Dear Editors
We read with great interest the study by De Bus et al. [1] regarding the impact of de-escalation of beta-lactamic antibiotics on emergence of resistance and we share their concerns about the pragmatic obstacles in implementing rational de-escalation. Their major finding was that de-escalation was associated with a statistically non-significant increase in the cumulative incidence of emergent resistance compared to continuation of empiric antibiotics. We are nonetheless concerned that their results reflect uncontrolled confounding bias and are prone to misinterpretation, especially when the study’s take-home message is summarized as “Do NOT de-escalate” [2]. Before such inferences on null (or even harmful) causal effects of de-escalation are drawn, we call for caution in interpreting observational evidence.
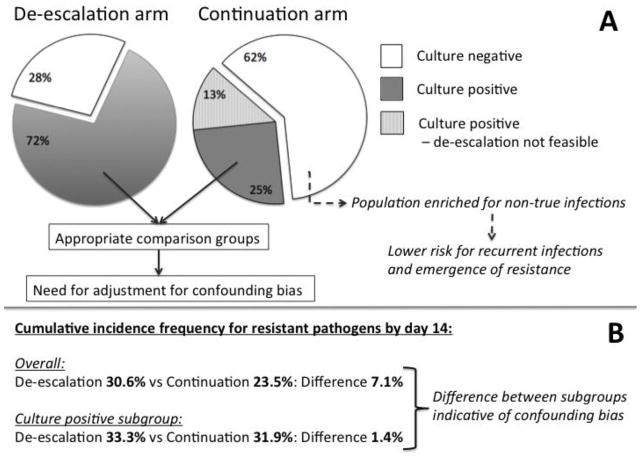
First, the compared groups of de-escalation and continuation are largely heterogeneous in terms of the infectious etiology of their underlying illness. As shown in the Figure (panel A), 62% of the continuation arm patients did not have microbiologic confirmation of infection. This population is likely enriched for patients without a true infection that received unnecessary antibiotics, thus by definition at lower risk for “recurrence” by resistant pathogens. This imbalance is evident when we note that the “harmful” effect of de-escalation disappeared in the subgroup comparison of patients with positive cultures only (Figure, panel B). Targeted de-escalation should thus be studied in those cases with identification of a causal pathogen, as performed in a recent controlled trial that randomized patients to de-escalation or continuation strategies, only after an antibiogram was available [3].
Figure.
Panel A: Proportion of patients with negative cultures (white), positive cultures able to guide de-escalation of antibiotics (dark grey), and positive cultures not able to allow de-escalation (shaded gray) in the de-escalation and continuation arms of therapy with anti-pseudomonal beta-lactam antibiotics. Panel B: Cumulative incidence frequencies of emergence of resistant pathogens by day 14. Results shown for the overall groups and for the subgroup with positive cultures. Cumulative incidence frequencies for the culture positive subgroup were extracted from Supplementary Figure 2c of [1] after digitization.
Furthermore, this subgroup comparison we propose as appropriate is still vulnerable to confounding by indication the way it was analyzed [1]. While the authors nicely demonstrate major determinants of de-escalation selection, no effort to control for confounding by such factors was made. This confounding is important, as 25% of patients in the continuation arm had culture data to permit de-escalation, but their physicians decided not to, suggesting that other factors influenced their decisions. The risks of assuming a causal effect of de-escalation when proper control for confounding has not taken place [4] is illustrated by the fact that escalation of antibiotics in this cohort was associated with near-doubling of the risk of subsequent infection compared to continuation (55% vs 33%, p=0.008). Should we then recommend withholding intensified antibiosis against confirmed resistant organisms due to perceived increased risk of subsequent infections? Obviously not, but this scenario provides an example of applying a confounded result.
The study by De Bus et al. represents a detailed depiction of what happens with de-escalation in clinical practice, but not due to de-escalation. It also highlights how problematic guidance by microbiologic cultures is, both in ruling out infections and in timely defining causal pathogens. More research is needed with culture-independent techniques that directly sequence, quantify and analyze bacterial DNA from patient samples. As such techniques evolve and mature for bedside applications [5], we believe that sequencing-guided de-escalation will allow for the realization of the well-shared ecological advantages of reducing antibiotic pressure, and promoting safe and rational antibiotic stewardship.
Footnotes
Conflict of Interest:
On behalf of all authors, the corresponding author states that there is no conflict of interest
Bibliography
- 1.De Bus L, Denys W, Catteeuw J, et al. Impact of de-escalation of beta-lactam antibiotics on the emergence of antibiotic resistance in ICU patients: a retrospective observational study. Intensive Care Med. 2016;42:1029–1039. doi: 10.1007/s00134-016-4301-z. [DOI] [PubMed] [Google Scholar]
- 2.Wong A. [Accessed 28 Jun 2016];Antimicrobial stewardship: Do NOT de-escalate. 2016 http://www.esicm.org/news-article/ARTICLE-REVIEW-antimicrobial-stewardship-ICM-JUNE%202016.
- 3.Leone M, Bechis C, Baumstarck K, et al. De-escalation versus continuation of empirical antimicrobial treatment in severe sepsis: a multicenter non-blinded randomized noninferiority trial. Intensive Care Med. 2014;40:1399–1408. doi: 10.1007/s00134-014-3411-8. [DOI] [PubMed] [Google Scholar]
- 4.Kitsios GD, Dahabreh IJ, Callahan S, et al. Can We Trust Observational Studies Using Propensity Scores in the Critical Care Literature? A Systematic Comparison With Randomized Clinical Trials. Crit Care Med. 2015;43:1870–1879. doi: 10.1097/CCM.0000000000001135. [DOI] [PubMed] [Google Scholar]
- 5.Judge K, Harris SR, Reuter S, et al. Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes. J Antimicrob Chemother. 2015;70:2775–2778. doi: 10.1093/jac/dkv206. [DOI] [PMC free article] [PubMed] [Google Scholar]