.22E–02 | 5.24E–03 |
| Kidneys | 5.11E–02 | 3.20E–02 | 5.81E–02 | 3.65E–02 |
| Liver | 1.49E–01 | 8.02E–02 | 1.84E–01 | 9.91E–02 |
| Lungs | 1.13E–02 | 4.53E–03 | 1.36E–02 | 5.34E–03 |
| Muscle | 3.67E–03 | 1.38E–03 | 4.46E–03 | 1.65E–03 |
| Ovaries | 8.32E–03 | 4.51E–03 | ||
| Pancreas | 1.40E–02 | 5.95E–03 | 1.70E–02 | 7.12E–03 |
| Red marrow | 6.89E–03 | 2.44E–03 | 7.20E–03 | 2.47E–03 |
| Osteogenic cells | 4.19E–03 | 1.55E–03 | 5.48E–03 | 2.00E–03 |
| Skin | 1.87E–03 | 7.46E–04 | 2.24E–03 | 8.82E–04 |
| Spleen | 5.76E–03 | 3.62E–03 | 7.36E–03 | 4.01E–03 |
| Testes | 1.26E–03 | 6.91E–04 | ||
| Thymus | 2.61E–03 | 1.16E–03 | 3.18E–03 | 1.41E–03 |
| Thyroid | 7.12E–04 | 2.83E–04 | 8.57E–04 | 3.57E–04 |
| Urinary bladder wall | 3.22E–02 | 3.76E–02 | 4.34E–02 | 5.11E–02 |
| Uterus | 6.24E–03 | 2.83E–03 | ||
| Total body | 7.93E–03 | 3.26E–03 | 9.56E–03 | 3.85E–03 |
| Effective dose equivalent | 3.22E–02 | 1.07E–02 | 3.88E–02 | 1.29E–02 |
| Effective dose | 2.28E–02 | 8.21E–03 | 2.69E–02 | 9.28E–03 |
Human Injection and Scan Parameters
The mean administered activity was 178 ± 8 MBq (range, 157–185 MBq). Injection and scan parameters are listed in Table 1. The injected dose and injected mass did not significantly differ between test and retest scans (paired t test, P > 0.05). We detected no relationship between BPND and injected mass, suggesting that we were working in the tracer range, because a negative correlation was not found.
Safety
There were no adverse events or clinically detectable pharmacologic effects reported in any of the 6 subjects. No significant changes in vital signs were observed. Each subject completed both scans without reporting discomfort that would warrant cessation of scanning.
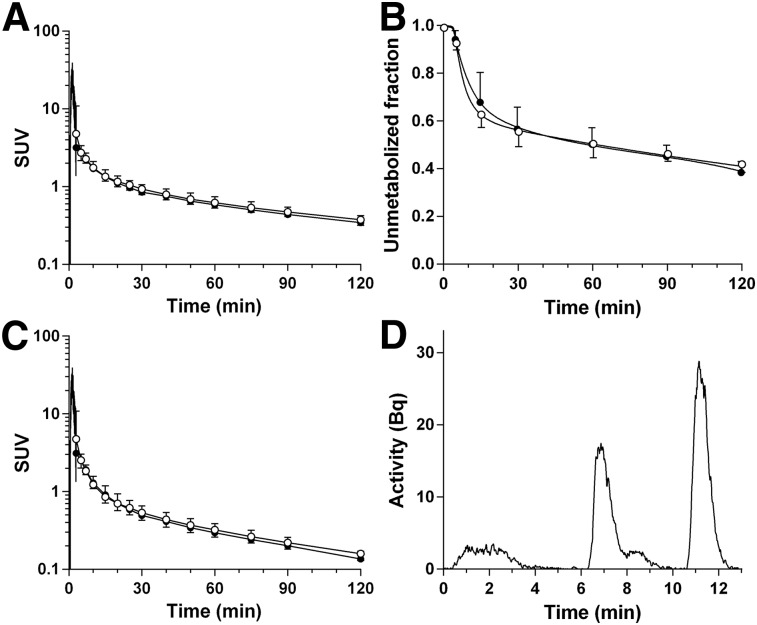
Plasma Analysis
Plasma data for the test and retest scans are displayed in Figures 2A–2C. The fraction of 18F-PF-05270430 plasma radioactivity decreased moderately over time, from 93% ± 3% at 5 min to 40% ± 4% at 120 min (n = 11). A typical example of a high-performance liquid chromatogram (90 min after injection) is shown in Figure 2D. The retention time was 11–12 min, and the HPLC profile revealed metabolites that were less lipophilic than parent tracer. The fP was 0.13 ± 0.03 (n = 10). There was no statistically significant difference in fP between test and retest scans (2-sided unpaired t test, P = 0.76). The TRV of fP was −4% ± 30% (n = 4).
FIGURE 2.
Mean ± SD of total plasma activity (A), plasma parent fraction (B), and metabolite-corrected plasma activity (C) in the test (●, n = 6) and retest (○, n = 5) scans. (D) HPLC plasma analysis at 90 min after injection. A and C are displayed in SUV units (concentration/[injected dose/body weight]).
Brain Distribution and Kinetics
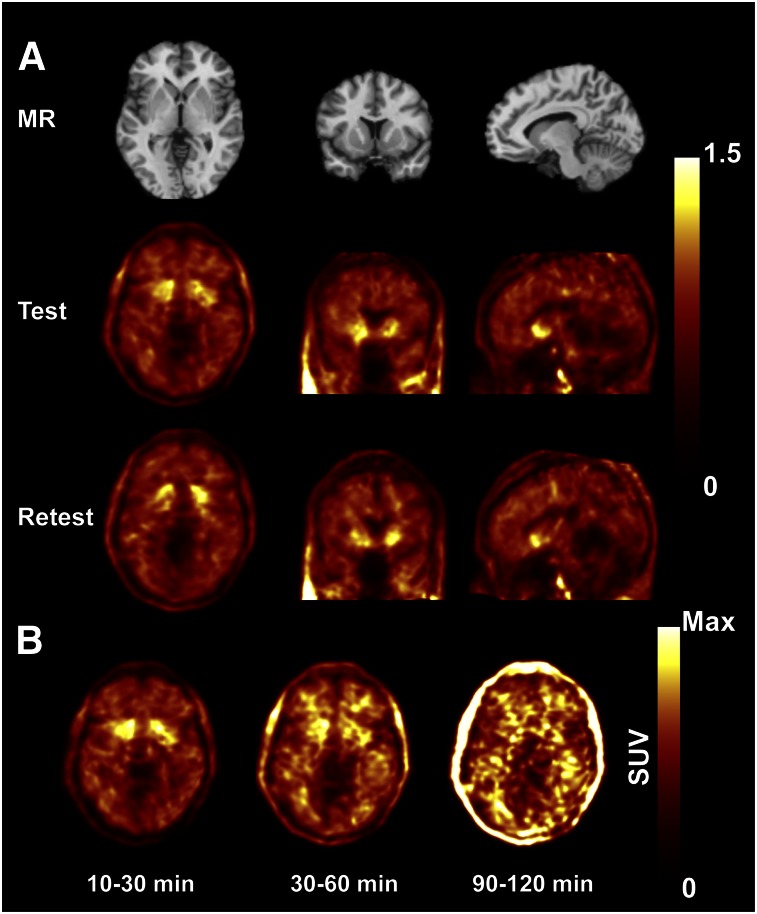
Figure 3 shows typical SUV images of 18F-PF-05270430. The highest uptake was seen in the putamen, caudate, and nucleus accumbens, followed by cortical regions and cerebellum. This was expected based on the known relative distribution of PDE2A in the central nervous system and on the nonhuman primate PET imaging studies performed in the companion paper (6). The radioactivity cleared from all ROIs relatively quickly, resulting in SUVs of less than 1 by 30 min after injection. In later frames, uptake in white matter was more pronounced than in gray matter (Fig. 3B) because of slower kinetics (Supplemental Fig. 1).
FIGURE 3.
(A) MR and coregistered typical PET images in test and retest conditions (10–60 min after injection of 18F-PF-05270430). PET images are displayed in SUV units. (B) PET images showing temporal distribution over 2 h, with each image scaled to its own maximum (SUVmax scales were 2, 1, and 0.4 for 10–30, 30–60, and 90–120 min, respectively). Max = maximum.
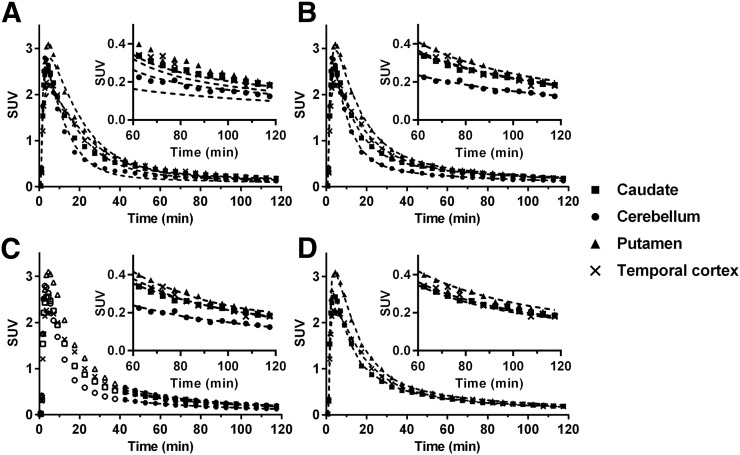
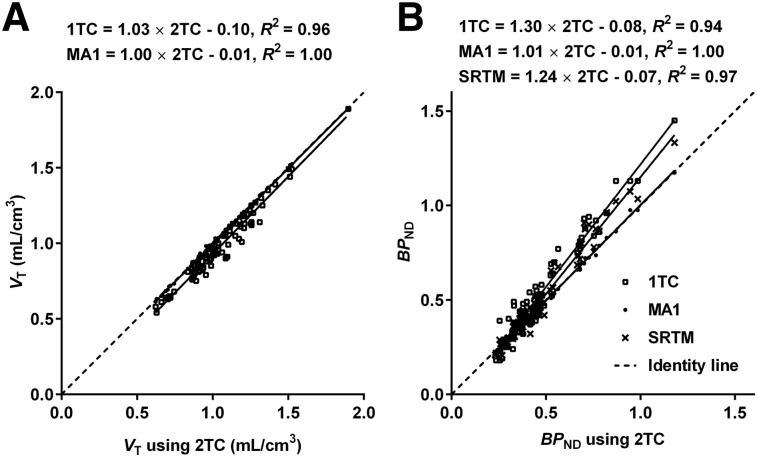
Typical time–activity curves and fitted curves are shown in Figure 4. Time–activity curves were fitted well with a 2TC model. 2TC K1 estimates (mL/min/cm3) ranged from 0.041 ± 0.004 (centrum semiovale) to 0.112 ± 0.010 (putamen). 2TC VT estimates without blood volume correction were similar to those with correction (VT [uncorrected] = 1.01 × VT [corrected] + 0.03, R2 = 1.00), whereas 2TC BPND estimates without blood volume correction were slightly lower than corrected values (BPND [uncorrected] = 0.96 × BPND [corrected] − 0.00, R2 = 1.00). MA1 (t* = 40 min) and SRTM fits were also excellent, whereas a moderate lack of fit was seen with a 1TC model. Although 2TC fits were excellent, 2TC parameters could not be reliably estimated in some cases (%SE > 25% in 6/99 fits). After estimates with large %SE were excluded, there was excellent correlation between 2TC and MA1 for VT and BPND (Fig. 5). SRTM BPND values were well correlated with the 2TC values but with a slope and intercept offset from the line of identity (Fig. 5B). The multilinear reference tissue model was also applied, but it produced many unstable BPND estimates in high-binding regions with all t* settings that were tested (data not shown).
FIGURE 4.
Time–activity curves from putamen, temporal cortex, caudate, and cerebellum of 18F-PF-05270430. Fits with 1TC (A), 2TC (B), MA1 (t* = 40 min) (C), and SRTM (D) are displayed as dotted lines. Insets show last 60 min.
FIGURE 5.
Comparison of 1TC and MA1 VT estimates against 2TC VT estimates (A) and 1TC, MA1, and SRTM BPND estimates against 2TC BPND estimates (B). Solid line = linear regression fit. All values were estimated with blood volume correction.
Given the quality of fit and comparison with 2TC estimates, the model of choice is MA1 (t* = 40 min), when arterial data are available. MA1 VT estimates using different t* values had small differences from the 40-min values, ranging from −2.1% ± 1.5% (t* = 10 min) to +1.0% ± 0.6% (t* = 60 min). Mean distribution volumes and binding potentials are shown in Table 4. VT was highest in the putamen and caudate, followed by neocortical regions and white matter, and lowest in the cerebellum. Intersubject variability was 7%–14% for VT, 9%–24% for BPND, and 8%–23% for BPP (n = 5) and 7%–15% for VT/fP and 6%–33% for BPF (n = 4).
TABLE 4.
Binding Parameters from MA1
|
VT (mL/cm3) (%COV) |
VT/fP (mL/cm3) (%COV) |
BPND (%COV) |
BPP (mL/cm3) (%COV) |
BPF (mL/cm3) (%COV) |
||||||
| Region | Test | Retest | Test | Retest | Test | Retest | Test | Retest | Test | Retest |
| Putamen | 1.27 (11%) | 1.31 (6%) | 10.07 (6%) | 10.14 (27%) | 0.79 (20%) | 0.80 (25%) | 0.52 (12%) | 0.54 (19%) | 3.96 (14%) | 4.11 (39%) |
| Nucleus accumbens | 1.21 (15%) | 1.22 (7%) | 9.69 (6%) | 9.49 (22%) | 0.70 (16%) | 0.67 (14%) | 0.46 (18%) | 0.45 (8%) | 3.57 (16%) | 3.46 (24%) |
| Caudate | 1.03 (13%) | 1.06 (6%) | 8.32 (6%) | 8.38 (26%) | 0.45 (18%) | 0.45 (30%) | 0.28 (15%) | 0.29 (23%) | 2.20 (23%) | 2.35 (46%) |
| Occipital cortex | 1.02 (16%) | 1.05 (8%) | 8.29 (3%) | 8.19 (22%) | 0.44 (12%) | 0.44 (12%) | 0.27 (19%) | 0.28 (6%) | 2.18 (13%) | 2.16 (24%) |
| Centrum semiovale | 0.97 (18%) | 1.02 (10%) | 7.84 (6%) | 7.79 (18%) | 0.36 (21%) | 0.39 (22%) | 0.22 (30%) | 0.25 (22%) | 1.72 (26%) | 1.76 (12%) |
| Temporal cortex | 0.95 (15%) | 0.99 (8%) | 7.64 (3%) | 7.64 (23%) | 0.34 (22%) | 0.36 (21%) | 0.20 (23%) | 0.22 (17%) | 1.53 (21%) | 1.61 (29%) |
| Frontal cortex | 0.95 (14%) | 0.98 (8%) | 7.59 (4%) | 7.59 (25%) | 0.34 (22%) | 0.35 (27%) | 0.20 (19%) | 0.21 (25%) | 1.47 (19%) | 1.56 (39%) |
| Parietal cortex | 0.93 (14%) | 0.95 (9%) | 7.50 (4%) | 7.35 (24%) | 0.32 (27%) | 0.30 (22%) | 0.18 (28%) | 0.18 (21%) | 1.38 (32%) | 1.32 (35%) |
| Cerebellum | 0.71 (17%) | 0.73 (12%) | 5.80 (3%) | 5.74 (22%) | ||||||
%COV = coefficient of variation (intersubject variability).
Values are mean of n = 5 subjects (n = 4 for VT/fP and BPF), with each value measured twice in test and retest scans.
The minimum scan duration required to satisfy stability criteria was 70 min for MA1 (t* = 40 min) (Supplemental Fig. 2). With increasing scan duration, VT estimates showed small increases (<10%, Supplemental Table 1). For higher-binding regions, the change in VT values (240 vs. 120 min) was small (1.0%–1.9%), with the largest change (9%) in the parietal cortex.
Test–Retest Variability and Reproducibility of Binding Parameters
Table 5 shows the test–retest statistics for VT and binding potentials, which were similar across regions. VT showed aTRV less than 10% in all regions, and the global mean of TRV was −4%. Both BPND and BPP showed moderate aTRV (∼15%), with a global mean TRV of 0% (BPND) and −5% (BPP). For VT, the ICC was good (>0.6) except in the putamen (0.23), and ICC tended to be lower for BPND except for the caudate, putamen, and frontal cortex. BPND using SRTM showed statistics similar to BPND using MA1. For BPP, the ICC was poor in most regions (<0.6). Scaling with fP (VT/fP, BPF) led to poor reproducibility with an aTRV of approximately 20% (VT/fP) and approximately 25% (BPF) and lower reliability.
TABLE 5.
Test–Retest Reproducibility of Volume of Distribution and Binding Potential
| MA1 (t* = 40 min) |
SRTM |
|||||||||||
|
VT (n = 5) |
BPND (n = 5) |
BPP (n = 5) |
BPND (n = 5) |
|||||||||
| Region | *aTRV | *TRV | †ICC | *aTRV | *TRV | †ICC | *aTRV | *TRV | †ICC | *aTRV | *TRV | †ICC |
| Putamen | 9% | −4% ± 11% | 0.23 [–0.64, 0.87] | 16% | 0% ± 23% | 0.58 [–0.33, 0.94] | 15% | −4% ± 24% | −0.01 [–0.76, 0.80] | 14% | −1% ± 22% | 0.64 [–0.24, 0.95] |
| Nucleus accumbens | 7% | −2% ± 10% | 0.66 [–0.20, 0.96] | 17% | 4% ± 22% | 0.15 [–0.69, 0.85] | 14% | 0% ± 23% | −0.17 [–0.83, 0.74] | 14% | 2% ± 18% | 0.17 [–0.68, 0.86] |
| Caudate | 7% | −3% ± 8% | 0.68 [–0.17, 0.96] | 18% | 2% ± 21% | 0.73 [–0.08, 0.97] | 14% | −3% ± 19% | 0.55 [–0.36, 0.94] | 15% | 2% ± 20% | 0.83 [0.19, 0.98] |
| Occipital cortex | 8% | −4% ± 8% | 0.77 [0.03, 0.97] | 12% | 0% ± 16% | 0.27 [–0.62, 0.88] | 13% | −5% ± 18% | 0.15 [–0.69, 0.85] | 15% | 3% ± 17% | 0.36 [–0.55, 0.91] |
| Centrum semiovale | 10% | −6% ± 11% | 0.71 [–0.12, 0.96] | 18% | −9% ± 19% | 0.63 [–0.26, 0.95] | 24% | −15% ± 22% | 0.53 [–0.38, 0.94] | 11% | −4% ± 14% | 0.71 [–0.11, 0.96] |
| Temporal cortex | 8% | −4% ± 9% | 0.71 [–0.12, 0.96] | 14% | −4% ± 24% | 0.57 [–0.34, 0.94] | 22% | −10% ± 28% | 0.15 [–0.69, 0.85] | 18% | −3% ± 26% | 0.51 [–0.41, 0.93] |
| Frontal cortex | 7% | −4% ± 7% | 0.75 [–0.03, 0.97] | 12% | −2% ± 16% | 0.85 [0.26, 0.98] | 13% | −7% ± 17% | 0.69 [–0.14, 0.96] | 15% | 5% ± 17% | 0.85 [0.26, 0.98] |
| Parietal cortex | 5% | −2% ± 7% | 0.84 [0.20, 0.98] | 21% | 6% ± 25% | 0.65 [–0.23, 0.96] | 22% | 2% ± 30% | 0.51 [–0.41, 0.93] | 21% | 9% ± 24% | 0.69 [–0.16, 0.96] |
| Cerebellum | 5% | −3% ± 6% | 0.90 [0.45, 0.99] | |||||||||
TRV = (test value – retest value)/(test value + retest value) × 2, and aTRV is absolute value of TRV.
ICC = (BSMSS – WSMSS)/(BSMSS + WSMSS) where BSMSS is mean sum of squares between subjects and WSMSS is mean sum of squares within subjects. ICC is presented with lower and upper bounds of 95% confidence interval in brackets.
DISCUSSION
The aims of this study were to determine the dosimetry of the novel PDE2A tracer 18F-PF-05270430 and perform a first-in-human study. To our knowledge, 18F-PF-05270430 is the first PDE2A radiotracer evaluated in humans. All subjects tolerated the tracer well without adverse effects. Modeling methods were evaluated, and regional brain test–retest variability was determined.
PDE2A is known to be involved in the pathophysiology of cognitive disorders (16) and other disease states. Thus, noninvasively imaging PDE2A enzymes could be used to facilitate the development of PDE2A inhibitors. This technique enables the capability to quantitatively measure PDE2A enzyme occupancy levels of drugs, which is a prerequisite for expression of PDE2-mediated pharmacology and target modulation (17). In turn, this allows for understanding the relationship between drug concentrations and PDE2A enzyme occupancy. In addition, differences in PDE2A enzyme levels in patients versus healthy controls can be established (18). A PET PDE2A ligand might also be useful for assessment of agents that increase cGMP levels, such as a PDE10 inhibitor, which raised cGMP, activating PDE2A through the GAF domain (19). Thus, a PDE2A PET ligand may be useful to identify signaling interactions between pathways that alter cGMP and cAMP.
This study compared 3 kinetic models using arterial input functions. Time–activity curves were fitted well by the 2TC model and MA1. The 2TC model and MA1 produced almost identical VT and BPND estimates. As is often the case, the 2TC model VT estimates were associated with large errors in some fits, whereas the MA1 VT estimates were stable in all fits. Further, MA1 also had the favorable property that VT estimates were little influenced by changing t*. Thus, MA1 is the model of choice for 18F-PF-05270430.
Blood volume correction was applied in this study. This correction is important when VT is low or the parent fraction in blood is low. VT values were low (0.7–1.4), and the parent fraction was in the medium range (40% at 120 min). The effect of blood volume correction was largest in the cerebellum, which had the lowest VT value; lack of correction led to smaller BPND values, because of larger overestimation of cerebellar VT. Assuming a 5% blood volume, the fraction of the cerebellum time–activity curve at 70 min accounted for by blood activity was 5% ± 1% (mean ± SD, n = 11).
The cerebellum was used as a reference region to calculate binding potentials. Western blot analysis of human brain extracts with anti-PDE2A antibodies revealed that PDE2A immunoreactivity was prominently observed in the striatum, cingulate cortex, prefrontal cortex, and hippocampus, with no detectable signal in the cerebellum (4). A blocking study in humans would be needed to validate the suitability of the cerebellum as a reference region.
The outcome measures corrected by fP, VT/fP, and BPF showed poorer reliability, because of variability of the fP measurement, which was larger than expected. BPND and BPP showed similar magnitudes of aTRV and TRV. However, the ICC of BPP was poor in most regions. Therefore, BPND appears to be the best binding potential outcome measure.
VT values were insensitive to scan duration. Over the 240-min scan, VT values in the striatum, nucleus accumbens, and centrum semiovale showed almost no change, whereas in neocortical regions and cerebellum, VT increased slightly (3.7%–9.1%). Potential factors contributing to these small increases include brain uptake of radiolabeled metabolites, or spill-in from extra-brain activity (Fig. 3B). However, the minimum scan time with MA1 was 70 min in all regions under relatively strict criteria. Thus, 18F-PF-05270430 has rapid kinetics, and long acquisitions are not needed to obtain robust parameter estimates.
For BPND, the test–retest variability with MA1 was acceptable with 16% of aTRV and 0% of TRV on average. Although often BPND has better TRV than VT, given the low BPND values (0.3–0.8), it is not surprising that the test–retest performance of VT was better than BPND. We compared the test–retest variability data of BPND across several radioligands (Supplemental Table 2). The aTRV of BPND with 18F-PF-05270430 was poorer than many other tracers. The ICC values were low in several regions, including the putamen (0.58) and nucleus accumbens (0.15). The VT and BPND estimates were low in all regions, indicating that 18F-PF-05270430 might only provide reliable data in the higher-binding regions, such as the putamen, caudate, and nucleus accumbens.
CONCLUSION
In this first-in-human PET study with the novel PDE2A radiotracer 18F-PF-05270430, the tracer showed rapid kinetics, providing reliable parameter estimates with a 70-min scan. MA1 is the most suitable model for describing the kinetics. Binding potential was estimated with the cerebellum as reference region, although this choice should be validated with blocking studies. 18F-PF-05270430 dosimetry showed that an effective dose was estimated as 0.023 mSv/MBq for a 70-kg male. On the basis of the results of this study, 18F-PF-05270430 is a suitable PDE2A PET ligand and can be reliably used in future clinical studies to assess enzyme occupancy levels of PDE2A inhibitors.
DISCLOSURE
The costs of publication of this article were defrayed in part by the payment of page charges. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734. This study was sponsored by Pfizer, Inc. This publication was also made possible by CTSA grant UL1 RR024139 jointly from the National Center for Research Resources (NCRR) and the National Center for Advancing Translational Sciences (NCATS), components of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH. No other potential conflict of interest relevant to this article was reported.
Supplementary Material
Acknowledgments
We appreciate the excellent technical assistance of the staff at the Yale University PET Center and the Pfizer CRU and the support of the Yale–Pfizer Bioimaging Alliance.
REFERENCES
- 1.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. [DOI] [PubMed] [Google Scholar]
- 2.Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS. Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the x-ray structure of a near full-length construct. Proc Natl Acad Sci USA. 2009;106:18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephenson DT, Coskran TM, Kelly MP, et al. The distribution of phosphodiesterase 2A in the rat brain. Neuroscience. 2012;226:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephenson DT, Coskran TM, Wilhelms MB, et al. Immunohistochemical localization of phosphodiesterase 2A in multiple mammalian species. J Histochem Cytochem. 2009;57:933–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Villalobos A, Beck EM, et al. Design and selection parameters to accelerate the discovery of novel central nervous system positron emission tomography (PET) ligands and their application in the development of a novel phosphodiesterase 2A PET ligand. J Med Chem. 2013;56:4568–4579. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Nabulsi N, Naganawa M, et al. Preclinical evaluation of 18F-PF-05270430, a novel PET radioligand for the phosphodiesterase 2A enzyme. J Nucl Med. May 19, 2016 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristy M, Eckerman KF. Specific Absorbed Fractions of Energy at Various Ages from Internal Photon Sources. I. Methods. Oak Ridge, TN: Oak Ridge National Laboratory; 1987. ORNL/TM-8381/V8381. [Google Scholar]
- 8.Stabin MG, Sparks RB, Crowe E. OLINDA/EXM: the second-generation personal computer software for internal dose assessment in nuclear medicine. J Nucl Med. 2005;46:1023–1027. [PubMed] [Google Scholar]
- 9.Hilton J, Yokoi F, Dannals RF, Ravert HT, Szabo Z, Wong DF. Column-switching HPLC for the analysis of plasma in PET imaging studies. Nucl Med Biol. 2000;27:627–630. [DOI] [PubMed] [Google Scholar]
- 10.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. [DOI] [PubMed] [Google Scholar]
- 11.Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22:324–333. [DOI] [PubMed] [Google Scholar]
- 12.Papademetris X, Jackowski M, Rajeevan N, Constable RT, Staib LH. Bioimage suite: an integrated medical image analysis suite. Insight J. 2006;2006:209. [PMC free article] [PubMed] [Google Scholar]
- 13.Gallezot JD, Nabulsi N, Holden D, et al. Development of a lysophosphatidic acid receptor 1 (LPA1) radioligand for lung imaging with PET [abstract]. J Nucl Med. 2014;55(suppl 1):96. [Google Scholar]
- 14.Frankle WG, Huang Y, Hwang DR, et al. Comparative evaluation of serotonin transporter radioligands 11C-DASB and 11C-McN 5652 in healthy humans. J Nucl Med. 2004;45:682–694. [PubMed] [Google Scholar]
- 15.Gallezot JD, Zheng MQ, Lim K, et al. Parametric imaging and test-retest variability of 11C-(+)-PHNO binding to D2/D3 dopamine receptors in humans on the high-resolution research tomograph pet scanner. J Nucl Med. 2014;55:960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez L, Breitenbucher JG. PDE2 inhibition: potential for the treatment of cognitive disorders. Bioorg Med Chem Lett. 2013;23:6522–6527. [DOI] [PubMed] [Google Scholar]
- 17.Morgan P, Van Der Graaf PH, Arrowsmith J, et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving phase II survival. Drug Discov Today. 2012;17:419–424. [DOI] [PubMed] [Google Scholar]
- 18.Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov. 2014;13:290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rombouts FJ, Tresadern G, Buijnsters P, et al. Pyrido[4,3-e][1,2,4]triazolo[4,3-a]pyrazines as selective, brain penetrant phosphodiesterase 2 (PDE2) inhibitors. ACS Med Chem Lett. 2015;6:282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.