Abstract
Fluorescence in situ hybridization (FISH) enables the detection of specific nucleic acid sequences within single cells. For example, RNA FISH provides information on both the expression level and localization of RNA transcripts and, when combined with detection of associated proteins and chromatin modifications, can lend essential insights into long noncoding RNA (lncRNA) function. Epigenetic effects have been postulated for many lncRNAs, but shown for only a few. Advances in in situ techniques and microscopy, however, now allow for visualization of lncRNAs that are expressed at very low levels or are not very stable. FISH-based detections of RNA and DNA coupled with immunological staining of proteins/histone modifications offer the possibility to connect lncRNAs to epigenetic effects. Here, we describe an integrated set of protocols to detect, individually or in combination, specific RNAs, DNAs, proteins, and histone modifications in single cells at a high level of sensitivity using conventional fluorescence microscopy.
Keywords: Immunofluorescence, Fluorescence in situ hybridization, RNA FISH, DNA FISH, Long noncoding RNAs, Epigenetic, Chromatin, Histone modifications
1 Introduction
LncRNAs are increasingly invoked as factors that establish epigenetic profiles [1, 2]. Some lncRNAs are hypothesized to regulate gene expression at specific loci [3]. Other lncRNAs are postulated to regulate gene expression chromosome-wide, as in the case of the inactive X-chromosome [1, 4]. Refinements of fluorescence in situ hybridization (FISH) techniques have resulted in the ability to visualize lncRNAs at their sites of synthesis on the chromosome or site of function in the genome [5]. Combined with immunofluorescence (IF) detection of epigenetic factors and histone modifications, FISH techniques can permit the formulation of specific hypotheses of lncRNA function. For example, colocalization of epigenetic modifiers or histone modifications with lncRNA transcripts can suggest the recruitment of epigenetic factors to discrete loci in the genome.
Here, we describe detection via FISH of lncRNAs both with probes that do not distinguish the DNA strand of synthesis and with strand-specific probes. We additionally describe methods for IF-based detection of proteins and histone modifications and DNA FISH procedure to mark specific DNA loci of interest in cultured and embryonic cells. These protocols can be used individually or in combination. We provide examples of cells profiled by IF and RNA FISH and RNA FISH followed by DNA FISH interrogating the X-linked Xist and Tsix lncRNAs that are expressed from and function in cis on the X-chromosome along with histone H3 lysine 27 trimethylation (H3-K27me3).
2 Materials
2.1 Sample Preparation: Cultured Cells
Sterile, gelatinized glass cover slips (see Note 1).
0.2 % Gelatin: 0.2 g Gelatin per 100 ml of ultrapure DNase/RNase-free water. Autoclave to dissolve and sterilize.
6-Well tissue culture dish.
Cytoskeletal buffer (CSK): 100 mM NaCl, 300 mM sucrose, 3 mM magnesium chloride, 10 mM PIPES (C8H18N2O6S2, pH 6.8; be sure to avoid using the sodium-salt version of PIPES). For PIPES, make 0.25–0.5 M stock; bring into solution by adding NaOH while measuring pH.
CSK buffer with 0.4 % Triton X-100: Make stock of 10–20 % Triton X-100 solution in ultrapure DNase/RNase-free water before adding to CSK.
4 % Paraformaldehyde (PFA; Electron Microscopy Science 16 % Paraformaldehyde in aqueous solution (Fisher, #50-980-487), diluted in ultrapure DNase/RNase-free water and with a final concentration of 1× phosphate-buffered saline [PBS]).
70 % Ethanol made with filtered ddH2O.
Plate sealer tape.
2.2 Sample Preparation: Mouse Embryos/Embryo Fragments
Sterile, gelatinized glass cover slips (see Note 1).
0.2 % Gelatin: 0.2 g Gelatin per 100 ml of ultrapure DNase/RNase-free water. Autoclave to dissolve and sterilize.
Pipette for plating embryos or embryo fragments: A glass Pasteur pipette pulled to a width only slightly larger than the embryos/embryo fragments.
1× PBS with 6 mg/ml of bovine serum albumin (BSA): To make 50 ml, combine 4 ml BSA (Invitrogen, 7.5 g /100 ml #15260037), 5 ml 10× PBS, and 41 ml ultrapure DNase/RNase-free water.
Fixation and permeabilization solution: 1 % PFA with 0.05 % Tergitol in a final concentration of 1× PBS.
1 % PFA.
70 % Ethanol made with filtered ddH2O.
6-Well dish, or similar container for storage (see Note 2).
Plate sealer tape.
2.3 Immunofluorescence (IF)
1× PBS.
6-Well dish, or similar chamber to be used for washing cover slips (see Note 2).
Blocking buffer: 1× PBS with 0.5 mg/ml BSA, 50 µg/ml tRNA, 80 units/ml RNase inhibitor (such as Invitrogen’s RNaseOUT), and 0.2 % Tween-20 (make and use a 10 % Tween-20 stock). Pre-warm to 37 °C.
Primary antibody of choice.
Small glass plate (see Note 3).
Parafilm.
Forceps.
IF chamber: A small humid chamber for incubating slides, humidity provided by 1× PBS (see Note 4).
Incubator set to 37 °C.
1× PBS with 0.2 % Tween-20.
Fluorescently conjugated secondary antibody: AlexaFluor (Invitrogen) secondary antibodies work well with this protocol; AF488, AF555, and AF647 have very similar absorption and emission spectra to the Fluorescein-12, Cy3, and Cy5 dyes used for FISH, respectively. These antibodies can be used in conjunction with FISH probes for multicolor imaging with the same set of fluorescence microscope filters.
4′,6-Diamidino-2-phenylindole, dihydrochloride (DAPI) (Life Technologies, 10 mg) (for IF without subsequent RNA FISH).
Mounting medium (for IF without subsequent RNA FISH, see Note 5).
Microscope slides (for IF without subsequent RNA FISH).
2 % PFA (for IF followed by RNA FISH only).
2.4 RNA and DNA FISH: Probe Labeling for Double-Stranded Probes (See Note 6)
BioPrime DNA Labeling System (Life Technologies, #18094011) (see Note 7).
Ultrapure DNase/RNase-free water.
TE buffer: 10 mM Tris–HCl, 1 mM EDTA, pH 8 (see Note 8).
DNA template for labeling: Large templates, such as fosmids or BACs, work best for labeling by random priming. BACs are recommended for DNA FISH probes, as the larger template size will maximize the signal despite low copy number of DNA compared to transcribed RNA in the cell. Ensure that the template preparation is extremely pure; contaminating bacterial DNA will lead to high levels of background fluorescent signal.
- Custom dNTP mixture:
- For use with Cy3 or Cy5 fluorescently labeled dCTP (see below): 2 mM dATP, 2 mM dGTP, 2 mM dTTP, 1 mM dCTP.
- For use with fluorescein-labeled dUTP (fluorescein-12-dUTP; see below): 2 mM dCTP, 2 mM dATP, 2 mM dGTP, 1 mM dTTP.
1 mM Fluorescently labeled nucleotide: Fluorescein-12-dUTP (Roche)-labeled probes excite maximally at 495 nm and emit maximally at 521 nm. Cy3-labeled dCTP (GE Healthcare)-labeled probes excite maximally at 550 nm and emit maximally at 570 nm, while Cy5-labeled dCTP (GE Healthcare)-labeled probes absorb maximally in the far-red end of the spectrum, at 649 nm, and emit maximally at 670 nm. The spectra for these dyes do not overlap significantly, and labeled probes can be combined with other FISH probes and used with DAPI, which absorbs maximally at 358 nm and emits maximally at 461 nm when bound to double-stranded DNA, or fluorescently conjugated antibodies for multicolor imaging.
G-50 ProbeQuant Micro Columns (GE Healthcare).
Tabletop centrifuge.
20 mg/ml Yeast tRNA: Reconstitute lyophilized yeast tRNA (Invitrogen, #15401-029) in ultrapure DNase/RNase-free water. Aliquot and store at −20 °C.
3 M Sodium acetate, pH 5.2 (see Note 8).
Molecular biology-grade 100 % ethanol.
Heat block set to 37 °C.
2.5 RNA FISH: Probe Labeling for Strand-Specific Probes (See Note 6)
MAXIscript T3/T7 Kit (Ambion, AM1326).
Linear DNA template, PCR amplified with T3 or T7 promoter sequence upstream of the sequence to be labeled: To detect RNA transcribed from the desired strand, the probes must be synthesized complementary to the target transcript; the T3 or T7 RNA polymerase promoter sequences must therefore be incorporated at the appropriate ends of the template DNA. See MAXIscript kit instructions for details on template preparation.
- Custom NTP mixture (see Note 9):
- For use with Cy3 or Cy5 fluorescently labeled CTP: 2 mM ATP, 2 mM GTP, 2 mM TTP, 1 mM CTP.
- For use with fluorescein fluorescently labeled UTP (fluorescein-12-UTP: 2 mM CTP, 2 mM ATP, 2 mM GTP, 1 mM TTP.
100 nmol Fluorescently labeled nucleotide: Fluorescein-12-UTP (Roche), Cy3-CTP (GE Healthcare), or Cy5-CTP (GE Healthcare) (see notes on emission spectra in Subheading 2.4).
0.5 M EDTA (see Note 8).
20 mg/ml Yeast tRNA: Reconstitute lyophilized yeast tRNA in ultrapure DNase/RNase-free water. Aliquot and store at −20 °C.
5 M Ammonium acetate (see Note 8).
100 % Molecular biology-grade ethanol.
Ultrapure DNase/RNase-free water.
37 °C Heat block.
Mini Quick Spin RNA Columns (Roche, #11814427001).
Tabletop centrifuge.
2.6 RNA and DNA FISH : Probe Precipitation
Fluorescently labeled probes in ethanol (see Subheadings 3.4 and 3.5): A combination of double-stranded and strand-specific probes may be used.
20 mg/ml Yeast tRNA: Reconstitute lyophilized yeast tRNA in ultrapure DNase/RNase-free water. Aliquot and store at −20 °C.
1 mg/ml COT-1 DNA (Invitrogen) (Optional, see Note 10).
10 mg/ml Salmon sperm DNA.
5 M Ammonium acetate (see Note 8).
Ultrapure DNase/RNase-free water.
100 % Molecular biology-grade ethanol.
Tabletop centrifuge.
70 % Ethanol: Dilute 100 % ethanol using filtered ddH2O.
Deionized formamide.
20× Sodium saline citrate (SSC) (Invitrogen).
2× Hybridization solution: Two parts ultrapure DNase/RNase-free water, one part 20× SSC, two parts 50 % dextran sulfate (see Note 11).
Vacuum centrifuge.
Heat block set to 90 °C.
2.7 RNA FISH : Sample Hybridization
100 % Molecular biology-grade ethanol: Use filtered ddH2O to make stocks of 70, 85, and 95 % ethanol.
Fluorescently labeled probes (see Subheadings 2.4–2.6).
6-Well dish, or similar chamber to be used for dehydration and for washing cover slips (see Note 2).
Small glass plate (see Note 3).
Parafilm.
Forceps.
Hybridization chamber: A small humid chamber for incubating slides, humidity provided by 2× SSC/50 % deionized formamide (see Note 4).
Incubator set to 37 °C, for overnight incubation.
2× SSC/50 % deionized formamide: 5 ml 20× SSC, 25 ml deionized formamide, 20 ml filtered ddH2O.
2× SSC: 5 ml 20× SSC, 45 ml filtered ddH2O.
1× SSC: 2.5 ml 20× SSC, 47.5 ml filtered ddH2O.
DAPI.
Incubator set to 39 °C, for washes.
Mounting medium (see Note 5).
Microscope slides.
2.8 DNA FISH : Sample Hybridization
1× PBS.
1 % PFA with 0.5 % Tergitol and 0.5 % Triton X-100.
100 % Molecular biology-grade ethanol: Use filtered ddH2O to make stocks of 70, 85, and 95 % ethanol.
6-Well dish, or similar chamber to be used for dehydration and for washing cover slips (see Note 2).
RNase A solution: 1.25 µg/µl RNase A, diluted in 2× SSC.
2× SSC/70 % deionized formamide: 5 ml 20× SSC, 35 ml deionized formamide, 10 ml filtered ddH2O.
Heat block set to 95 °C.
Fluorescently labeled double-stranded probe (see Subheadings 2.5 and 2.6).
Small glass plate (see Note 3).
Parafilm.
Forceps.
Hybridization chamber: A small humid chamber for incubating slides, humidity provided by 2× SSC/50 % deionized formamide (see Note 4).
Incubator set to 37 °C, for overnight incubation.
2× SSC/50 % deionized formamide: 5 ml 20× SSC, 25 ml deionized formamide, 20 ml filtered ddH2O.
2× SSC: 5 ml 20× SSC, 45 ml filtered ddH2O.
1× SSC: 2.5 ml 20× SSC, 47.5 ml filtered ddH2O.
DAPI.
Incubator set to 39 °C, for washes.
Mounting medium (see Note 5).
Microscope slides.
3 Methods
3.1 Sample Preparation: Cultured Cells
Grow cells to desired confluency on sterile, gelatinized glass cover slips (see Note 1) in the bottom of a 6-well tissue culture dish.
Prepare reagents for fixation and permeabilization: Make and sterile filter CSK and CSK with 0.4 % Triton X-100 buffers and chill to 4 °C. Prepare 4 % PFA. All subsequent steps should be performed in the tissue culture hood. The CSK and CSK + 0.4 % Triton X-100 buffers should be kept chilled on ice during the procedure.
Aspirate media from all six wells of a 6-well dish
Pipette 2 ml of ice-cold CSK buffer in each well of the 6-well dish.
After 30 s, aspirate the CSK buffer: By the time CSK buffer has been pipetted into each of the six wells, the first well has incubated in the CSK buffer for ~30 s, so simply pipette CSK buffer into the six wells and then aspirate right afterward from the first well onward.
Pipette 2 ml of ice-cold CSK + 0.4 % Triton X-100 buffer in each well of the 6-well dish.
After 30 s, aspirate the CSK + 0.4 % Triton X-100. See details in step 3.
Pipette 2 ml of ice-cold CSK buffer in each well of the 6-well dish.
After 30 s, aspirate the CSK buffer. See details in step 3.
Pipette 2 ml of 4 % PFA in each well of the 6-well dish. Incubate the cells in PFA for 10 min.
Aspirate the PFA and pipette in 5 ml of cold 70 % ethanol in each well. Repeat the ethanol wash a total of three times, to remove all traces of PFA.
Seal dish with plate sealer tape and store cells at −20 °C (see Note 12).
3.2 Sample Preparation: Mouse Embryos
Plate embryos or dissected embryo fragments on sterile, gelatinized glass cover slips (22 × 22 cm square cover slips, see Note 1) in 1× PBS 6 mg/ml BSA using finely pulled Pasteur pipette.
Aspirate excess 1× PBS 6 mg/ml BSA and let dry for about 20–30 min.
Fix and permeabilize with 50 µl of 1 % PFA in 1× PBS with 0.05 % Tergitol for 5 min (to get rid of excess solution on cover slip after the 5-min permeabilization/fixation, simply tap solution off onto a paper towel).
Fix again with 50 µl of 1 % PFA (no Tergitol) in 1× PBS for 5 min. Drain the excess solution onto a paper towel.
Place cover slips with plated embryos in a 6-well dish. Rinse with three changes of 70 % ethanol. Seal dish with plate sealer tape and store in 70 % ethanol at −20 °C until use (see Note 12).
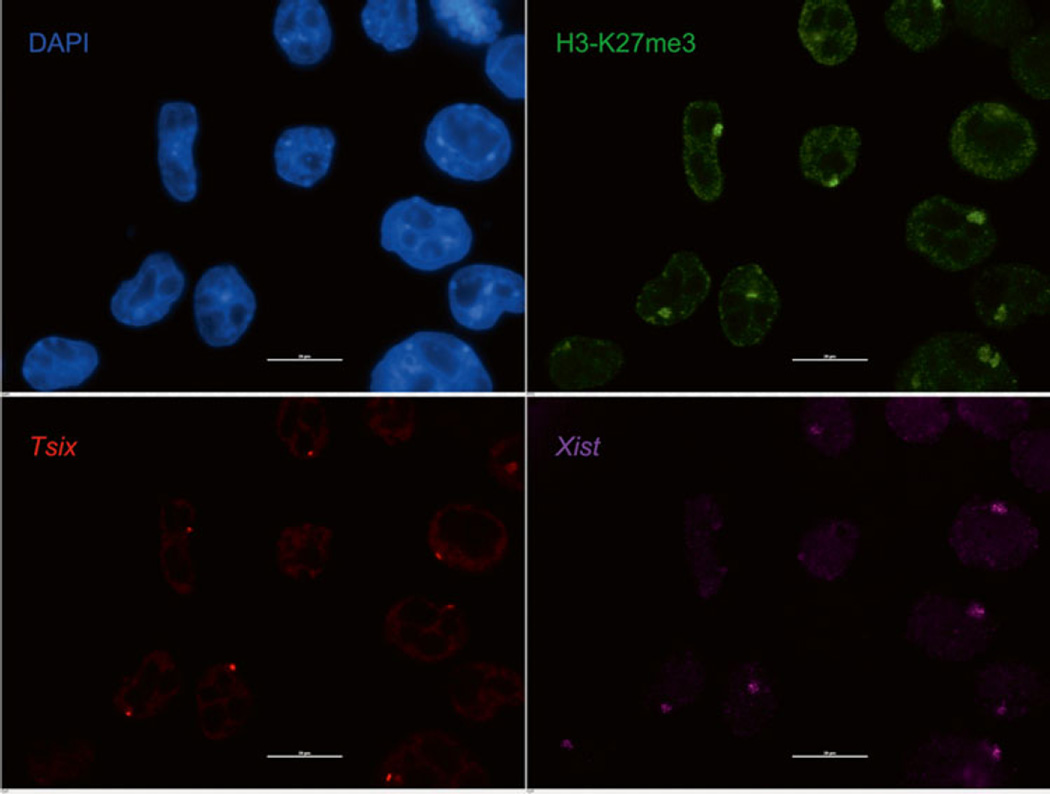
3.3 Immunofluorescence (IF) (Fig. 1)
Fig. 1.
Immunofluorescence followed by strand-specific RNA FISH in female mouse trophoblast stem cells. Histone H3 lysine 27 trimethylation (H3-K27me3; in green) is enriched on the inactive X-chromosome that is marked by Xist lncRNA accumulation (purple). The active X-chromosome is marked by nascent expression of the Xist antisense Tsix lncRNA (red pinpoints). Nuclei are stained blue with DAPI
Begin with fixed, permeabilized cells or embryo samples, plated on gelatinized glass cover slips and stored in 70 % ethanol (see Subheadings 3.1 and 3.2).
Make blocking buffer and warm to 37 °C.
Place sample cover slip in a 6-well dish that contains 2 ml of 1× PBS in each well (see Note 2).
Wash briefly with three changes of 1× PBS to remove ethanol.
Wash with 1× PBS three times, 3 min each on a rocker.
Wrap a glass plate tightly with parafilm for incubating cover slips for subsequent steps.
Block slides for 30 min at 37 °C in 50 µl pre-warmed blocking buffer in a humid chamber: Place a 50 µl drop of blocking buffer on the parafilm-wrapped glass plate and invert the cover slip, sample side down, into the blocking buffer. Place the parafilm-wrapped plate in the humid chamber, and incubate for 30 min at 37 °C. All incubations in blocking buffer, primary antibody, or secondary antibody should be set up in this manner.
Carefully lift cover slip from blocking buffer with forceps and place into a 50 µl droplet of diluted primary antibody on a parafilm-wrapped plate. Incubate with 50 µl primary antibody diluted in pre-warmed blocking buffer (dilution based on primary antibody you are using) in a humid chamber at 37 °C for 1 h.
Remove cover slip from primary antibody solution and place, sample-side up, in a 6-well dish. Wash three times with 1× PBS/0.2 % Tween-20 for 3 min on a rocker.
Incubate in 50 µl pre-warmed blocking buffer on a parafilm-wrapped plate in a humid chamber for 5 min at 37 °C.
Incubate with 50 µl secondary antibody diluted in pre-warmed blocking buffer in humid chamber at 37 °C for 30 min. Antibody dilution depends on secondary antibody used; AlexaFluor-conjugated secondary antibodies should be used at a 1:300 dilution.
- Remove cover slip from secondary antibody and wash three times with 1× PBS/0.2 % Tween-20 for 3 min each on a rocker.
- If processing samples only for IF, the first wash of step 11 should contain a 1:100,000 to 1:200,000 dilution of DAPI (see Note 13). Then, rinse once briefly with PBS/0.2 % Tween-20 and wash two more times for 5–7 min each while rocking to remove excess DAPI. Remove cover slip from dish, tap off excess liquid, and then mount on a slide, sample-side down, in mounting medium. Image samples or store at −20 °C for later imaging.
- If samples will be stained by RNA FISH probes following IF, pipette a 100 µl drop of 2 % PFA on a glass plate wrapped in parafilm. Place the cover slip sample-side down in PFA and place in the humid chamber. Incubate the sample for 10 min at room temperature, and then proceed to RNA FISH hybridization (Subheading 3.7).
3.4 RNA FISH: Probe Labeling for Double-Stranded Probes
Dissolve 50–100 ng DNA template in TE buffer in a final volume of 19 µl.
On ice, add 20 µl 2.5× random primer solution (supplied with BioPrime labeling kit).
Denature DNA by heating for 5 min in a 90 °C heat block.
Immediately cool on ice for 5 min.
Add 5 µl of custom dNTP mixture.
Add 5 µl of 1 mM fluorescently labeled nucleotide.
Add 2 µl (80 units) Klenow fragment (supplied with the BioPrime Labeling Kit, see Note 14).
Incubate at 37 °C overnight. Cover the tube with aluminum foil to protect the probe from light during this step.
Add 5 µl of stop buffer.
Prepare G-50 column: Vortex briefly to mix the Sephadex beads. Twist off the bottom and place the column in one of the provided collection tubes. Spin at 750 × g for 1 min.
Move column to a new microcentrifuge tube and pipette your probe into the center of the column. Spin at 750 × g for 1 min. Measure eluted volume by pipette (see Note 15).
Add 10 µl of 20 mg/ml yeast tRNA.
Add 3 M sodium acetate to a final concentration of 0.3 M.
Add 2.5 volumes of 100 % ethanol.
Spin at 4 °C for 20 min at top speed, ~21,100 × g.
Carefully aspirate the supernatant using the vacuum apparatus (see Note 16).
Resuspend the pellet in 360 µl of RNase/DNase-free ultrapure ddH2O. Make sure that the pellet is fully dissolved before moving to the next step.
Add 40 µl 3 M sodium acetate.
Add 1 ml 100 % ethanol.
Store labeled probe at −20 °C in the dark (see Note 17).
3.5 RNA FISH: Probe Labeling for Strand-Specific Probes
Start with 1 µg template DNA.
Add RNase/DNase-free ultrapure ddH2O so that the total volume at the end of step 6 will be 20 µl.
Add 2 µl 10× transcription buffer (supplied with MAXIscript kit).
Add 5 µl NTP mixture (supplied with MAXIscript kit, see Note 9).
Add 2 µl of fluorescently labeled NTP.
Add 2 µl T3 (60 units) or T7 (30 units) enzyme mix (supplied with MAXIscript kit, see Note 9).
Mix thoroughly.
Incubate for 2 h at 37 °C.
Add 1 µl of TURBO DNase (supplied with MAXIscript kit) and mix well.
Add 1 µl of 0.5 mM EDTA.
Prepare Quick Spin RNA Column: Vortex briefly to mix the Sephadex beads. Remove cap, twist off the bottom tip, and then place the column in a microcentrifuge tube. Spin at 1000 × g for 1 min.
Move column to a new microcentrifuge tube and pipette your probe into the center of the column (see Note 15). Spin at 1000 × g for 1 min. Measure your new volume by pipette.
Add 10 µl of 20 mg/ml yeast tRNA.
Add 5 M ammonium acetate to a final concentration of 0.5 M.
Add 2.5 volumes of 100 % ethanol.
Spin at 4 °C for 20 min at top speed, 21,100 × g.
Carefully aspirate the supernatant using the vacuum apparatus (see Note 16).
Resuspend the pellet in 360 µl RNase/DNase-free ultrapure ddH2O. Make sure that the pellet is fully dissolved before moving to the next step.
Add 40 µl of 5 M ammonium acetate.
Add 1 ml of 100 % ethanol.
Store labeled probe at −20 °C in the dark (see Note 17).
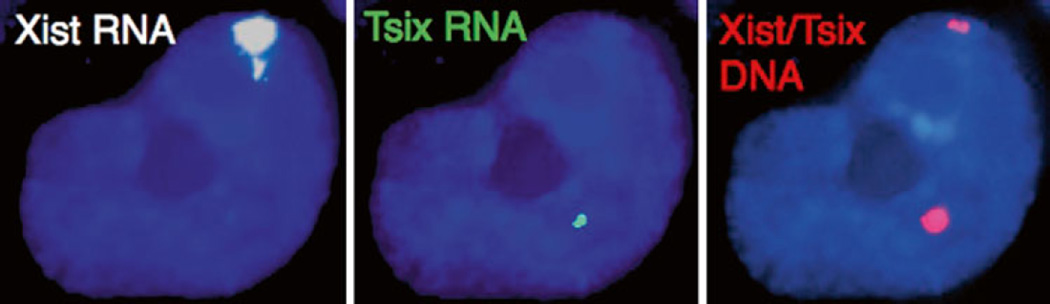
3.6 RNA FISH: Probe Precipitation (Figs. 1 and 2)
Fig. 2.
RNA followed by DNA FISH in female mouse epiblast stem cells. Strand-specific RNA FISH detection of Xist lncRNA (white) and the Xist antisense Tsix lncRNA (green) followed by DNA FISH detection of the Xist/Tsix locus. Nuclei are stained blue with DAPI
All spin steps should be completed at top speed, ~21,100 × g.
Aliquot 100 µl of fluorescently labeled probe in ethanol. A combination of double-stranded and strand-specific probes may be precipitated together (see Note 18). Vortex all probe stocks before aliquoting.
Add 15 µl of 20 mg/ml tRNA (300 µg). Mix thoroughly by pipetting (see Note 19).
Add 15 µl of 1 mg/ml mouse COT-1 DNA (15 µg). Mix thoroughly by pipetting. (Optional: See Notes 10 and 19. If not using COT-1 DNA, add 15 µl of RNase/DNase-free ultrapure water.)
Add 15 µl of 10 mg/ml sheared boiled salmon sperm DNA (150 µg). Mix thoroughly by pipetting (see Note 19).
Add 20 µl 5 M ammonium acetate. Vortex briefly to mix (see Note 20).
Add 45 µl of DNase/RNase-free ultrapure water. Vortex briefly to mix (see Note 20).
Add 250 µl of 100 % ethanol. Vortex briefly to mix (see Note 20).
Spin for 20–30 min at 4 °C to precipitate probe.
Carefully aspirate the supernatant using a vacuum apparatus (see Note 16).
Add 500 µl of 70 % ethanol. Vortex thoroughly. Spin at room temp for 4 min.
Carefully aspirate the supernatant using a vacuum apparatus (see Note 16).
Add 500 µl of 100 % ethanol. Vortex thoroughly. Spin at room temp for 4 min.
Carefully aspirate the supernatant using a vacuum apparatus (see Note 16).
Dry in a speed vacuum without heat for 5 min, or alternatively air-dry. No ethanol should remain in the tube for the next step.
Add 50 µl of 100 % deionized formamide to the tube. Make sure that the pellet is submerged in the formamide.
Denature in a heat block at 80–90 °C for 10 min. At ~5 min, pipette the formamide up and down to make sure that the pellet goes into solution. Place back into the heat block.
Immediately cool on ice for 5 min.
Add 50 µl 2× hybridization solution. Mix thoroughly by pipetting up and down.
Preanneal at 37 °C for 1–1.5 h. (Optional: See Note 10. Preannealing is not required for strand-specific probes or for double-stranded probes precipitated without COT-1 DNA.)
Store probe at −20 °C in the dark.
3.7 RNA FISH: Sample Hybridization (Fig. 2)
Start with permeabilized and fixed samples plated on cover slips in 70 % ethanol (see Subheading 3.1 or 3.2), or samples previously processed for IF (see Subheading 3.3).
Dehydrate the cover slips by moving them through a room-temperature ethanol series (85, 95, and 100 % ethanol) for 2 min each.
Remove the cover slips from the well and air-dry at room temperature for 15 min (see Note 21).
Set up the FISH hybridization. Use 8–10 µl of probe per 22 × 22 mm cover slip. Pipette the probe onto a parafilm-wrapped glass plate (see Note 3), invert the cover slip onto the droplet of probe, and tap lightly with forceps to help the probe spread across the cover slip.
Hybridize overnight at 37 °C in a humid chamber (humidity provided by 2× SSC/50 % formamide) (see Note 4).
Make all wash solutions (2× SSC/50 % formamide, 2× SSC, and 1× SSC), and warm the solutions to 39 °C.
Carefully peel the cover slip off of the parafilm, re-invert, and place in a well containing pre-warmed 2× SSC/50 % formamide. Be sure that the cover slip goes into the well with the sample-side up.
Wash with pre-warmed 2× SSC/50 % formamide at 39 °C, three times for 7 min each.
Wash with pre-warmed 2× SSC at 39 °C, three times for 7 min each. Add DAPI into the last 2× SSC wash at a 1:100,000–1:200,000 dilution (see Note 13).
Rinse once quickly with pre-warmed 1× SSC.
Wash with pre-warmed 1× SSC at 39 °C, two times for 7 min each.
Use mounting medium to mount the cover slip onto a labeled microscope slide. Invert the slide onto a paper towel and press gently but firmly to remove excess mounting medium from under the cover slip.
Seal the cover slip and let dry thoroughly before viewing under a microscope.
Slides can be stored in the dark at −20 °C to preserve the fluorescent signal.
3.8 DNA FISH: Sample Hybridization
Begin with fixed, permeabilized cells or embryo samples, plated on gelatinized glass cover slips (22 × 22 cm square cover slips) and stored in 70 % ethanol (see Subheadings 3.1 and 3.2) or with samples previously stained for RNA FISH (see Subheading 3.7). For samples previously stained by RNA FISH, RNA FISH signals will be degraded during DNA FISH, so samples should be imaged prior to beginning this protocol. Take note of the visual fields imaged, so that RNA FISH images can be aligned with DNA stains later.
If using samples previously stained for RNA FISH, use a razor blade to cut away the nail polish used to seal the cover slip to the slide. Then submerge the entire slide with the cover slip still attached in a solution of 2× SSC. While the sample is still submerged gently peel off the cover slip by letting the solution of 2× SSC infiltrate between the cover slip and the slide. For samples not previously processed for RNA FISH, proceed to step 5.
Wash the cover slip with 1× PBS three times quickly and then incubate in 1× PBS for 5 min at room temperature.
Refix the cell/embryos with 1 % PFA containing 0.5 % Tergitol and 0.5 % Triton X-100. Fix for 10 min at room temperature.
Dehydrate the cover slips by moving them through a room-temperature ethanol series (70, 85, and 100 % ethanol) for 2 min each.
Remove the cover slips from the well and air-dry at room temperature for 15 min (see Note 21).
RNase treatment: Treat with 1.25 µg/µl RNase A in 2× SSC. Invert slides onto a 100 µl drop of RNase A solution on parafilm stretched over a glass slide. Incubate at 37 °C for 30 min. RNase treatment allows the probe to gain access to the DNA.
Dehydrate the cover slips by moving them through a room-temperature ethanol series (85, 95, and 100 % ethanol) for 2 min each.
Remove the cover slips from the well and air-dry at room temperature for 15 min (see Note 21).
Denature the samples in a pre-warmed solution of 2× SSC/70 % formamide on a glass slide or glass plate stationed on top of a heat block set at 95 °C for 11 min.
Immediately dehydrate through a −20 °C ethanol series (70, 85, 95, and 100 % ethanol) for 2 min each.
Remove the cover slips from the well and air-dry at room temperature for 15 min (see Note 21).
Set up the FISH hybridization. Use 8–10 µl of probe per 22 × 22 mm cover slip. Pipette the probe onto parafilm-wrapped glass plate (see Note 3), invert the cover slip onto the droplet of probe, and tap lightly with forceps to help the probe spread across the cover slip. Be sure to avoid air bubbles that may result in uneven distribution of the probe.
Hybridize overnight at 37 °C in a humid chamber (humidity provided by 2× SSC/50 % formamide, see Note 4).
Make all wash solutions (2× SSC/50 % formamide, 2× SSC), and warm the solutions to 39 °C.
Carefully peel the cover slip off of the parafilm, re-invert, and place in a well containing pre-warmed 2× SSC/50 % formamide. Be sure that the cover slip goes into the well with the sample side up.
Wash with 2× SSC/50 % formamide at 39 °C twice, 7 min each.
Wash with pre-warmed 2× SSC plus a 1:100,000–1:200,000 dilution of DAPI for 7 min at 39 °C (see Note 13).
Wash with pre-warmed 2× SSC without DAPI.
Use mounting medium to mount the cover slip onto a microscope slide. Invert the slide onto a paper towel and press gently but firmly to remove excess mounting medium from under the cover slip.
Seal the cover slip with clear nail polish and let dry thoroughly before viewing under a microscope.
Image and store the slide at −20 °C. When combining RNA FISH with DNA FISH, RNA FISH-stained samples must be imaged first and the coordinates of the cells on cover slip recorded. Following DNA FISH, the same cells should be imaged, to allow for analysis of overlaid images.
Acknowledgments
This work was funded by an NIH National Research Service Award #5-T32-GM07544 from the National Institute of General Medicine Sciences to E.M.; an NIH Director’s New Innovator Award (DP2-OD-008646-01) to S.K.; a March of Dimes Basil O’Connor Starter Scholar Research Award (5-FY12-119); and the University of Michigan Endowment for Basic Sciences.
Footnotes
- For tissue culture cells, sterilize cover slips by dipping in 100 % ethanol and running through a flame. Once the cover slip is sterilized in this manner, place it in a well of the 6-well dish. In the tissue culture hood, pipette 0.5–1 ml sterile 0.2 % gelatin onto the cover slip and spread around. Keep the gelatin on the surface of the cover slip and avoid letting the gelatin infiltrate between the cover slip and the bottom of the well, or the cover slip may stick to the tissue culture surface. Aspirate excess gelatin and let dry.
- For embryos, place a 25–50 µl droplet of 0.2 % sterile gelatin solution on the cover slip, spread liberally around with pipette tip, and let dry.
Use 22 × 22 mm cover slips, which fit well within a single well of a 6-well dish. All the dehydration and washing steps can be performed in the wells of this dish.
Short glass plates designed for casting protein gels are a good size for the hybridization. For example, BioRad’s Mini-PROTEAN Short Plates fit well in the humid chamber described in Note 4.
For humid chambers, a microscope slide box (i.e., one that holds 100 slides) works well. Place paper towels soaked in 1× PBS in the bottom of the box to create a humid chamber for immunofluorescence. For RNA and DNA FISH hybridization procedures, create a humid chamber by placing paper towels soaked in 2× SSC/50 % formamide in the bottom of the box.
For mounting medium, use Vectashield (Vector Labs) or similar anti-fade mounting medium and seal cover slips with clear nail polish after mounting on slides.
For “double-stranded” probes (Subheadings 2.4 and 3.4), while the labeled probes are single stranded, the probes are created from a double-stranded fosmid or BAC template using random primers. As such, these probes will hybridize to both strands of DNA or RNA. Double-stranded probes can be used for DNA FISH and for RNA FISH for most genes. With strand-specific probes (Subheadings 2.5 and 3.5), only one strand is labeled, and the probe will detect RNA from only the complementary strand. These probes can be used to distinguish sense and antisense transcription from a single locus.
This kit was originally designed for the preparation of biotinylated probes. The protocol outlined above modifies the BioPrime labeling procedure to make fluorescently labeled probes. Do not use the supplied 10× dNTP mixture, which includes biotinylated nucleotides. Prepare your own dNTP mixture, and add in the fluorescently labeled nucleotides separately (Subheading 3.4, steps 5 and 6). Alternative random primer solutions have been tested, but do not work as well as the 2.5× solution provided with this kit.
While these solutions can be prepared from scratch, purchasing them is preferred to ensure their RNase/DNase-free purity.
A set of 10 mM NTPs and the T3 and T7 enzymes are included in the MAXIscript kit. Both the NTPs and enzymes can be replaced with other commercially available substitutes with no apparent loss of enzymatic activity.
COT-1 DNA can be added to double-stranded probes to help reduce background by hybridizing to repetitive sequences, if necessary. COT-1 DNA should not be used in strand-specific probes, as these probes are specific to the target sequence. While we do not routinely use mouse COT-1 in the precipitation of mouse probes, we do usually add species-specific COT-1 to double-stranded probes for other species (i.e., human, rat; but have not tested whether background staining would increase if COT-1 were omitted). If COT-1 is omitted, preannealing is not required.
50 % Dextran sulfate is very viscous. Make the 2× hyb solution in a tube with graduations. If you add the dextran sulfate last, instead of pipetting it you can use a pipette tip to scoop it into the tube up to the appropriate measurement marking on the side of microtube. Vortex thoroughly to mix.
Fixed cells and embryos on cover slips in 70 % ethanol can be stored in −20 °C for at least 1 year. Seal with plate sealer tape to minimize evaporation of ethanol, and replace the ethanol in the wells occasionally (once every 2 months or so), as it will evaporate over time despite the plate sealer.
Dilute DAPI 1:500 in RNase/DNase-free ultrapure water, and store in the dark at −20 °C. Add 6–10 µl of this dilution into each well while washing samples.
The Klenow Fragment included with the BioPrime kit can be replaced with other commercially available Klenow enzymes with no apparent loss of enzymatic activity.
Be sure to pipet your probe into the center of the beads in the column during column purification. The probe must travel through the beads, not down the side.
The pellet is usually attached to the bottom of the tube, but be careful during aspiration of the supernatant. You can put a 10 µl pipette tip on the end of your aspiration hose to slow down the suction.
Ethanol stocks of probes should last up to 1 year when stored sealed and protected from light.
At this stage, multiple probes that were labeled with different fluorophores can be combined. To start, precipitate 100 µl ethanol stock of each probe together. If any of the probes are too bright or too faint, the initial starting volume of the ethanol stock of the probes can be adjusted.
The volumes of tRNA, Cot-1 DNA, and salmon sperm DNA (Subheading 3.6, steps 2–4) are determined by the final volume of probe; these will stay constant whether you are precipitating one or multiple probes together.
The volumes of ammonium acetate, H2O, and 100 % ethanol (Subheading 3.6, steps 5–7) are determined by the starting volume of ethanol; these will need to be scaled up proportionately if you are precipitating multiple probes together. For example, if precipitating two probes together and using 100 µl of each ethanol stock, then the volumes of ammonium acetate, H2O, and ethanol need to be doubled.
Do not simply aspirate the 100 % ethanol out and leave the cover slip to dry in the well; the glass cover slip may stick to the bottom of the well. Find a safe place to set the cover slip to dry. Laying them across the inside of a empty pipette tip box (for example, a p1000 tip box) works well.
References
- 1.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JD, Mitchell SE, O’Neill RJ. Making a long story short: noncoding RNAs and chromosome change. Heredity. 2012;108:42–49. doi: 10.1038/hdy.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maclary E, Hinten M, Harris C, Kalantry S. Long nonoding RNAs in the X-inactivation center. Chromosome Res. 2013;21:601–614. doi: 10.1007/s10577-013-9396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maclary E, et al. Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation. Nat Commun. 2014;5:4209. doi: 10.1038/ncomms5209. [DOI] [PMC free article] [PubMed] [Google Scholar]