Abstract
Background
Cecal ligation and puncture (CLP) is considered the gold standard for inducing abdominal sepsis in mice. However, the model lacks source control, a component of sepsis management in humans. Using a CLP-excision model, we characterized peritoneal cytokines and cells and hypothesized these analyses would allow us to predict survival.
Methods
Fifty-eight mice were first subjected to CLP. Twenty hours later, the necrotic cecums were debrided, abdominal cavity lavaged, and intraperitoneal antibiotics administered. Peritoneal cytokines and leukocytes collected from the peritoneal lavage were analyzed. These immune parameters were used to generate receiver operator characteristic curves. In separate experiments, the accuracy of the model was verified with a survival cohort. Finally, we collected the peritoneal lavage and analyzed both serum and peritoneal cytokines, bacterial load, and leukocyte functionality.
Results
Peritoneal interleukin (IL)-6 levels and neutrophil CD11b intensity were observed to be significantly different in mice that lived versus those who died. In separate experiments, mice predicted to live (P-LIVE) had decreased bacterial loads, systemic IL-10, and neutrophil oxidative burst and increased peritoneal inflammatory monocyte numbers and phagocytosis.
Conclusions
This study couples a clinically relevant sepsis model with methodology to limit pathogen spread. Using surgical waste, stratification of the mice into groups P-LIVE and predicted to die was possible with a high degree of accuracy and specificity. In mice P-LIVE, increased inflammatory monocyte recruitment and phagocytosis were associated with decreased systemic IL-10 and bacterial loads.
Keywords: Cecal ligation and puncture, Neutrophil activation, IL-6, Inflammation, Bacterial clearance, Leukocyte
1. Introduction
Despite advances in technology and medical care, mortality from sepsis remains relatively constant at 18%–30% [1–3]. Severe sepsis and septic shock are leading causes of death in the United States, accounting for 215,000 deaths annually [1,4]. Over the past decade, the Surviving Sepsis Campaign, with its of hallmarks of early broad-spectrum antibiotics, early source control, and administration and maintenance of tissue perfusion, has demonstrated modest improvements in survival with strict adherence to the guidelines [5,6]. Recent randomized controlled trials, however, have illustrated that these hallmarks of sepsis management have likely reached an asymptote for improving survival [2,3]. In general, these treatments largely lack disease and patient specificity.
The pathophysiology of sepsis revolves around a complex immunological balance between an initial phase dominated by hyperinflammation followed by a prolonged period of profound immunologic compromise or paralysis [7–9]. Countless attempts to improve survival through the modulation of this ever-changing immune response have largely been unsuccessful [10,11]. Improvements in expeditious diagnosis, determination of the immune status and tailoring immunomodulation therapies to a specific patient, and disease populations appear to be crucial in advancing the management of this complex disease [12,13].
Previous research in mice subjected to cecal ligation and puncture (CLP) has demonstrated that interleukin (IL)-6 levels in the blood could be used to predict survival in mice and guide immunomodulation therapies [14]. Although CLP is the gold standard for inducing sepsis in experimental murine models, the lack of source control is a limitation when extrapolating to sepsis management in humans [15]. Therefore, we used a CLP-excision (CLP-E) model in which cecal excision, peritoneal wash, and antibiotic treatment were performed after CLP [16]. Using this model, we hypothesized that we could use the surgical waste from the peritoneal washes to predict survival, and in mice predicted to live (P-LIVE), we would observe an initial increased inflammatory response and decreased bacterial loads.
2. Materials and methods
2.1. Mice
Outbred CF-1 male mice were purchased from Charles River Laboratory (Spencerville, OH). All mice were 6-wk-old when purchased from Charles River Laboratory and allowed to acclimate for 1–3 wk. All mice were housed in standard environmental conditions and were fed with a commercial pellet diet and water ad libitum.
2.2. Cecal ligation and puncture
Male mice aged between 6 and 10 wk (28–35 g) were used. All experiments involving animals were performed under protocols approved by the Institutional Animal Care and Use Committee of the University of Cincinnati. Polymicrobial sepsis was induced similarly as described [17]. Briefly, the CLP operations were always performed between 8 AM and 1 PM. Mice were anesthetized to effect by 2% isoflurane in oxygen via a face mask. The skin was shaved and disinfected. After a 1–2-cm laparotomy, the latter 50% of the cecum was ligated with a 3-0 silk tie (Ethicon, Cincinnati, OH) and a full thickness through and through puncture was made on the anti-mesenteric side with a 20-gauge needle. A small amount of the bowel contents was extruded through the puncture holes to assure a full thickness perforation. The cecum was replaced in its original location, and the midline incision was closed by two-layer suture with 4-0 silk (Henry Schein, Melville, NY). The animals were resuscitated with 1 mL of sterile saline administered subcutaneously and kept on a heating blanket for 1 h [17]. Survival studies were carried out for 10 d.
2.3. Pathogen source control
At 20 h after CLP, the mice were anesthetized identically to the procedure mentioned previously. The laparotomy incisions were opened, and their abdomens were explored. All filmy adhesions were lysed, and the latter 50% of the cecum, now 100% necrotic, was debrided at the previous ligation site. The peritoneal cavity was washed with 35 mL of warm sterile phosphate-buffered saline and collected [18,19]. Finally, a single intraperitoneal dose of imipenem (Merck, Whitehouse Station, NJ) was administered at a dose of 2.5 mg/kg [20]. The abdomens are then closed, and the mice resuscitated in manner consistent with the CLP procedure.
2.4. Enzyme-linked immunosorbent assay
Peritoneal fluid was harvested from mice by peritoneal wash and centrifuged at 450g for 10 min. The supernatant fluid was used for cytokine analysis, and pelleted cells were analyzed by flow cytometry. For serum collection, blood collected by cardiac puncture was placed in serum separator tubes (BD Biosciences, San Jose, CA), centrifuged at 1000g for 10 min with isolated serum subsequently transferred to a new sterile tube. The IL-6 and IL-10 concentrations in the peritoneal fluid and serum were analyzed by enzyme-linked immunosorbent assay (BD Biosciences) as previously described [17].
2.5. Bacterial counts
Bacterial counts were performed on aseptically harvested peritoneal wash. Aerobic samples were serially diluted in sterile saline and cultured on Tryptic Soy Agar plates (BD Biosciences). Aerobic plates were incubated at 37°C for 24 h, and colony counts were performed [21]. Anaerobic samples were serially diluted in sterile saline and cultured on PRAS Brucella plates (Anaerobe Systems, Morgan Hill, CA). Anaerobic plates were incubated in BD GasPak Anaerobic Containers with three GasPak sachets (BD Biosciences) incubated at 37°C for 48 h, and colony counts were performed [22]. All septic mice had positive cultures resulting from fecal spillage into the peritoneum.
2.6. Flow cytometry
Analyses of cell surface antigen expression and of cytokine expression in situ were performed. The cells were labeled with Phycoerythrin–anti-mouse Ly-6G (clone: 1A8) and Allophycocyanin–anti-mouse CD11b (clone M1/70), and FITC anti-mouse CD54 (clone: 3E2). Using flow analysis, Ly-6G/CD11b positive cells were identified as neutrophils. CD11b and CD54 were used as markers of neutrophil activation [23–25]. All antibodies used for flow cytometry were obtained from BD Pharmingen [26]. Flow cytometry acquisition and analysis were performed on an Attune Flow Cytometer (Life Technologies, Foster City, CA).
2.7. Oxidative burst measurement
Measurement of spontaneous hydrogen peroxide was determined by measuring the oxidation of dihydrorhodamine (DHR) to fluorescent rhodamine. Cells collected from peritoneal lavages were first incubated with DHR (2.5 × 10−5 M) and subsequently stimulated with formyl-methionyl-leucyl-phenylalanine (fMLP) (10−5 M) or the phosphate-buffered saline vehicle. This reaction was stopped by putting cells on ice. Before antibody labeling, FcgII/III receptors were blocked by anti-mouse CD16/CD32 (10 min; 1 mg/mL). Without washing, cells were labeled with PE–anti-mouse Ly-6G (clone: 1A8) and APC–anti-mouse CD11b (clone M1/70). Using flow analysis, Ly-6G/CD11b positive cells were identified as neutrophils. Intracellular reactive oxygen species (ROS) production was indirectly detected on the fluorescence-1 channel by rhodamine intensity [21].
2.8. Phagocytosis assay
Cells from the peritoneal wash were incubated with a bacterial suspension consisting of Fluorescein isothiocyanate– labeled Escherichia coli BioParticles (Molecular Probes, Eugene, OR) at 37°C for 10 min. The cells were then fixed with paraformaldehyde and washed twice. Cells were stained with PE–anti-mouse Ly-6G (clone:1A8) and APC–anti-mouse CD11b (clone: M1/70). Flow cytometric analysis was performed without trypan blue to determine bacteria associated with and ingested by the cell of interest. Subsequently, trypan blue was added with the samples reanalyzed to determine the amount of bioparticles ingested by the cells of interest [26].
2.9. Statistics
Statistical comparisons were performed using Kaplan–Meier logrank (survival) or Student t-test (two groups, Prism 6; GraphPad Software, La Jolla, CA). The receiver operator characteristic (ROC) curve was generated using Statistical Analysis System software (Cary, NC). A value of P < 0.05 was considered statistically significant. Subsequently, specificity and sensitivity values were determined using a cutoff value of 200,000 for neutrophil CD11b mean fluorescence intensity (MFI).
3. Results
3.1. ROC curve generation associating survival with immune parameters
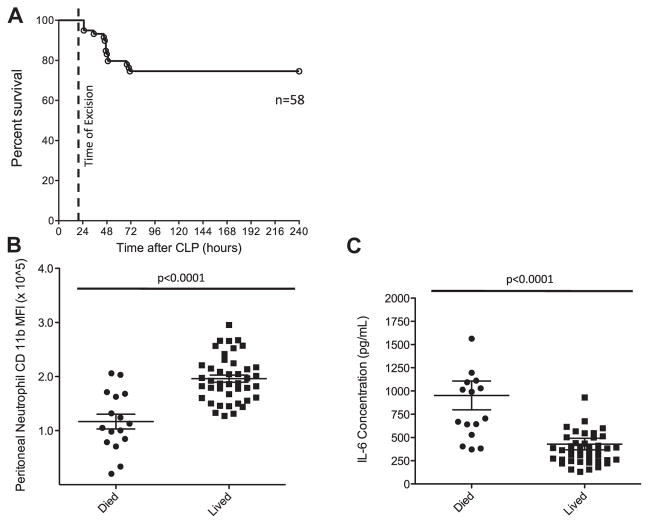
Fifty-eight mice were subjected to CLP and subsequent source control as described in the methods. Survival was monitored over 10 d. The overall mortality associated with the combined procedures was approximately 25% (Fig. 1A). At the time of excision, surgical waste in the form of peritoneal wash was collected from all mice. After centrifugation, the cell-free supernatant was analyzed for IL-6, whereas the pelleted cells were analyzed by flow cytometry. From these analyses, we observed that peritoneal neutrophil CD11b intensity and wash IL-6 differed significantly between the mice that lived and mice that died. Neutrophil CD11b was noted to be 40% more elevated in mice that lived (196,065 CD11b MFI versus 116,878 CD11b MFI, P < 0.0001; Fig. 1B). Wash IL-6 levels were decreased by 2.2-fold in mice that lived compared with mice that died (953 versus 430 pg/mL, P < 0.001; Fig. 1C). In addition, we observed no significant differences in neutrophil frequency, total number, or CD54 intensity. With these five determinants, we formulated ROC curves with area under the curve (AUC) [27] as a diagnostic measure outcome (Table). The AUCs for CD11b and IL-6 were 0.8676 and 0.883, respectively, with an AUC of 1.0 signifying a perfect test, 0.8 signifying a good test, and 0.5 signifying a worthless test. Altogether, neutrophil CD11b intensity and peritoneal IL-6 levels were observed to predict death with a good degree of accuracy (AUC > 0.80).
Fig. 1.
Survival and surgical waste analysis after CLP-E. (A) Septic mouse survival was monitored for 10 d. (B) Neutrophils isolated that form the peritoneal wash at the time of excision were analyzed for CD11b expression. (C) Cell-free peritoneal wash was analyzed for IL-6 by enzyme-linked immunosorbent assay.
Table 1.
Accuracy of neutrophil CD11b and IL-6 levels from peritoneal wash for predicting survival.
| Marker | Sensitivity (%) | Specificity (%) | AUC | SEM | 95% confidence interval | P value | |
|---|---|---|---|---|---|---|---|
| CD11ba | 56 | 100 | 0.8676 | 0.057 | 0.7551 | 0.9800 | <0.0001 |
| IL-6b | 50 | 98 | 0.883 | 0.047 | 0.7982 | 0.9768 | <0.0001 |
| % Neutc | 31 | 97 | 0.631 | 0.094 | 0.4451 | 0.8168 | 0.125 |
| Total Neutd | 6 | 97 | 0.608 | 0.083 | 0.4441 | 0.7731 | 0.204 |
| CD54e | 6 | 100 | 0.631 | 0.083 | 0.4682 | 0.7937 | 0.126 |
SEM = standard error of measurement.
neutrophil CD11b expression,
peritoneal IL-6 concentration,
neutrophil frequency in peritoneal wash,
neutrophil number from peritoneal wash,
neutrophil CD54 expression.
3.2. ROC verification using neutrophil CD11b expression to predict survival
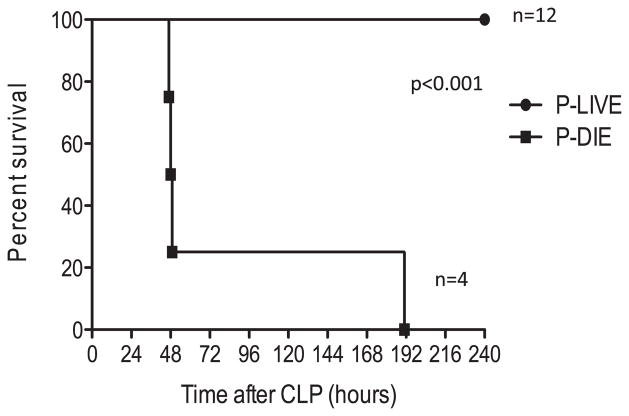
We next tested whether neutrophil CD11b expression could be used to predict survival in an independent cohort of mice. Sixteen mice were subjected to CLP-E, peritoneal washes characterized, and survival monitored for 10 d. Based on a CD11b level of >200,000 from the peritoneal wash, mice were stratified into two groups. Four mice were predicted to die (P-DIE), and 12 mice were P-LIVE. All 12 mice in the P-LIVE group survived compared with the 4 mice in the P-DIE group (P < 0.001, Fig. 2). Thus, we verified that CD11b expression on peritoneal neutrophils accurately predicted survival by analyzing cells obtained during the source control procedure.
Fig. 2.
Survival curve from the test cohort. Mice were subjected to cecal ligation and puncture, and after 20 h, mice underwent excision, peritoneal lavage, and administration of antibiotics. The peritoneal lavage was analyzed for neutrophil CD11b expression, mice categorized as either P-LIVE or P-DIE, and survival monitored for 10 d.
3.3. Characterization of host response of mice P-LIVE
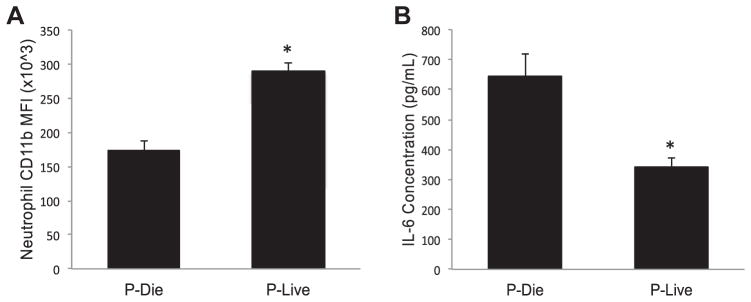
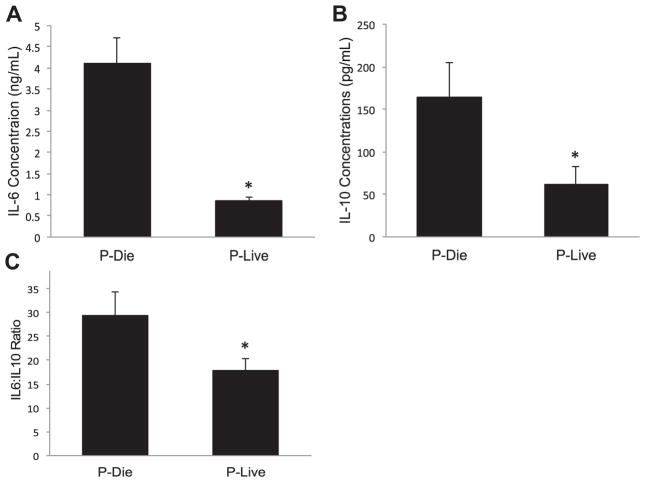
Taking advantage of being able to accurately predict mouse survival, we next investigated how the host response of mice P-LIVE differed from those P-DIE. Twenty-four hours after being subjected to CLP, peritoneal washes were performed with peritoneal neutrophil CD11b and wash IL-6 levels assessed (Fig. 3). Based on these analyses, the mice were stratified into P-LIVE and P-DIE groups. In addition to local immune parameter analysis, systemic levels of IL-6 and IL-10 were determined. Serum IL-6 levels were approximately 5-fold lower in P-LIVE mice compared with those in P-DIE mice (859 versus 4098 pg/mL, P < 0.0001), whereas IL-10 levels were more than 2.5-fold lower in the P-LIVE mice compared with those in the P-DIE mice (61 versus 165 pg/mL, P = 0.03, Fig. 4A and B). Finally, the IL-6:IL-10 ratio was noted to be 42% lower in the P-LIVE mice compared with that in the P-DIE mice (17 versus 29, P = 0.03, Fig. 4C).
Fig. 3.
Mice P-LIVE display less peritoneal inflammation. (A) Peritoneal neutrophil CD11b expression was determined and used to predict survival. (B) Cell-free supernatant was used to determine peritoneal IL-6 levels. Total sample size was 20: 7 in the P-DIE cohort and 13 in the P-LIVE cohort. *P < 0.05 as determined by student t test.
Fig. 4.
Stratification reveals differences in systemic cytokine levels. (A) Serum IL-6 and (B) serum IL-10 levels were determined by enzyme-linked immunosorbent assay as described in the methods. (C) The IL-6:IL-10 ratio was determined to evaluate overall systemic inflammation. Total sample size was 20: 7 in the P-DIE cohort and 13 in the P-LIVE cohort. *P < 0.05 as determined by student t test.
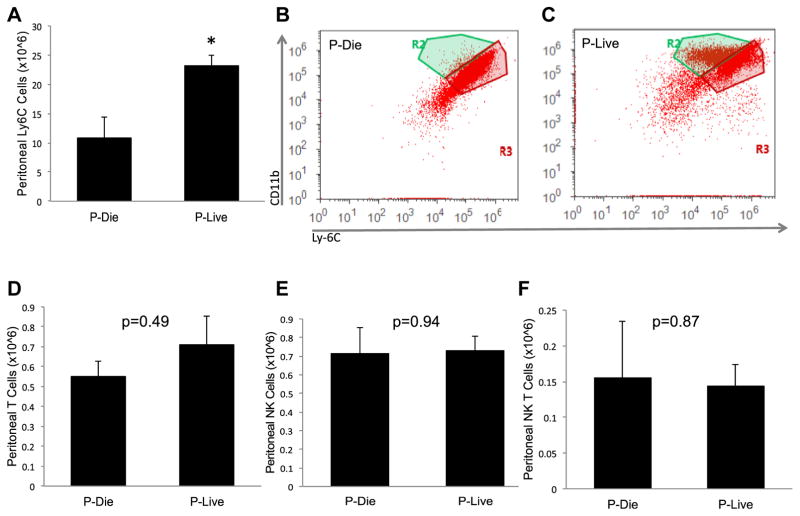
To further characterize the local leukocyte response, we examined myeloid cell, T cell, Natural Killer (NK) cell, and NK T cellpopulations in micethatwere stratified into P-LIVE and P-DIE groups. We observed in the P-LIVE group that inflammatory monocyte numbers increased approximately 2-fold compared with number of cells isolated from the P-DIE group (2.3 × 106 versus 1.1 × 106 cells, P < 0.05, Fig. 5A–C). Notably, there were no changes noted in other leukocyte groups to include T cells, NK cells, or NK T cells between the P-LIVE and P-DIE cohorts (Fig. 5D–F).
Fig. 5.
Peritoneal leukocyte analysis demonstrates increased inflammatory monocytes in mice P-LIVE. (A) Peritoneal inflammatory monocytes from the stratified cohorts were determined. Representative flow cytometric dot plots from (B) mice P-DIE and (C) mice P-LIVE. Peritoneal cell types and numbers were determined for (D) T cells, (E) NK cells, and (F) NK T cells. Total sample size was 20: 7 in the P-DIE cohort and 13 in the P-LIVE cohort. *P <0.05 as determined by student t test. (Color version of figure is available online.)
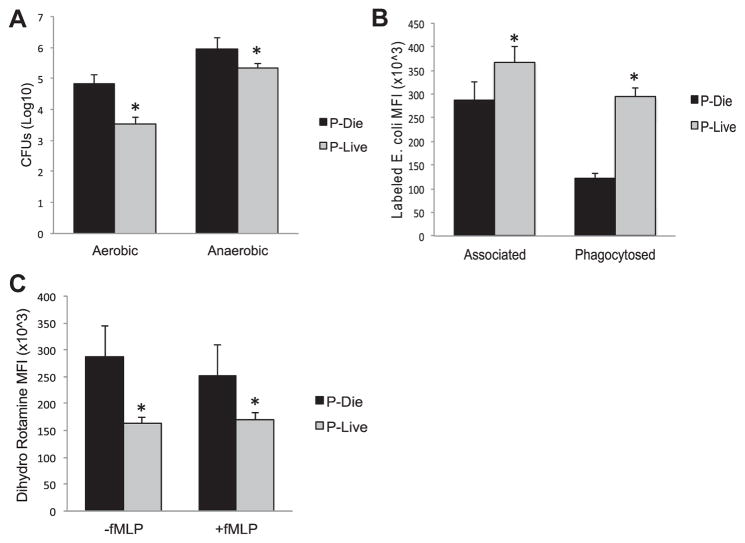
To characterize local bacterial loads, peritoneal wash was plated and incubated under both aerobic and anaerobic conditions. As observed in Figure 6A, there was a 1.3 log decrease in aerobic bacteria (4.8 versus 3.5 CFU, P < 0.05) and a 0.6 log decrease in anaerobic bacteria in the mice P-LIVE (6.0 versus 5.3 CFU, P < 0.05). To elucidate the underpinnings of this disparity, myeloid cells were analyzed for their phagocytic ability and oxidative burst as described in the methods. Peritoneal cells isolated from mice P-LIVE demonstrated a 2.4-fold increase in phagocytosis compared with those from mice P-DIE (295,000 versus 121,000 MFI, P < 0.001, Fig. 6B). In contrast, peritoneal cells from the mice P-LIVE showed an almost 1.5-fold decrease in reactive oxygen species generation compared with those from the mice P-DIE (163,000 versus 287,000 DHR MFI, P < 0.02, Fig. 6C).
Fig. 6.
Divergent bacterial load and antimicrobial function in stratified cohorts. (A) Bacteria from the peritoneal wash were analyzed for aerobic and anaerobic bacterial loads. Neutrophils were analyzed for (B) bacteria association and phagocytosis and (C) oxidative burst by flow cytometry. Total sample size was 201: 7 in the P-DIE cohort and 13 in the P-LIVE cohort. *P < 0.05 as determined by student t test.
4. Discussion
The first goal of these experiments was to develop a model capable of accurately predicting survival after sepsis. We first tailored the mortality severity of the murine CLP-E model (25%) to that observed from human septic patients, 18%–30% [1–3] (Fig. 1). From the necrotic tissue debridement and peritoneal wash, we characterized a number of immunological and bacterial parameters from the resulting surgical waste taken from the infection site. Predominantly, among these variables tested, we observed decreased IL-6 levels and increased expression of neutrophil CD11b in mice P-LIVE compared with mice P-DIE (Fig. 1B and C). These two variables demonstrated high accuracy and specificity on the generation of ROC curves to serve as a prognostic test to predict survival from sepsis (Table). When we repeated CLP-E on a test cohort of mice, the CD11b expression on peritoneal neutrophils accurately predicted all 12 surviving and 4 nonsurviving mice (Fig. 2). Our second goal was to determine the differences of mice P-LIVE and P-DIE. Interestingly, systemic inflammation was increased in mice P-DIE (Fig. 4). In contrast, among a number of leukocytes analyzed, inflammatory monocytes were significantly increased in mice P-LIVE. Furthermore, peritoneal leukocytes collected from mice P-LIVE displayed enhanced phagocytosis capability (Fig. 6B) but displayed diminished oxidative burst when compared with mice P-DIE (Fig. 6C). Finally, both aerobic and anaerobic bacterial counts were decreased in these mice (Fig. 6A). These data suggest that among sepsis survivors, productive inflammation is limited to the site of infection such that bacterial clearance is enhanced and bystander tissue injury from oxidative burst is reduced.
Mice P-LIVE display more activated neutrophils and increased number of macrophages with enhanced phagocytosis ability and a resultant improved bacterial clearance at the surgical site. IL-6 has long been used as a surrogate marker for inflammation during sepsis [28–30]. Furthermore, previous research has demonstrated that this initial proinflammatory phase is vital to overcoming the acute infectious insult [26,31,32]. Unfortunately, an overly exuberant proinflammatory or a hyperinflammatory response often leads to unintended tissue injury and organ dysfunction [33]. In our model, despite improved bacterial clearance, there appeared to be less local and systemic inflammation, as evidenced by IL-6 and the IL-6:IL-10 ratio, as well as diminished oxidative burst at the site of injury. In contrast, the mice that died displayed diminished bacterial clearance and more inflammation systemically and locally or in short, the mice that died developed a hyperinflammatory response rather than an appropriate proinflammatory response. The mechanism or machinery responsible for these differences in the immune response remains unclear, but recent studies have investigated the genetic polymorphisms associated with patients developing sepsis as a likely suspect [34–37]. Further elucidating these mechanisms and tailoring immune modulating therapies to sepsis are at the forefront of current research [37,38].
A recent study by Osuchowski et al. demonstrated tailoring immune modulating therapy based on a predictive model similar to ours. In that study, they developed a predictive model based on serum cytokine levels and then tested the effects of corticosteroid therapy in mice P-LIVE versus those P-DIE [14]. Of interest, the study generated that without using a stratification model; corticosteroid therapy was not efficacious. However, when the authors stratified the mice, they observed that corticosteroid therapy was beneficial in mice P-DIE and had no impact on mice P-LIVE. Our stratification study differs from this in a couple of important aspects. First, we used the CLP-E model with antibiotic treatment for pathogen source control. Source control and antibiotic therapy are hallmarks of sepsis management in humans [6]. By including this into our experiments, we speculate that this increases the clinical relevancy. We further postulate that using surgical waste in addition to serum could improve early diagnosis and management of sepsis when extrapolated to clinical scenarios when surgical waste is available.
Sepsis mortality remains unacceptably high despite advancement in medication, protocols, and diagnostic equipment and tests [39]. Several studies have championed the role of determining immune status and even ex vivo modalities to test the response to immune modulating therapies before intervention [28,40,41]. This study advances our ability to stratify patients and guide any future intervention. Although this study does only directly correlate with abdominal sepsis and scenarios where we have access to the surgical site and lavage, we would argue that these data could be extrapolated to include early diagnosis and stratification of pneumonia, the leading cause of sepsis nationally [34]. Bronchoalveolar lavage, a form of surgical waste, remains the hallmark of diagnosis and would theoretically provide the same opportunity as peritoneal lavage.
This study has some limitations. This article does not include animal vital statistics to better characterize the systemic response to sepsis. The ability to monitor temperature, heart rate, or blood pressure might allow a more timely sepsis diagnosis. As such, we were forced to determine that 20 h was sufficient to elicit intra-abdominal sepsis in our model. Second, we observed that in the mice P-LIVE, bacteria were effectively cleared with more local inflammation but less systemic inflammation. Currently, the underlying mechanism(s) for this variable immune response between mice remain unclear. Third, this model uses surgical waste and accurately predicts death from abdominal sepsis; therefore, it can only represent a fraction of sepsis cases in which there is access to the site of infection. Finally, some sepsis studies using animal models, when applied to human clinical trials, have not resulted in measurable benefit. Recent research suggests that in some cases there is little similarity between rodent and human inflammatory responses to severe injury or sepsis [42] and that caution must be used in directly extrapolating rodent data to the human condition. Indeed, there is an ongoing and robust debate on the usefulness of animal studies in sepsis and trauma research [43]. As such, we recognize that there may be limitation of extrapolating rodent research to human sepsis.
5. Conclusions
An ongoing goal is to use immune modulating strategies to minimize sepsis mortality and morbidity. Two difficulties that still persist in using this approach are determining whether the septic patient will succumb and which potential immune modulator to use. The data presented here suggest that early immune enhancing therapies would likely be efficacious in those predicted not to survive. However, further understanding of the disease and patient differences will be necessary before utilization of immune modulating therapies in the future.
Acknowledgments
Authors’ contributions: J.W.K. and C.C.C. contributed to the study design and drafting the article. J.W.K. and E.F.M. did the acquisition of data. J.W.K., E.F.M., T.C.R., and C.C.C. did the analysis of data and approval of the final version. E.F.M., T.C.R., and C.C.C. did the critical revisions.
Footnotes
Disclosure
The authors reported no proprietary or commercial interest in any product mentioned or concept discussed in the article.
References
- 1.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Yealy DM, Kellum JA, Huang DT, et al. A randomized trial of protocol-based care for early septic shock. N Engl J Med. 2014;370:1683. doi: 10.1056/NEJMoa1401602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peake SL, Delaney A, Bailey M, et al. Goal-directed resuscitation for patients with early septic shock. N Engl J Med. 2014;371:1496. doi: 10.1056/NEJMoa1404380. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Kochanek KD, Murphy SL, Arias E. Mortality in the United States, 2012. NCHS Data Brief. 2014:1. [PubMed] [Google Scholar]
- 5.Dellinger RP, Levy MM, Carlet JM, et al. Surviving Sepsis Campaign: international guidelines for management of severe sepsis and septic shock: 2008. Crit Care Med. 2008;36:296. doi: 10.1097/01.CCM.0000298158.12101.41. [DOI] [PubMed] [Google Scholar]
- 6.Levy MM, Rhodes A, Phillips GS, et al. Surviving sepsis campaign: association between performance metrics and outcomes in a 7. 5-year study. Crit Care Med. 2015;43:3–12. doi: 10.1097/CCM.0000000000000723. [DOI] [PubMed] [Google Scholar]
- 7.Hotchkiss RS, Coopersmith CM, McDunn JE, Ferguson TA. The sepsis seesaw: tilting toward immunosuppression. Nat Med. 2009;15:496. doi: 10.1038/nm0509-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Poll T, Opal SM. Host-pathogen interactions in sepsis. Lancet Infect Dis. 2008;8:32. doi: 10.1016/S1473-3099(07)70265-7. [DOI] [PubMed] [Google Scholar]
- 9.Limaye AP, Kirby KA, Rubenfeld GD, et al. Cytomegalovirus reactivation in critically ill immunocompetent patients. JAMA. 2008;300:413. doi: 10.1001/jama.300.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angus DC. The search for effective therapy for sepsis: back to the drawing board? JAMA. 2011;306:2614. doi: 10.1001/jama.2011.1853. [DOI] [PubMed] [Google Scholar]
- 11.Opal SM, Dellinger RP, Vincent JL, Masur H, Angus DC. The next generation of sepsis clinical trial designs: what is next after the demise of recombinant human activated protein C?*. Crit Care Med. 2014;42:1714. doi: 10.1097/CCM.0000000000000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samraj RS, Zingarelli B, Wong HR. Role of biomarkers in sepsis care. Shock. 2013;40:358. doi: 10.1097/SHK.0b013e3182a66bd6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barati M, Alinejad F, Bahar MA, et al. Comparison of WBC, ESR, CRP and PCT serum levels in septic and non-septic burn cases. Burns. 2008;34:770. doi: 10.1016/j.burns.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Osuchowski MF, Connett J, Welch K, Granger J, Remick DG. Stratification is the key: inflammatory biomarkers accurately direct immunomodulatory therapy in experimental sepsis. Crit Care Med. 2009;37:1567. doi: 10.1097/CCM.0b013e31819df06b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejager L, Pinheiro I, Dejonckheere E, Libert C. Cecal ligation and puncture: the gold standard model for polymicrobial sepsis? Trends Microbiol. 2011;19:198. doi: 10.1016/j.tim.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard WJ, Choudhry M, Schwacha MG, et al. Cecal ligation and puncture. Shock. 2005;24(Suppl 1):52. doi: 10.1097/01.shk.0000191414.94461.7e. [DOI] [PubMed] [Google Scholar]
- 17.Tschop J, Kasten KR, Nogueiras R, et al. The cannabinoid receptor 2 is critical for the host response to sepsis. J Immunol. 2009;183:499. doi: 10.4049/jimmunol.0900203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker CC, Chaudry IH, Gaines HO, Baue AE. Evaluation of factors affecting mortality rate after sepsis in a murine cecal ligation and puncture model. Surgery. 1983;94:331. [PubMed] [Google Scholar]
- 19.Chaudry IH, Hirasawa H, Baue AE. Effect of adenosine triphosphate-glucose administration following sepsis. J Surg Res. 1980;29:348. doi: 10.1016/0022-4804(80)90068-2. [DOI] [PubMed] [Google Scholar]
- 20.Enoh VT, Fairchild CD, Lin CY, Varma TK, Sherwood ER. Differential effect of imipenem treatment on wild-type and NK cell-deficient CD8 knockout mice during acute intra-abdominal injury. Am J Physiol Regul Integr Comp Physiol. 2006;290:R685. doi: 10.1152/ajpregu.00678.2005. [DOI] [PubMed] [Google Scholar]
- 21.Martignoni A, Tschop J, Goetzman HS, et al. CD4-expressing cells are early mediators of the innate immune system during sepsis. Shock. 2008;29:591. doi: 10.1097/SHK.0b013e318157f427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tschop J, Martignoni A, Goetzman HS, et al. Gammadelta T cells mitigate the organ injury and mortality of sepsis. J Leukoc Biol. 2008;83:581. doi: 10.1189/jlb.0707507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris ES, McIntyre TM, Prescott SM, Zimmerman GA. The leukocyte integrins. J Biol Chem. 2000;275:23409. doi: 10.1074/jbc.R000004200. [DOI] [PubMed] [Google Scholar]
- 24.Kuijpers TW, Tool AT, van der Schoot CE, et al. Membrane surface antigen expression on neutrophils: a reappraisal of the use of surface markers for neutrophil activation. Blood. 1991;78:1105. [PubMed] [Google Scholar]
- 25.Li Z. The alphaMbeta2 integrin and its role in neutrophil function. Cell Res. 1999;9:171. doi: 10.1038/sj.cr.7290015. [DOI] [PubMed] [Google Scholar]
- 26.Kasten KR, Tschop J, Goetzman HS, et al. T-cell activation differentially mediates the host response to sepsis. Shock. 2010;34:377. doi: 10.1097/SHK.0b013e3181dc0845. [DOI] [PubMed] [Google Scholar]
- 27.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 28.Gogos CA, Drosou E, Bassaris HP, Skoutelis A. Pro- versus anti-inflammatory cytokine profile in patients with severe sepsis: a marker for prognosis and future therapeutic options. J Infect Dis. 2000;181:176. doi: 10.1086/315214. [DOI] [PubMed] [Google Scholar]
- 29.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 30.Blackwell TS, Christman JW. Sepsis and cytokines: current status. Br J Anaesth. 1996;77:110. doi: 10.1093/bja/77.1.110. [DOI] [PubMed] [Google Scholar]
- 31.Caldwell CC, Martignoni A, Leonis MA, Ondiveeran HK, Fox-Robichaud AE, Waltz SE. Ron receptor tyrosine kinase-dependent hepatic neutrophil recruitment and survival benefit in a murine model of bacterial peritonitis. Crit Care Med. 2008;36:1585. doi: 10.1097/CCM.0b013e318170a8c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiel M, Caldwell CC, Kreth S, et al. Targeted deletion of HIF-1alpha gene in T cells prevents their inhibition in hypoxic inflamed tissues and improves septic mice survival. PloS One. 2007;2:e853. doi: 10.1371/journal.pone.0000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remick DG. Pathophysiology of sepsis. Am J Pathol. 2007;170:1435. doi: 10.2353/ajpath.2007.060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esposito S, Zampiero A, Pugni L, et al. Genetic polymorphisms and sepsis in premature neonates. PloS One. 2014;9:e101248. doi: 10.1371/journal.pone.0101248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christaki E, Giamarellos-Bourboulis EJ. The beginning of personalized medicine in sepsis: small steps to a bright future. Clin Genet. 2014;86:56. doi: 10.1111/cge.12368. [DOI] [PubMed] [Google Scholar]
- 36.Tang BM, Huang SJ, McLean AS. Genome-wide transcription profiling of human sepsis: a systematic review. Crit Care. 2010;14:R237. doi: 10.1186/cc9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin MT, Albertson TE. Genomic polymorphisms in sepsis. Crit Care Med. 2004;32:569. doi: 10.1097/01.CCM.0000110878.49476.42. [DOI] [PubMed] [Google Scholar]
- 38.Boomer JS, To K, Chang KC, et al. Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA. 2011;306:2594. doi: 10.1001/jama.2011.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bone RC, Grodzin CJ, Balk RA. Sepsis: a new hypothesis for pathogenesis of the disease process. Chest. 1997;112:235. doi: 10.1378/chest.112.1.235. [DOI] [PubMed] [Google Scholar]
- 40.Kuethe JW, Mintz-Cole R, Johnson BL, 3rd, Midura EF, Caldwell CC, Schneider BS. Assessing the immune status of critically ill trauma patients by flow cytometry. Nurse Res. 2014;63:426. doi: 10.1097/NNR.0000000000000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedemann NC, Guo RF, Ward PA. The enigma of sepsis. J Clin Invest. 2003;112:460. doi: 10.1172/JCI19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osuchowski MF, Remick DG, Lederer JA, et al. Abandon the mouse research ship? Not just yet! Shock. 2014;41:463. doi: 10.1097/SHK.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]