Abstract
Background
Deletions in the long arm of chromosome 1 have been described in patients with a phenotype consisting primarily of obesity, intellectual disability and autism-spectrum disorder. The minimal region of overlap comprises two genes: DPYD and MIR137.
Case presentation
We describe a 10-year-old boy with syndromic obesity who carries a novel 1p21.3 deletion overlapping the critical region with the MIR137 gene only.
Conclusions
This study suggests that MIR137 is the mediator of the obesity phenotype of patients carrying 1p21.3 microdeletions.
Keywords: Obesity, Array-CGH, Genetics, MIR137, 1p21.3, Intellectual disability, Autism spectrum disorder
Background
Obesity is a complex disease that results from the interactions of a variety of hereditary and environmental factors. Heritability for obesity is high (typically 0.4 to 0.7) [1], comparable to that of other complex, polygenic diseases such as autism [2]. In most individuals, the genetic basis for obesity is complex and likely to involve the interaction of multiple genes and gene-by-environment interactions. However, there are rare examples of monogenic causes for obesity that provide insights to pathways that may account for more common causes of obesity and indicate targets for therapeutic intervention. These disorders include obesity syndromes such as Prader Willi syndrome and Bardet–Biedl syndrome and microdeletion/duplication syndromes such as 1p36 and 17p11.2, in which affected patients are usually identified by additional phenotypes, such as intellectual disability (ID), dysmorphic features or other developmental abnormalities. Gaining insight into genetic causes through the study of obesity-related disorders has provided a more comprehensive picture of the mechanisms that control food intake and energy balance related to the development of obesity (e.g., SIM1).
Deletions in chromosome 1p21.3 have been reported in a total of 12 individuals presenting with a variable combination of ID, autism spectrum disorders (ASD) and obesity [3–6]. The shortest region of overlap (SRO) has been restricted to two genes: dihydropyrimidine dehydrogenase (DPYD) and microRNA 137 (MIR137). The DPYD encodes for the rate-limiting enzyme in the catabolism of the pyrimidine bases, and mutations in this gene cause DPYD deficiency (OMIM ♯274270), an autosomal recessive disease of variable severity typically characterized by the presence of childhood onset neurological problems, such as developmental delay and convulsions. MIR137 encodes for miR-137, a microRNA (miRNA) molecule involved in the post-transcriptional regulation of genes [7].
In this study, we report an additional case carrying a 1p21.3 microdeletion that presented with ID, ASD, obesity and additional clinical features. We propose the MIR137 gene as the mediator of the obesity phenotype that presents in patients carrying chromosome 1p21.3 microdeletions.
Case presentation
The proband is a ten-year-old male and the first-born child of healthy non-consanguineous parents. The family history was unremarkable for neurological disorders, behavioral problems, and congenital anomalies.
He was born at 39 weeks of gestation via elective caesarian section. His birth weight was 2800 g (10-25th centile) and his length, occipital frontal circumference and APGAR scores were not reported but were said to be normal. At birth, no medical problems were recorded. Postnatally, his growth was normal; global psychomotor delay was noted: the boy took his first steps at the age of 20 months and said his first words at the age of 36 months. In infancy, he had several episodes of febrile convulsions, which spontaneously resolved at the age of 3 years. During a routine heart examination, a heart murmur was noticed, which prompted further tests. An echocardiogram showed a bicuspid aortic valve, with mild stenosis of the aortic root.
At the age of 9, he attended the fifth year of primary school due to speech disorder and anxiety. Neuropsychiatric assessment revealed anxiety, normal non verbal cognitive function, specific language impairment and fine motor skills disturbances, requiring weekly psychomotricity sessions. His growth parameters were as follows: weight was 39.3 kg (75th centile), height 147.2 cm (97th centile), body mass index (BMI) was 18.13 kg/m2. At the age of 10, the boy weighted 48 kg (>97th centile), height was 154 cm (>97th centile), head circumference was 58 cm (>97th centile) and BMI was 20.6. Pubic hair appearance, growth spurt and weight gain were noted. The boy was hospitalized, and further tests were performed, confirming precocious puberty of central origin: a radiograph of the hand and wrist, which showed advanced bone age (bone age of 13 years, biological age 9 years and 5 months); renal echography, adrenal glands and tests, which were normal, and the LHRH test, which showed an LH response. The boy started agonist LHRH therapy. Brain magnetic resonance imaging (MRI) showed increased cerebrospinal fluid (CSF) spaces posteriorly to the cerebellum and a moderate focal increase of CSF in the anterior temporal horns and in the lamina of the quadrigeminal cistern. The pituitary gland was normal.
Chromosome analysis by array-CGH (a-CGH) revealed a 5.2-Mb deletion on chromosome 1p21.3p21.1 (chromosome 1:98,456,293-103,682,084 – hg19). a-CGH was performed using a Cytochip ISCA 180 K Oligo platform (BlueGnome Ltd.). a-CGH analysis of the parents revealed the de novo origin of the deletion.
Conclusions
Deletions at 1p21.3 have been recognized as a cause of childhood obesity syndrome. Twelve patients carrying 1p21.3 deletions have been described in the literature to date [3–6], presenting with mild to moderate ID (100 %), obesity or a tendency to obesity, with a weight 3 or more standard deviations above the mean (92 %), ASD (92 %) and mild dysmorphic features (67 %) (Fig. 1 and Table 1). The minimal overlapping region was restricted to a chromosomal segment of 1.22 Mb, which includes the DPYD and MIR137 genes. DPYD was proposed as the gene contributing to the phenotype [3, 5], based on the assumption that its haploinsufficiency would contribute to the development of a neuropsychiatric phenotype, which also leads to obesity. However, the case herein presented and the patient recently reported by Pinto et al. [6] carried a deletion overlapping in the critical region only at the MIR137, which suggests that this is the gene underlying the phenotype of patients carrying deletions at 1p21.3.
Fig. 1.
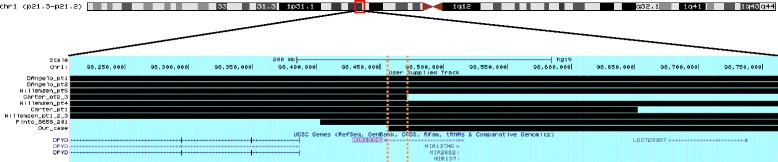
Chromosome 1p21.3 microdeletions described to date. Minimal region of overlap highlighted by the dashed orange lines. The figure was drawn according to the UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) assembly
Table 1.
Clinical features of patients carrying overlapping 1p21.3 deletions
| Carter pt 1 | Carter pt 2, pt 3 | Willemsen pt 1, pt2, pt 3 | Willemsen pt 4 | Willemsen pt 5 | D’Angelo pt 1 | D’Angelo pt 2 | Pinto pt 8658_201 | Our case | |
|---|---|---|---|---|---|---|---|---|---|
| Chromosome Position on chr 1 (hg19) | 97,559,579–98,652,079 | 96,969,562–98,471,225 | 97,727,412–99,477,412 | 97,547,412–98,957,412 | 96,497,412–98,947,412 | 93,919,217–99,846,176 | 95,696,444–107,755,879 | 98,403,034–101,151,364 | 98,456,293–103,682,084 |
| Age | 13 years 9 months | 7 years, 5 years | 42 years, 38 years, NA | 33 years | 18 years | 15 years | 8 years 8 months | NA | 10 years |
| Gender | M | M, F | M, M, F | M | F | F | F | F | |
| Intellectual disability | Severe language delay | Severe language delay | Borderline, Mild-moderate | Mild | Moderate | Language delay | Severe language delay | Severe language delay | Mild |
| Autism spectrum disorder | + | + | + | + | + | NA | + | + | + |
| Weight | NA | > > 97th centile 50th centile, | 90th, >98th centile | 98th centile | >98th centile | >95th centile | >95th centile | Overweight | Overweight |
| Dysmorphic features | NA | + | + | + | + | + | - | NA | - |
| Ocular problems | NA | NA | Myopia, astigmatism | Myopia, astigmatism | / | Myopia | / | NA | Hypermetropy |
| Precocious puberty | NA | NA | - | - | - | - | + | NA | + |
| Others | / | Small joint hypermobility, macrocephaly | / | / | / | / | / | High pain intolerance | Bicuspic aortic valve |
miRNAs are noncoding 21–23 nucleotide small RNAs that are highly conserved and involved in several biological processes, such as cell proliferation, apoptosis, cell differentiation and morphogenesis [8]. They regulate posttranscriptional processes via sequence-specific interactions with the untranslated regions (UTRs) of cognate messenger RNA targets [7, 9]. Each individual miRNA can regulate many mRNAs, and conversely, each mRNA can be targeted by a number of miRNAs. The involvement of miR-137 in obesity, ID and ASD could be explained at two different levels. First, miR-137 regulates a multitude of genes in the central nervous system (CNS) [10]. Specifically, miR-137 has been shown to be involved in modulating neurogenesis, with a dose-dependent effect [11]. Moreover, the miR137 targets Ezh2, a H3-K27 methyltransferase involved in the regulation of neuroprogenitor cell maintenance and differentiation [12]. Notably, mutations in the EZH2 gene cause Weaver syndrome, another syndromic overgrowth disorder characterized by ID, dysmorphic features and obesity [13]. Although the role of neuronal noncoding RNAs in the energy control of the body is not fully understood, recently, the hypothalamic miR-103 has been shown to protect against hyperphagic obesity in mice [14]. Therefore, the role of miR-137 in the control of food intake or energy accumulation and expenditure in the CNS can be speculated.
At a peripheral level, recent studies have shown that miRNAs are dysregulated in obese adipose tissue [15]. Furthermore, during adipogenesis, miRNAs regulate fat cell development by accelerating or inhibiting adipocyte differentiation. In addition, miRNAs may regulate the commitment to the adipogenic lineage in multipotent stem cells and therefore govern the numbers of fat cells [16].
Ninety-two percent of the cases carrying 1p21.3 deletion present with ASD. miR-137 is a brain expressed miRNA [17, 18] that regulates several protein coding genes that are important for brain function, and involved in the pathogenesis of neuropsychiatric disorders including schizophrenia [17, 19]. Functional studies indicate that miR-137 is involved in controlling neuronal differentiation and maturation [20, 21] as well as synapse development [22], all of which have been implicated on the pathogenesis of autism [20, 23]. Of note, lymphoblastoid cell lines from patients carrying 1p21.3 deletion involving the miR-137 gene have reduced levels of miR-137 [4]. miR-137 is highly expressed in the hippocampus, occipital cortex, and frontal cortex in human post-mortem tissue, as well as in the synaptosomal fractions in mouse brain preparations, providing further evidence that miR-137 plays a role in synapse formation during brain development [4].
Several studies have implicated miRNA in autism. A number of miRNAs have shown differing expression levels in samples from autistic individuals compared to matched controls [24, 25], suggesting that dysregulation of miRNAs could be related to autism. Furthermore, miR-137 has been recently implicated in ASD via a genome-wide meta-analysis that considered single nucleotide polymorphism data across five psychiatric disorders (ASD, attention deficit-hyperactivity disorder, bipolar disorder, major depressive disorder, and schizophrenia), and found an association between mir-137 and both schizophrenia and ASD [26].
Lastly, Devanna and Vernes provided support for miR-137 as an autism candidate by showing that the RORa gene, a novel autism candidate gene, is regulated directly by miR-137 through targeting its 3 UTR in a site-specific manner [27].
It is therefore likely that the core clinical features of obesity and ASD in patients carrying 1p21.3 deletion are both mediated by miR-137.
The case herein described presents with clinical features not previously described in the 1p21.3 microdeletion syndrome, such as precocious puberty and bicuspid aortic valve, with mild stenosis of the aortic root. The boy carries a 1p21.3 deletion, which extends 5 Mb proximally to the SRO and includes a number of genes. PALMD and S1PR1 encode for proteins that are expressed during normal embryonic heart development and morphogenesis. Specifically, PALMD has been associated with aortic root size [28], while S1PR1 has an important role in the regulation of sprouting angiogenesis and vascular maturation. It inhibits sprouting angiogenesis to prevent excessive sprouting during blood vessel development [29]. With regard to precocious puberty, we found two DECIPHER patients that carried deletions partially overlapping with our case and precocious puberty: case 260349, which presented with precocious puberty and specific learning disability and carrying a deletion at chr1:97,000,749–99,813,665; case 254871 presented with ASD, ID, precocious puberty and spotty hyperpigmentation, carrying the deletion at chr1:97019762–102447536. The SRO among these microdeletions includes the SNX7, PLPPR4 and PLPPR5 genes. Both PLPPR4 and PLPPR5 encode for a lipid phosphate phosphatase enzyme specifically expressed in neurons and located in the membranes of outgrowing axons, important for axonal outgrowth during development and regenerative sprouting [30, 31].
In summary, with this study, we present a case carrying a novel 1p31.3 microdeletion, and we propose that the miRNA miR137 is the mediator of the obesity, ASD and ID phenotype. Further studies are warranted to elucidate the molecular mechanisms by which miR-137 mediates the phenotype, both in the CNS and the adipose tissue.
Acknowledgements
We thank the patient and his parents for their kind cooperation.
Funding
Not applicable.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors’ contributions
AT performed the paediatric evaluation of the patient, wrote the manuscript and analysed the SNP array result; CC and GS analysed the SNP array result and e and critically read the manuscript; SE revised the manuscript critically; DM initiated the study, performed the paediatric evaluation of the patient, made substantial contribution to study design and revised the manuscript critically. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Written informed consent was obtained from the patient’s parents for publication of this case report.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ASD
Autism-spectrum disorders
- BMI
Body mass index
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- ID
Intellectual disability
- MRI
Magnetic resonance imaging
- SRO
Shortest region of overlap
- UTRs
Untraslated regions
Contributor Information
Arianna Tucci, Email: a.tucci@ucl.ac.uk.
Claudia Ciaccio, Email: claudia.ciaccio@unimi.it.
Giulietta Scuvera, Email: giulietta.scuvera@policlinico.mi.it.
Susanna Esposito, Email: susanna.esposito@unimi.it.
Donatella Milani, Phone: +39-02-55032412, Email: donatella.milani@policlinico.mi.it.
References
- 1.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–51. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 2.Santangelo SL, Tsatsanis K. What is known about autism: genes, brain, and behavior. Am J Pharmacogenomics Genomics. 2005;5(2):71–92. doi: 10.2165/00129785-200505020-00001. [DOI] [PubMed] [Google Scholar]
- 3.D’Angelo CS, Moller Dos Santos MF, Alonso LG, Koiffmann CP. Two new cases of 1p21.3 deletions and an unbalanced translocation t(8;12) among individuals with syndromic obesity. Mol Syndromol. 2015;6(2):63–70. doi: 10.1159/000371600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willemsen MH, Vallès A, Kirkels LAMH, Mastebroek M, Olde Loohuis N, Kos A, Wissink-Lindhout WM, de Brouwer APM, Nillesen WM, Pfundt R, Holder-Espinasse M, Vallée L, Andrieux J, Coppens-Hofman MC, Rensen H, Hamel BCJ, van Bokhoven H, Aschrafi A, Kleefstra T. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48(12):810–8. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 5.Carter MT, Nikkel SM, Fernandez BA, Marshall CR, Noor A, Lionel AC, Prasad A, Pinto D, Joseph-George AM, Noakes C, Fairbrother-Davies C, Roberts W, Vincent J, Weksberg R, Scherer SW. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin Genet. 2011;80(5):435–43. doi: 10.1111/j.1399-0004.2010.01578.x. [DOI] [PubMed] [Google Scholar]
- 6.Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, Vorstman JAS, Thompson A, Regan R, Pilorge M, Pellecchia G, Pagnamenta AT, Oliveira B, Marshall CR, Magalhaes TR, Lowe JK, Howe JL, Griswold AJ, Gilbert J, Duketis E, Dombroski BA, De Jonge MV, Cuccaro M, Crawford EL, Correia CT, Conroy J, Conceição IC, Chiocchetti AG, Casey JP, Cai G, Cabrol C, Bolshakova N, Bacchelli E, Anney R, Gallinger S, Cotterchio M, Casey G, Zwaigenbaum L, Wittemeyer K, Wing K, Wallace S, van Engeland H, Tryfon A, Thomson S, Soorya L, Rogé B, Roberts W, Poustka F, Mouga S, Minshew N, McInnes LA, McGrew SG, Lord C, Leboyer M, Le Couteur AS, Kolevzon A, Jiménez González P, Jacob S, Holt R, Guter S, Green J, Green A, Gillberg C, Fernandez BA, Duque F, Delorme R, Dawson G, Chaste P, Café C, Brennan S, Bourgeron T, Bolton PF, Bölte S, Bernier R, Baird G, Bailey AJ, Anagnostou E, Almeida J, Wijsman EM, Vieland VJ, Vicente AM, Schellenberg GD, Pericak-Vance M, Paterson AD, Parr JR, Oliveira G, Nurnberger JI, Monaco AP, Maestrini E, Klauck SM, Hakonarson H, Haines JL, Geschwind DH, Freitag CM, Folstein SE, Ennis S, Coon H, Battaglia A, Szatmari P, Sutcliffe JS, Hallmayer J, Gill M, Cook EH, Buxbaum JD, Devlin B, Gallagher L, Betancur C, Scherer SW. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94(5):677–94. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Plasterk RHA. Micro RNAs in animal development. Cell. 2006;124(5):877–81. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Guella I, Sequeira A, Rollins B, Morgan L, Torri F, van Erp TGM, Myers RM, Barchas JD, Schatzberg AF, Watson SJ, Akil H, Bunney WE, Potkin SG, Macciardi F, Vawter MP. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. 2013;47(9):1215–21. doi: 10.1016/j.jpsychires.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189(1):127–41. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sher F, Rössler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells Dayt Ohio. 2008;26(11):2875–83. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- 13.Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte SDV, Ramsay E, Snape K, Murray A, Perdeaux ER, Seal S, Loveday C, Banka S, Clericuzio C, Flinter F, Magee A, McConnell V, Patton M, Raith W, Rankin J, Splitt M, Strenger V, Taylor C, Wheeler P, Temple KI, Cole T, Douglas J, Rahman N, Childhood Overgrowth Collaboration Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2(12):1127–33. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinnikov IA, Hajdukiewicz K, Reymann J, Beneke J, Czajkowski R, Roth LC, Novak M, Roller A, Dörner N, Starkuviene V, Theis FJ, Erfle H, Schütz G, Grinevich V, Konopka W. Hypothalamic miR-103 protects from hyperphagic obesity in mice. J Neurosci Off J Soc Neurosci. 2014;34(32):10659–74. doi: 10.1523/JNEUROSCI.4251-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie H, Sun L, Lodish HF. Targeting microRNAs in obesity. Expert Opin Ther Targets. 2009;13(10):1227–38. doi: 10.1517/14728220903190707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11(4):304–16. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forero DA, vanderVen K, Callaerts P, Del-Favero J. miRNA genes and the brain: implications for psychiatric disorders. Hum Mutat. 2010;31(11):1195–204. doi: 10.1002/humu.21344. [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright C, Calhoun VD, Ehrlich S, Wang L, Turner JA, Perrone-Bizzozero NI. Meta gene set enrichment analyses link miR-137-regulated pathways with schizophrenia risk. Front Genet. 2015;6:147. doi: 10.3389/fgene.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, Vaccarino FM. FOXG1-dependent dysregulation of GABA/Glutamate neuron differentiation in autism spectrum disorders. Cell. 2015;162(2):375–90. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, Jin P, Zhao X. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28(6):1060–70. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins AL, Kim Y, Bloom RJ, Kelada SN, Sethupathy P, Sullivan PF. Transcriptional targets of the schizophrenia risk gene MIR137. Transl Psychiatry. 2014;4:e404. doi: 10.1038/tp.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–93. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of posttranscriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med. 2010;2(4):23. doi: 10.1186/gm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abu-Elneel K, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9(3):153–61. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- 26.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devanna P, Vernes SC. A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137. Sci Rep. 2014;4:3994. doi: 10.1038/srep03994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, Watzinger N, Larson MG, Smith NL, Dehghan A, Grosshennig A, Schillert A, Teumer A, Schmidt R, Kathiresan S, Lumley T, Aulchenko YS, König IR, Zeller T, Homuth G, Struchalin M, Aragam J, Bis JC, Rivadeneira F, Erdmann J, Schnabel RB, Dörr M, Zweiker R, Lind L, Rodeheffer RJ, Greiser KH, Levy D, Haritunians T, Deckers JW, Stritzke J, Lackner KJ, Völker U, Ingelsson E, Kullo I, Haerting J, O’Donnell CJ, Heckbert SR, Stricker BH, Ziegler A, Reffelmann T, Redfield MM, Werdan K, Mitchell GF, Rice K, Arnett DK, Hofman A, Gottdiener JS, Uitterlinden AG, Meitinger T, Blettner M, Friedrich N, Wang TJ, Psaty BM, van Duijn CM, Wichmann HE, Munzel TF, Kroemer HK, Benjamin EJ, Rotter JI, Witteman JC, Schunkert H, Schmidt H, Völzke H, Blankenberg S. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302(2):168–78. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendelson K, Zygmunt T, Torres-Vázquez J, Evans T, Hla T. Sphingosine 1-phosphate receptor signaling regulates proper embryonic vascular patterning. J Biol Chem. 2013;288(4):2143–56. doi: 10.1074/jbc.M112.427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sciorra VA, Morris AJ. Roles for lipid phosphate phosphatases in regulation of cellular signaling. Biochim Biophys Acta. 2002;1582(1–3):45–51. doi: 10.1016/S1388-1981(02)00136-1. [DOI] [PubMed] [Google Scholar]
- 31.Broggini T, Nitsch R, Savaskan NE. Plasticity-related gene 5 (PRG5) induces filopodia and neurite growth and impedes lysophosphatidic acid- and nogo-A-mediated axonal retraction. Mol Biol Cell. 2010;21(4):521–37. doi: 10.1091/mbc.E09-06-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


