Abstract
Folate is an essential B vitamin required for the de novo synthesis of purines, thymidylate and methionine. Folate deficiency can lead to mutations and genome instability, and has been shown to exacerbate the genotoxic potential of environmental toxins. We hypothesized that a folic acid (FA) deficient diet would induce genotoxicity in mice as measured by the Pig-a mutant phenotype (CD24−) and micronuclei (MN) in reticulocytes (RET) and red blood cells/normochromatic erythrocytes (RBC/NCE). Male Balb/c mice were fed a FA deficient (0 mg/kg), control (2 mg/kg) or supplemented (6 mg/kg) diet from weaning for 18 wk. Mice fed the deficient diet had 70% lower liver folate (p < 0.001), 1.8 fold higher MN-RET (p < 0.001), and 1.5 fold higher MN-NCE (p < 0.001) than mice fed the control diet. RETCD24− and RBCCD24− frequencies were not different between mice fed the deficient and control diets. Compared to mice fed the FA supplemented diet, mice fed the deficient diet had 73% lower liver folate (p < 0.001), a 2.0 fold increase in MN-RET (p < 0.001), a 1.6 fold increase in MN-NCE (p < 0.001) and 3.8 fold increase in RBCCD24− frequency (p = 0.011). RETCD24− frequency did not differ between mice fed the deficient and supplemented diets. Our data suggest that FA adequacy protects against mutagenesis at the Pig-a locus and MN induction in the red blood cells of mice.
Keywords: Folic acid, Genome stability, Mutation, Micronucleus, Pig-a gene, Genotoxicity
1. Introduction
Folate, an essential B vitamin, is required for the de novo synthesis of purines and thymidylate, and the remethylation of homocysteine to methionine. In conditions of folate deficiency, impaired folate metabolism can have genotoxic consequences. The rate of DNA synthesis is dependent on de novo purine synthesis. Reduced purine synthesis can result in cell cytostasis and cytotoxicity, as well as aberrant DNA synthesis, repair and mutagenesis [1,2]. Reduced thymidylate synthesis can result in uracil incorporation into DNA. The DNA repair machinery attempts to remove the uracil, but in the presence of a high dUTP to dTTP ratio, uracil can be incorporated again. This futile cycle of uracil incorporation and repair can ultimately lead to double DNA strand breaks and genome instability [3]. Finally, impaired folate-dependent methionine synthesis can reduce S-adenosylmethionine (1AdoMet) synthesis. AdoMet is the main cellular methyl donor and perturbations in its availability influence methylation reactions, including that of DNA, RNA and histones, which can alter gene expression and genome stability. The genotoxic consequences of folate insufficiency, either through reduced folate intake or functional folate deficiency due to polymorphisms in folate-related genes, may contribute to cancer susceptibility and other folate-dependent pathologies.
The presence of micronuclei is indicative of chromosome damage that results in partial chromosome deletion, addition or rearrangement (clastogenicity), or whole chromosome gain/loss (aneugenicity). Micronuclei represent extranuclear chromosome fragments or whole chromosomes that lag behind during anaphase that have not been included in the nucleus when the nuclear envelope reforms [4,5]. Higher dietary folate intake has been associated with lower micronucleus (MN) frequency in an Australian population [6]. Human lymphocytes cultured in folate deficient media exhibit DNA double strand breaks [3], reduced DNA repair [3] and MN formation [7–9]. Further, functional folate deficiency due to single nucleotide polymorphisms (SNPs) in folate-related genes has been associated with MN formation in lymphocytes [10].
Although the evidence for a relationship between folate and chromosome stability is strong, there is less evidence for its direct effect on gene mutations in vivo. Most studies have focused on the exacerbating effect of folate deficiency on mutagen-induced mutations. For example, folate deficiency was shown to exacerbate the mutagenic potential of alkylating agents at the hprt locus in human lymphoblastoid cell lines [11], Chinese hamster ovary cells [12] and in mice [13]. There is limited and contrasting evidence for the effect of folate deficiency alone. For example, folate deficiency was shown to cause an increased mutation frequency at the aprt locus in cultured Chinese hamster ovary cells, an effect that was increased further in DNA-repair deficient cells [14]. However, Balb/c mice fed a diet lacking both choline and FA for eight weeks exhibited no increase in Expanded Simple Tandem Repeat (ESTR) mutation frequencies in sperm [15]. Analysis of the cII mutation reporter gene in Big Blue mice fed a FA deficient diet from 3 months of age for 8 months also did not reveal significant increases in mutation frequency in colons [16]. Furthermore, mutant frequency in the Muta™Mouse lacZ reporter gene was not increased in the colons of mice whose mothers were fed a low FA versus high FA diet during pregnancy [17]. Thus, our understanding of the repercussions of folate deficiency or supplementation on gene mutations, especially in vivo or in a range of somatic cell types, is limited.
In an attempt to elucidate the effect of altered folate metabolism on genotoxicity, we recently showed that male Balb/c mice fed a folic acid (FA) deficient diet for 12 weeks followed by a three week wash-out period during which the mice were fed a FA sufficient diet had a higher MN frequency in normochromatic erythrocytes (MN-NCE), but not reticulocytes (RET) [18]. Balb/c mice fed a FA deficient diet for multiple generations had an amplified MN-NCE frequency and a higher MN-RET frequency than mice fed a control diet [18]. These mice also had a higher DNA fragmentation index and a 2-fold higher ESTR mutation frequency in sperm [19]. We found no difference in ESTR mutation frequency in sperm between mice fed a FA supplemented diet and a control diet, which to our knowledge, is the only study to examine the effect of FA supplementation on mutation.
Here we have used a modified study design in which mice were fed FA defined diets from weaning for 18 weeks without a control diet wash-out period to determine the direct genotoxic potential of FA deficiency on Pig-a mutant (CD24−) phenotype and MN frequencies in the red blood cells (RBC) of male Balb/c mice. The Pig-a gene is found on the X chromosome and encodes for the phosphatidylinositol glycan-class A protein that is required for glycosylphosphatidylinosital (GPI) anchored protein expression on the surface of erythrocytes. Pig-a mutations therefore result in GPI anchor deficiency and the frequency of RET and RBC with a Pig-a mutant phenotype (absence of the surface marker) can be assessed by flow cytometry using antibodies for GPI-anchored cell surface markers such as CD24 [20]. Simultaneous sampling for the Pig-a and MN assays allows for the determination of two genotoxic endpoints representing mutagenicity, and clastogenicity or aneugenicity, respectively, and their relationship to folate status and uracil content in DNA. The FA content of the diets used in our study was physiologically relevant: the deficient diet does not result in obvious clinical manifestations such as weight loss [18]; the control diet represents adequate dietary FA intake; and the supplemented diet approximates the combined intake of FA from fortified foods and supplements available on the Canadian market.
2. Materials and methods
2.1. Mice
All mice were cared for in accordance with the Guidelines of the Canadian Council on Animal Care, described in their Guide to the Care and Use of Experimental Animals [21]. The study was approved by the Health Canada Animal Care Committee. Eighteen three week old male weanling Balb/c mice were purchased from Charles River Laboratories (Saint Constant, QC). They were fed one of three FA-defined diets (n = 6/group) based on the AIN-93G formula [22], as published earlier [18]; deficient, adequate/control, or supplemented. The FA deficient diet contained 0 mg FA/kg diet, the FA control diet contained 2 mg FA/kg diet and the FA supplemented diet contained 6 mg FA/kg diet (Dyets, Inc, Bethlehem, PA). Mice were fed the diets for 18 weeks ad libitum.
A single 12-wk-old male C57BL/6 mouse that had been fed a fixed-formula, nonpurified diet (Teklad Diets, Madison WI) was injected intraperitoneally with 80 mg/kg ethylnitrosourea (ENU) (Sigma–Aldrich, Oakville, ON) 20 days before being killed to serve as a positive control for the Pig-a mutation assay.
2.2. Sample collection
Mice were anesthetized under isoflurane and blood was drawn by cardiac puncture. A needle and syringe were pre-coated with 200–300 μl of anticoagulant solution provided by Litron Laboratories (Rochester, NY) prior to drawing the blood. The whole blood-anticoagulant mix was collected in EDTA coated Vacutainer® tubes and shipped overnight on ice to Litron Laboratories for Pig-a and MN mutation analysis (performed in-kind by Litron Laboratories). One Pig-a analysis sample was lost during shipping and was therefore not analysed. Livers were immediately dissected and snap-frozen in liquid nitrogen.
2.3. Liver folate
Liver folate was measured using the Lactobacillus casei microbiological assay [23]. Folate content was normalized to total protein, which was determined by the modified Lowry assay [24].
2.4. Micronucleus detection
Blood was fixed in methanol and cells prepared for flow cytometric analysis using a three-color labeling method, as previously described [25]. The mean number of cells analyzed was 1.08 × 106 cells/sample with a minimum of 0.85 × 106 cells/sample.
2.5. Pig-a assay
Whole blood samples were processed using an immunomagnetic separation step and cells were stained with anti-CD24-PE and anti-CD61-PE for flow cytometric analysis as previously described [26,27]. The mean number of cells analyzed for %RET was 1.58 × 105 cells/sample with a minimum of 1.45 × 105 cells/sample, for RBCCD24− the mean number of cells analyzed was 1.51 × 108 cells/sample with a minimum of 1.39 × 108 cells/sample, and for RETCD24− the mean number of cells analyzed was 4.09 × 106 cells/sample with a minimum of 3.39 × 106 cells/sample.
2.6. Uracil content in DNA
DNA was extracted from liver tissue using a High Pure PCR Template Preparation Kit (Roche Diagnostics, Indianapolis, IN) per manufacturer’s instructions and including RNase treatment. This was followed by a second purification with the High Pure PCR Product Purification Kit (Roche Diagnostics). Uracil content in DNA was analyzed by GC–MS, as previously described using 4 μg DNA per sample [28].
2.7. Statistics
Data are presented as mean ± SEM. The p value cut-off for significance was <0.05 for all analyses. Body weight gain, liver folate and DNA uracil content were analyzed using One-way ANOVA with the Holm-Sidak post-hoc test for all-ways comparison. Body weight over time was analyzed using a Two-way ANOVA with Holm-Sidak post-hoc test for all-ways comparison. Correlations were determined using the Pearson Product Moment correlation test. These analyses were performed in SigmaPlot for Windows, version 11.0 (Systat Software, Inc.).
For %RET, MN-RET, MN-NCE, RETCD24−, and RBCCD24−, generalized estimating equations (GEEs) [29,30] assuming a Poisson distribution for the error was applied using the geepack library [31–33] in R [34]. GEEs are semi-nonparametric, and provide an alternative to generalized linear models. GEEs only require specification of the first two moments, the mean and the variance. In this analysis, a log link function was used and the results were back transformed to the original scale. The delta method was used to estimate the back transformed standard errors. The Grubbs test on the residuals was used to identify potential outlier values.
3. Results
3.1. Body weight
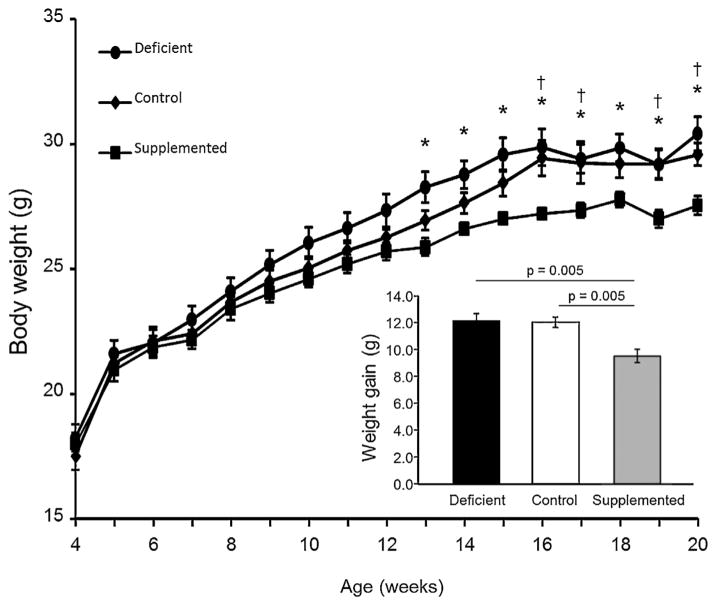
No differences were observed in body weight gain between the mice fed the control and FA deficient diets from weaning until the end of study (Fig. 1). However, by the end of study, mice fed the FA supplemented diet gained approximately 2 g less than mice fed the FA deficient and control diets over the study period (p = 0.005) (Fig. 1, inset). The difference in weight between the groups was significant from 13 to 16 wk of age (p < 0.05).
Fig. 1.
Body weight and weight gain of male mice fed folic acid defined diets. Circles, deficient diet; diamonds, control diet; squares, supplemented diet. Body weight change over time was compared among the diet groups by Two-way ANOVA, Holm-Sidak post-hoc analysis all-ways comparisons. An asterisk (*) indicates a significant difference between the deficient and supplemented diets at that time point (p ≤ 0.002) and a dagger (†) indicates a significant difference between the control and supplemented diets (p ≤ 0.04). Inset, total body weight gain. Closed bars, deficient diet; open bars, control diet; grey bars, supplemented diet. Weight gain was compared among the diet groups by One-way ANOVA, Holm-Sidak post-hoc analysis for all-ways comparison; p values are indicated for comparisons that were significantly different (p < 0.05). Data are presented as mean ± SEM. n = 6/diet group.
3.2. Liver folate and DNA uracil content
Liver folate was 70 and 73% lower in the mice fed the FA deficient diet compared to those fed the control (p < 0.001) and supplemented (p < 0.001) diets, respectively (Table 1). The 12% lower liver folate in the mice fed the control diet compared to mice fed the supplemented diet was not significant (p = 0.17). We did not observe a significant difference in liver DNA uracil content among the dietary groups. Liver folate and liver DNA uracil content did not correlate significantly.
Table 1.
Liver folate and uracil content in liver DNA in mice fed folic acid defined diets.
| Diet8 | n | Folate (fmol/μg protein) | Liver uracil (pg/μg DNA) |
|---|---|---|---|
| Deficient | 6 | 20.2 ± 2.2a | 0.312 ± 0.023 |
| Control | 6 | 66.7 ± 5.9b | 0.371 ± 0.032 |
| Supplemented | 6 | 75.2 ± 3.5b | 0.351 ± 0.039 |
| ANOVA p value* | < 0.001 | 0.48 |
One-way ANOVA and Holm-Sidak test for all-ways comparison. Values in a column that do not share a letter are significantly different, p < 0.05.
3.3. Pig-a mutant frequency
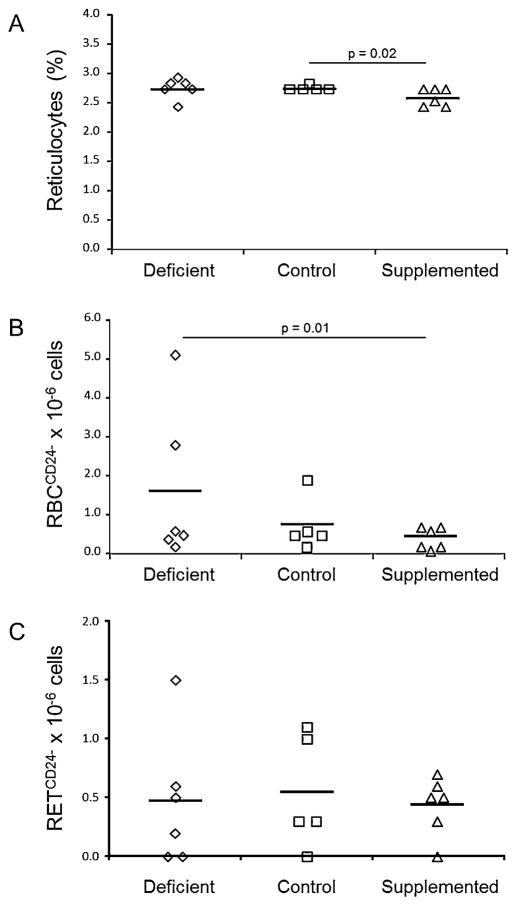
The %RET, as measured by the In Vivo MutaFlow® Pig-a mutation assay, did not differ between mice fed the deficient and control diets; %RET was 6% lower in mice fed the supplemented diet compared to mice fed the control diet (p = 0.015)(Fig. 2, panel A). Mice fed the deficient diet had 2.2 fold higher RBCCD24− frequency compared to mice fed the control diet, although this was not statistically significant (Fig. 2, panel B; p = 0.23). The RBCCD24− frequency was 3.8 fold higher in mice fed the deficient diet compared to mice fed the supplemented diet (p = 0.011). The RBCCD24− frequency was 2.2 fold higher in mice fed the deficient compared to those fed the supplemented diet (p = 0.17) with the removal of one potential outlier mouse (Grubbs outlier test, p = 0.05) from the deficient diet group with a high RBCCD24− frequency. RETCD24− frequency did not differ among the diet groups (Fig. 2) %RET, RBCCD24− and RETCD24− did not correlate significantly with either liver folate or DNA uracil content. The positive control, a single mouse injected with 80 mg/kg ENU, had 3.2% RET, 46 × 10−6 RBCCD24−, and 159.1 × 10−6 RETCD24−, representing a 1.2, 62 and 295 fold increase relative to control diet fed mice.
Fig. 2.
Percent reticulocytes and blood cell Pig-a mutant frequency in male mice fed folic acid defined diets. Panel A, % reticulocytes, Panel B, Pig-a mutant RBC frequency (RBCCD24−), and Panel C, Pig-a mutant reticulocyte frequency (RETCD24−). Diamonds, squares and triangles represent mice fed the folic acid deficient (n = 6), control (n = 5) or supplemented (n = 6) diets, respectively. The line represents the mean for each group. Differences among the diet groups were assessed using generalized estimating equations assuming a Poisson distribution for the error. P values are indicated for comparisons that were significantly different (p < 0.05).
RBCCD24− and MN-NCE frequencies correlated (Correlation coefficient: 0.55, p = 0.024); however, RETCD24− and MN-RET frequencies did not correlate. %RET measured by the In Vivo MicroFlow® and In Vivo MutaFlow® assays correlated (Correlation coefficient: 0.70, p = 0.0016).
3.4. Micronucleus frequency
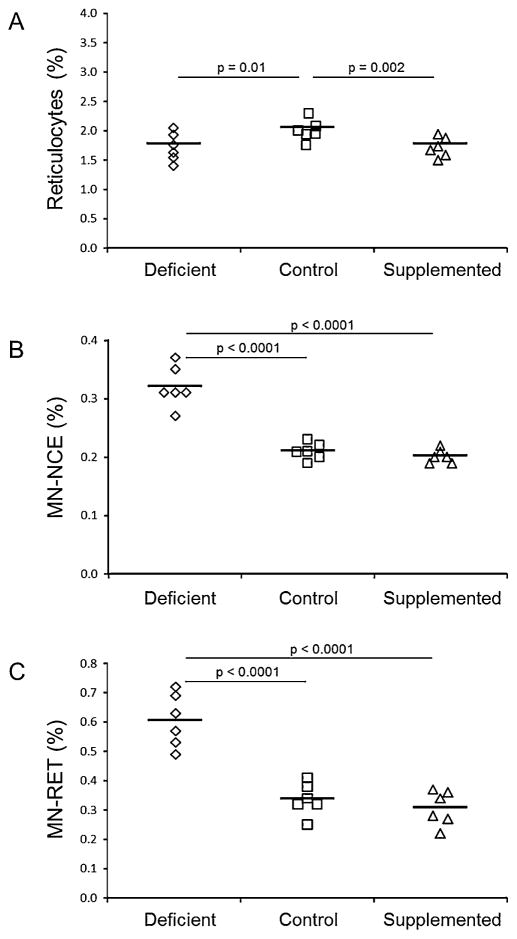
The %RET, as measured with the In Vivo MicroFlow® Micronucleus assay, was 14% lower in mice fed either the deficient (p = 0.0099) or supplemented (p = 0.0019) diets compared to mice fed the control diet (Fig. 3, panel A). %RET was not different between the deficient and supplemented mice. The frequency of MN-NCE was 1.5 and 1.6 fold higher in mice fed the deficient diet compared to mice fed either the control (p < 0.0001) or supplemented (p < 0.0001) diets, respectively (Fig. 3, panel B). Similarly, the frequency of MN-RET was 1.8 fold higher in mice fed the deficient diet compared to mice fed the control diet (p < 0.0001) and 2.0 fold higher than those fed the supplemented (p < 0.0001) diet (Fig. 3, panel C). There were no differences in MN-NCE and MN-RET between the mice fed the control and supplemented diets. MN-RET (Correlation coefficient: −0.83, p < 0.0001) and MN-NCE (Correlation coefficient: −0.88, p < 0.0001) were negatively correlated with liver folate concentration, but not with liver DNA uracil content.
Fig. 3.
Percent reticulocytes and micronucleus frequency in male mice fed folic acid defined diets. Panel A, % reticulocytes, Panel B, normochromatic erythrocyte micronucleus frequency (MN-NCE), and Panel C, reticulocyte micronucleus frequency (MN-RET). Diamonds, squares and triangles represent mice fed the folic acid deficient (n = 6), control (n = 6) or supplemented (n = 6) diets, respectively. The line represents the mean for each group. Differences among the diet groups were assessed using generalized estimating equations assuming a Poisson distribution for the error. P values are indicated for comparisons that were significantly different (p < 0.05).
4. Discussion
Folate deficiency is associated with increased risk for neural tube defects (NTDs) and many cancers. Its role in nucleotide synthesis and cellular methylation potential, and thereby its genotoxic potential, provide plausible mechanisms by which folate modifies disease risk. Our study design allowed for the simultaneous measurement of two key genotoxic endpoints, gene mutation and chromosomal aberrations (clastogenicity or aneugenicity). Our data suggest that, at minimum, an adequate FA intake reduces the modestly mutagenic potential of folate deficiency, as demonstrated by a significant reduction in RBCCD24− frequency in mice fed the FA supplemented diet compared to mice fed a deficient diet. We also clearly demonstrate that the induction of somatic cell genome instability by folate deficiency in mice can be prevented by adequate FA intake, as demonstrated by a >50% higher MN-NCE and an 80% higher MN-RET frequency in mice fed the FA deficient diet compared to mice fed a control diet. Compared to the control diet, FA supplementation did not provide additional reduction in RBCCD24− or MN frequency, but neither was it detrimental.
Because of its important role in the de novo synthesis of nucleotides, altered folate metabolism has been hypothesized to cause mutations. We show that mice with at least adequate FA intake were protected from Pig-a mutations, as indicated by a lower RBCCD24− frequency compared to mice fed the deficient diet. However, the effect of folate deficiency on RBCCD24− frequency was modest, being lower than many known chemical mutagens. For example, our positive control mouse injected with the strong mutagen ENU had 46 × 10−6 RBCCD24− compared to 1.6 × 10−6 RBCCD24− in folate deficient mice. Control diet fed mice had 0.7 × 10−6 RBCCD24−. Similarly, mice exposed to 25 mg benzo(a) pyrene/kg/day for 28 days had ~25 × 10−6 RBCCD24− [35]. Eight weak mutagens were found to induce what were described as modest increases in Pig-a mutant cell frequency in Sprague Dawley rats in a 28 day sub-acute exposure study [27], a design that is comparable to our dietary intervention. For example, rats exposed to cyclophosphamide for 28 d had a 4.6 fold higher RBCCD24− frequency compared to vehicle [27]. In comparison, a FA deficient diet resulted in, at best, a ~2–3 fold higher RBCCD24− frequency relative to a FA control or supplemented diet.
We did not detect an effect of folate deficiency on RETCD24− frequency, despite a significant increase in RBCCD24− frequency. This observation could be explained by the limits of the Pig-a MutaFlow® assay with respect to the relatively low number of RETs that are evaluated per sample compared to RBCs; the MutaFlow® assay analyzes approximately 40 fold more RBCs than RETs. As such, the effect size of folate deficiency (~2–3 fold) on RBCCD24− frequency is likely near the detection limit of the RET-based analysis given sample sizes of 5–6 per group. Thus, the larger number of RBCs analyzed coupled with their longer half-life allows for the detection of modest increases in Pig-a mutant frequency.
Folate deficiency can result in uracil incorporation into DNA, as well as genome hypomethylation. Both of these circumstances can lead to DNA double strand breaks and chromosome instability. We found that the FA deficient diet induced a 1.8–2.5 fold and 1.5–1.6 fold higher MN-RET and MN-NCE frequencies, respectively, compared to mice fed the control or supplemented diets. These frequencies are of a similar order of magnitude as exposure to sub-acute 28 d exposures to known environmental clastogens. For example, Muta™Mouse exposed to 25 mg benzo(a) pyrene/kg/day had ~3 fold higher [35] and Sprague Dawley rats exposed to 62.5 or 125 mg hydroxyurea/kg/day had a ~2.5 or 7 fold higher MN-RET frequency [27].
Although we demonstrated that MN frequency correlated with liver folate concentration, it was not correlated with liver DNA uracil content. These results can be interpreted in two ways: MN frequency was not dependent on DNA uracil content leaving changes in genome methylation as a potential causal mechanism for genome instability; or, liver DNA uracil content is not representative of DNA uracil content in other tissues, perhaps due to comparatively lower cellular turnover. While this study cannot exclude a role for genome methylation in MN formation, we favor the latter interpretation for a number of reasons. The first is that in our previous analysis we found that MN-RET and MN-NCE frequency correlated with DNA uracil content in bone marrow cells in a mouse model of long term, trans-generational folate deficiency [18]. Second, we showed that while bone marrow DNA uracil content was correlated with MN frequency, global DNA methylation was not [18]. Third, we and others have shown that the rate and degree of folate depletion due to low folate intake varies by tissue and that tissues characterised by rapid cell proliferation, such as the colon, deplete faster and to a lower relative concentration than liver [36–38]. Finally, while MN-RET and MN-NCE frequencies correlated with liver folate, liver folate did not correlate with liver DNA uracil content. Therefore, the association of liver DNA uracil content with MN frequency in RBC may not be expected to correlate. Ideally, we would have measured uracil in the DNA of bone marrow cells to specifically examine this relationship.
In our previous study, we observed that MN-NCE, but not MN-RET frequency, was significantly higher in mice fed a FA deficient diet compared to those fed a control diet followed by a 3 week wash-out period on a control diet [18]. In contrast, we report here that both MN-RET and MN-NCE frequency were higher in mice fed a FA deficient diet in the absence of a wash-out period compared to mice fed the control diet. These differing observations can be explained by the short lifespan of RET compared to RBC (days vs. weeks). RET exposed to the deficient or control diet would have matured to RBC leaving the RET pool during the 3 week control diet wash-out period. In addition, MN-RET would not be replenished during the 3 week period because bone marrow precursors would no longer be exposed to the dietary intervention. MN formation in progenitor bone marrow cells can induce senescence [39] or apoptosis [40], which would lead to their eventual elimination from the circulating RET, and later RBC, pool. The data indicate that the MN-RET or MN-NCE frequency is dependent on folate status, as has been observed previously [41], but that the effect is transient and can be corrected with adequate or supplemental FA intake [42].
Finally, we observed a modest effect of dietary FA on the %RET in circulation, an index of bone marrow toxicity. As might be expected because of its negative effect on genome stability and nucleotide synthesis, folate deficiency was associated with a small but significant decrease in the %RET compared to folate adequacy, as measured by the In Vivo MicroFlow® assay, but not the In Vivo MutaFlow® assay. These discrepancies may result from differences in the RET assays; the MicroFlow® assay measures cell surface expression of CD71, whereas the MutaFlow® assay measures RET based on RNA content. Perhaps unexpectedly, FA supplementation also resulted in 5–14% lower %RET compared to control mice, in both assays. The presence of unmetabolized FA in circulation, as can occur in cases of high FA intake, has been hypothesized to negatively affect folate metabolism [43–45]. The enzyme dihydrofolate reductase (DHFR) sequentially reduces FA to DHF and tetrahydrofolate (THF); DHF is an inhibitor of methyleneTHF reductase (MTHFR) and thymidylate synthase (TYMS) [46,47]. As such, its accumulation could reduce the synthesis of 5-methylTHF by MTHFR, which is required for homocysteine remethylation and maintenance of cellular methylation capacity, or decrease the capacity for TYMS-dependent de novo thymidylate synthesis. Therefore, the accumulation of DHF could theoretically lead to chromosome breaks and genome instability through a “functional” folate deficiency. However, we did not observe an increase in MN frequency in mice fed the FA supplemented diet in this or our previous study [18]. The FA supplemented mice did, however, demonstrate lower weight gain compared to mice fed the FA deficient or control diet, a finding that we have not observed previously [18]. Whether the lower %RET in FA supplemented mice is reproducible, or is due to their lower weight gain or a toxic effect on the bone marrow will need to be further explored.
Our study has a number of strengths and limitations. Strengths of the study include the use of the In Vivo MicroFlow® and In Vivo MutaFlow® assays to simultaneously determine RET and RBC MN and Pig-a mutant frequencies using a very small sample volume. These sensitive flow cytometry-based analytical methods allow for the detection of low numbers of MN and Pig-a mutant frequencies, as well as the identification of small differences between experimental groups due to the large number of blood cells examined. A limitation of the study includes the relatively small sample sizes, which could have limited our power to detect a modest effect size of folate deficiency, as may have been the case for RETCD24−. Also, the assessment of folate status and DNA uracil content in a tissue that was different than in which the Pig-a mutant and MN frequencies were measured likely limited our interpretation of the relationship between these factors.
In conclusion, we have demonstrated that folate deficiency is genotoxic; it is modestly mutagenic at the Pig-a locus, and induces blood cell MN at frequencies comparable to known chemical mutagens. Conversely, adequate FA intake protects somatic cells from these genotoxic effects, while FA supplementation provides little additional benefit. These findings provide mechanistic insights into the association between altered folate metabolism and DNA mutagenesis. Whether these associations can be extended to explain folate-dependent processes that underlie folate-dependent NTDs or cancers will require further investigation.
Acknowledgments
We would like to thank Stephen Dertinger for advice on methodology and data interpretation, Dorothea Torous for technical assistance, Cina Aghazadeh Sanaei for mouse colony maintenance, and Francesco Marchetti and Jayadev Raju for review of the manuscript. Litron Laboratories performed the micronucleus and Pig-a mutant frequency analyses in-kind with support from the National Institute of Environmental Health Sciences (grant R44ES018017). This work was funded by the Food and Consumer Safety Action Plan at Health Canada. The funding source had no involvement in the study design, the collection, analysis and interpretation of data, or in the writing of the report. Health Canada approved the manuscript for publication.
Abbreviations
- AdoMet
S-adenosylmethionine
- DHF
dihydrofolate
- DHFR
DHF reductase
- ENU
ethylnitrosourea
- ESTR
Expanded Simple Tandem Repeat
- FA
folic acid
- MTHFR
5,10-methylenetetrahydrofolate reductase
- MN
micronucleus
- CD24−
Pig-a mutant phenotype
- NCE
normochromatic erythrocyte
- RBC
red blood cell
- RET
reticulocyte
- THF
tetrahydrofolate
- TYMS
thymidylate synthase
Footnotes
Author disclosure
The authors declare no conflicts of interest.
References
- 1.Kondo M, Yamaoka T, Honda S, Miwa Y, Katashima R, Moritani M, Yoshimoto K, Hayashi Y, Itakura M. The rate of cell growth is regulated by purine biosynthesis via ATP production and G(1) to S phase transition. J Biochem. 2000;128:57–64. doi: 10.1093/oxfordjournals.jbchem.a022730. [DOI] [PubMed] [Google Scholar]
- 2.Collins AR, Black DT, Waldren CA. Aberrant DNA repair and enhanced mutagenesis following mutagen treatment of Chinese hamster Ade-C cells in a state of purine deprivation. Mutat Res. 1988;193:145–155. doi: 10.1016/0167-8817(88)90045-4. [DOI] [PubMed] [Google Scholar]
- 3.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. Faseb J. 1998;12:1491–1497. [PubMed] [Google Scholar]
- 4.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 5.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenech M, Baghurst P, Luderer W, Turner J, Record S, Ceppi M, Bonassi S. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability?results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis. 2005;26:991–999. doi: 10.1093/carcin/bgi042. [DOI] [PubMed] [Google Scholar]
- 7.Lu L, Ni J, Zhou T, Xu W, Fenech M, Wang X. Choline and/or folic acid deficiency is associated with genomic damage and cell death in human lymphocytes in vitro. Nutr Cancer. 2015;64:481–487. doi: 10.1080/01635581.2012.660671. [DOI] [PubMed] [Google Scholar]
- 8.Fenech M, Crott JW. Micronuclei nucleoplasmic bridges and nuclear buds induced in folic acid deficient human lymphocytes-evidence for breakage-fusion-bridge cycles in the cytokinesis-block micronucleus assay. Mutat Res. 2002;504:131–136. doi: 10.1016/s0027-5107(02)00086-6. [DOI] [PubMed] [Google Scholar]
- 9.Leopardi P, Marcon F, Caiola S, Cafolla A, Siniscalchi E, Zijno A, Crebelli R. Effects of folic acid deficiency and MTHFR C677T polymorphism on spontaneous and radiation-induced micronuclei in human lymphocytes. Mutagenesis. 2006;21:327–333. doi: 10.1093/mutage/gel031. [DOI] [PubMed] [Google Scholar]
- 10.Dhillon V, Thomas P, Fenech M. Effect of common polymorphisms in folate uptake and metabolism genes on frequency of micronucleated lymphocytes in a South Australian cohort. Mutat Res. 2009;665:1–6. doi: 10.1016/j.mrfmmm.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Branda RF, O’Neill JP, Brooks EM, Trombley LM, Nicklas JA. The effect of folate deficiency on the cytotoxic and mutagenic responses to ethyl methanesulfonate in human lymphoblastoid cell lines that differ in p53 status. Mutat Res. 2001;473:51–71. doi: 10.1016/s0027-5107(00)00138-x. [DOI] [PubMed] [Google Scholar]
- 12.Branda RF, Lafayette AR, O’Neill JP, Nicklas JA. The effect of folate deficiency on the hprt mutational spectrum in Chinese hamster ovary cells treated with monofunctional alkylating agents. Mutat Res. 1999;427:79–87. doi: 10.1016/s0027-5107(99)00095-0. [DOI] [PubMed] [Google Scholar]
- 13.Branda RF, O’Neill JP, Brooks EM, Powden C, Naud SJ, Nicklas JA. The effect of dietary folic acid deficiency on the cytotoxic and mutagenic responses to methyl methanesulfonate in wild-type and in 3-methyladenine DNA glycosylase-deficient Aag null mice. Mutat Res. 2007;615:12–17. doi: 10.1016/j.mrfmmm.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 14.James SJ, Basnakian AG, Miller BJ. In vitro folate deficiency induces deoxynucleotide pool imbalance, apoptosis, and mutagenesis in Chinese hamster ovary cells. Cancer Res. 1994;54:5075–5080. [PubMed] [Google Scholar]
- 15.Voutounou M, Glen CD, Dubrova YE. The effects of methyl-donor deficiency on mutation induction and transgenerational instability in mice. Mutat Res. 2012;734:1–4. doi: 10.1016/j.mrfmmm.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Linhart HG, Troen A, Bell GW, Cantu E, Chao WH, Moran E, Steine E, He T, Jaenisch R. Folate deficiency induces genomic uracil misincorporation and hypomethylation but does not increase DNA point mutations. Gastroenterology. 2009;136:227–235.e3. e223. doi: 10.1053/j.gastro.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trentin GA, Moody J, Heddle JA. Effect of maternal folate levels on somatic mutation frequency in the developing colon. Mutat Res. 1998;405:81–87. doi: 10.1016/s0027-5107(98)00147-x. [DOI] [PubMed] [Google Scholar]
- 18.Swayne BG, Behan NA, Williams A, Stover PJ, Yauk CL, MacFarlane AJ. Supplemental dietary folic acid has no effect on chromosome damage in erythrocyte progenitor cells of mice. J Nutr. 2012;142:813–817. doi: 10.3945/jn.112.157750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swayne BG, Kawata A, Behan NA, Williams A, Wade MG, MacFarlane AJ, Yauk CL. Investigating the effects of dietary folic acid on sperm count, DNA damage and mutation in Balb/c mice. Mutat Res. 2012;737:1–7. doi: 10.1016/j.mrfmmm.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Kimoto T, Suzuki K, Kobayashi XM, Dobrovolsky VN, Heflich RH, Miura D, Kasahara Y. Manifestation of Pig-a mutant bone marrow erythroids and peripheral blood erythrocytes in mice treated with N-ethyl-N-nitrosourea: direct sequencing of Pig-a cDNA from bone marrow cells negative for GPI-anchored protein expression. Mutat Res. 2011;723:36–42. doi: 10.1016/j.mrgentox.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Olfert ED, Cross BM, McWilliam AA, editors. Guide to the Care and Use of Experimental Animals. Canadian Council on Animal Care; Ottawa: 1993. [Google Scholar]
- 22.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 23.Horne DW, Patterson D. Lactobacillus casei microbiological assay of folic acid derivatives in 96-well microtiter plates. Clin Chem. 1988;34:2357–2359. [PubMed] [Google Scholar]
- 24.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 25.Dertinger SD, Camphausen K, Macgregor JT, Bishop ME, Torous DK, Avlasevich S, Cairns S, Tometsko CR, Menard C, Muanza T, Chen Y, Miller RK, Cederbrant K, Sandelin K, Ponten I, Bolcsfoldi G. Three-color labeling method for flow cytometric measurement of cytogenetic damage in rodent and human blood. Environ Mol Mutagen. 2004;44:427–435. doi: 10.1002/em.20075. [DOI] [PubMed] [Google Scholar]
- 26.Dertinger SD, Bryce SM, Phonethepswath S, Avlasevich SL. When pigs fly: immunomagnetic separation facilitates rapid determination of Pig-a mutant frequency by flow cytometric analysis. Mutat Res. 2011;721:163–170. doi: 10.1016/j.mrgentox.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dertinger SD, Phonethepswath S, Avlasevich SL, Torous DK, Mereness J, Bryce SM, Bemis JC, Bell S, Weller P, Macgregor JT. Efficient monitoring of in vivo Pig-a gene mutation and chromosomal damage: summary of 7 published studies and results from 11 new reference compounds. Toxicol Sci. 2012;130:328–348. doi: 10.1093/toxsci/kfs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, Stover PJ. Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem. 2008;283:25846–25853. doi: 10.1074/jbc.M802671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 30.Prentice RL, Zhao LP. Estimating equations for parameters in means and covariances of multivariate discrete and continuous responses. Biometrics. 1991;47:825–839. [PubMed] [Google Scholar]
- 31.Højsgaard S, Halekoh U, Yan J. The R Package geepack for Generalized Estimating Equations. J Stat Software. 2005;15:1–11. [Google Scholar]
- 32.Yan J, Fine J. Estimating equations for association structures. Stat Med. 2004;23:859–874. doi: 10.1002/sim.1650. discussion 875–857, 879–880. [DOI] [PubMed] [Google Scholar]
- 33.Yan J. Geepack: Yet another package for Generalized estimating equations. R-News. 2002;2:12–14. [Google Scholar]
- 34.Development Core Team. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. [Google Scholar]
- 35.Lemieux CL, Douglas GR, Gingerich J, Phonethepswath S, Torous DK, Dertinger SD, Phillips DH, Arlt VM, White PA. Simultaneous measurement of benzo[a]pyrene-induced Pig-a and lacZ mutations, micronuclei and DNA adducts in Muta Mouse. Environ Mol Mutagen. 2011;52:756–765. doi: 10.1002/em.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacFarlane AJ, Anderson DD, Flodby P, Perry CA, Allen RH, Stabler SP, Stover PJ. Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J Biol Chem. 2012;286:44015–44022. doi: 10.1074/jbc.M111.307629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacFarlane AJ, Perry CA, McEntee MF, Lin DM, Stover PJ. Mthfd1 is a modifier of chemically induced intestinal carcinogenesis. Carcinogenesis. 2011;32:427–433. doi: 10.1093/carcin/bgq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacFarlane AJ, Perry CA, McEntee MF, Lin DM, Stover PJ. Shmt1 heterozygosity impairs folate-dependent thymidylate synthesis capacity and modifies risk of Apcmin-mediated intestinal cancer risk. Cancer Res. 2011;71:2098–2107. doi: 10.1158/0008-5472.CAN-10-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Nguyen T, Puebla-Osorio N, Pang H, Dujka ME, Zhu C. DNA damage-induced cellular senescence is sufficient to suppress tumorigenesis: a mouse model. J Exp Med. 2007;204:1453–1461. doi: 10.1084/jem.20062453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attardi LD. The role of p53-mediated apoptosis as a crucial anti-tumor response to genomic instability: lessons from mouse models. Mutat Res. 2005;569:145–157. doi: 10.1016/j.mrfmmm.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 41.MacGregor JT, Wehr CM, Hiatt RA, Peters B, Tucker JD, Langlois RG, Jacob RA, Jensen RH, Yager JW, Shigenaga MK, Frei B, Eynon BP, Ames BN. ‘Spontaneous’ genetic damage in man: evaluation of interindividual variability, relationship among markers of damage, and influence of nutritional status. Mutat Res. 1997;377:125–135. doi: 10.1016/s0027-5107(97)00070-5. [DOI] [PubMed] [Google Scholar]
- 42.Everson RB, Wehr CM, Erexson GL, MacGregor JT. Association of marginal folate depletion with increased human chromosomal damage in vivo: demonstration by analysis of micronucleated erythrocytes. J Natl Cancer Inst. 1988;80:525–529. doi: 10.1093/jnci/80.7.525. [DOI] [PubMed] [Google Scholar]
- 43.Sauer J, Mason JB, Choi SW. Too much folate: a risk factor for cancer and cardiovascular disease? Curr Opin Clin Nutr Metab Care. 2009;12:30–36. doi: 10.1097/MCO.0b013e32831cec62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelly P, McPartlin J, Goggins M, Weir DG, Scott JM. Unmetabolized folic acid in serum: acute studies in subjects consuming fortified food and supplements. Am J Clin Nutr. 1997;65:1790–1795. doi: 10.1093/ajcn/65.6.1790. [DOI] [PubMed] [Google Scholar]
- 45.Bailey RL, Mills JL, Yetley EA, Gahche JJ, Pfeiffer CM, Dwyer JT, Dodd KW, Sempos CT, Betz JM, Picciano MF. Unmetabolized serum folic acid and its relation to folic acid intake from diet and supplements in a nationally representative sample of adults aged > or =60 y in the United States. Am J Clin Nutr. 2010;92:383–389. doi: 10.3945/ajcn.2010.29499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matthews RG, Daubner SC. Modulation of methylenetetrahydrofolate reductase activity by S-adenosylmethionine and by dihydrofolate and its polyglutamate analogues. Adv Enzyme Regul. 1982;20:123–131. doi: 10.1016/0065-2571(82)90012-7. [DOI] [PubMed] [Google Scholar]
- 47.Baram J, Chabner BA, Drake JC, Fitzhugh AL, Sholar PW, Allegra CJ. Identification and biochemical properties of 10-formyldihydrofolate, a novel folate found in methotrexate-treated cells. J Biol Chem. 1988;263:7105–7111. [PubMed] [Google Scholar]