Abstract
Encoding of sensory inputs in the cortex is characterized by sparse neuronal network activation. Optogenetic stimulation has previously been combined with fMRI (ofMRI) to probe functional networks. However, for a quantitative optogenetic probing of sensory-driven sparse network activation, the level of similarity between sensory and optogenetic network activation needs to be explored. Here, we complement ofMRI with optic fiber-based population Ca2+ recordings for a region-specific readout of neuronal spiking activity in rat brain. Comparing Ca2+ responses to the blood oxygenation level-dependent signal upon sensory stimulation with increasing frequencies showed adaptation of Ca2+ transients contrasted by an increase of blood oxygenation level-dependent responses, indicating that the optical recordings convey complementary information on neuronal network activity to the corresponding hemodynamic response. To study the similarity of optogenetic and sensory activation, we quantified the density of cells expressing channelrhodopsin-2 and modeled light propagation in the tissue. We estimated the effectively illuminated volume and numbers of optogenetically stimulated neurons, being indicative of sparse activation. At the functional level, upon either sensory or optogenetic stimulation we detected single-peak short-latency primary Ca2+ responses with similar amplitudes and found that blood oxygenation level-dependent responses showed similar time courses. These data suggest that ofMRI can serve as a representative model for functional brain mapping.
Keywords: Optogenetic fMRI, calcium recordings, optical neurophysiology, sparse network activation, light propagation
Introduction
Neurons of the mammalian cortex form a highly interconnected functional network, modulated by intracortical and subcortical excitatory and inhibitory drivers. Yet, processing of sensory inputs within this network is characterized by sparse spiking activity, the average spiking frequency of an individual excitatory neuron averages below 1 Hz, and the relative proportion of neurons of the local network directly driven by sensory input ranges typically at 5–20%, depending on the sensory modality.1,2 The complexity of the neural network renders specific stimulation of network components challenging. Electrical microstimulation has served as the classical tool for network probing, yet it generates anti- and orthodromic activation, it nonspecifically stimulates fibers of passage and consequently activates distant networks, such as cortico-thalamic and thalamo-cortical fibers upon cortical microstimulation. The discovery of genetically encoded light-gated cation channels such as channelrhodopsin-2 (ChR2) enabled the optical induction of suprathreshold spiking of genetically defined neurons with millisecond precision and within a spatially defined region—optogenetics.3 Spatial specificity can be achieved by a region-specific viral transduction of opsins, in combination with a targeted light delivery, for example via implanted optic fibers.4 The use of optogenetics in studying network connectivity has proven to be highly informative, allowing for a causal interrogation of aberrant functional connectivity, e.g. in Parkinson's disease,5 depression, and schizophrenia.6 To study spatial and temporal patterns of network recruitment, optogenetic network activation has recently been probed by combining optogenetic stimulation with blood oxygenation level-dependent (BOLD) fMRI, which represents a brain-wide readout capable of monitoring effects of large-scale network activation. Following first reports on the causal link between optogenetically induced neuronal activity and BOLD signals,7,8 the combination of optogenetics and BOLD fMRI has been explored in rodents, both anesthetized and awake, as well as in primates.9–18 Different aspects of optogenetically evoked hemodynamic responses as well as the suitability of optogenetic stimulation to recruit functional networks of the brain7,8,11,15–17 and to reproduce sensory stimulation18 have been assessed previously.
However, BOLD fMRI is an indirect measure of neuronal activity, based on the principle of neurovascular coupling.19,20 Due to the vascular origin of the BOLD signal, the spatial and temporal precision of neuronal network activity assessment is limited. Therefore, ofMRI studies are routinely combined with electrophysiological population recordings.7,8,11,12,15–17 Since, furthermore, neither the BOLD signal nor electrophysiological population recordings such as LFP are sensitive to spatially defined spiking activity only,21 these methods cannot resolve whether network activation induced by optogenetic stimulation is limited to ChR2-expressing neurons that are primarily activated. The spatiotemporal extent of optogenetic network activation could be significantly larger due to an immediate secondary recruitment of non-ChR2 expressing neurons within the heavily interconnected network.22,23 This is certainly the case upon sensory stimulation, yet optogenetic stimulation is creating an unavoidably artificial spatial pattern of activity, as we currently cannot selectively transduce exclusively those network components activated upon sensory stimulation.
Ideally, to match the spatiotemporal precision of optogenetics, a brain-wide measure of neuronal network activity as BOLD fMRI would be combined with a readout allowing for an assessment of local optogenetic network activation with high specificity toward neuronal spiking and high spatiotemporal resolution. Optic fiber-based Ca2+ recordings upon region-specific staining with fluorescent Ca2+ indicators meet those needs.4,24,25 Monitoring somatic Ca2+ of a small neuronal population—typically between 30 and 1000 cells—detects the spiking activity of this neuronal ensemble.26 These population-based Ca2+ recordings have already been combined with fMRI,25 unperturbed by the magnetic field. In the context of our study, one additional advantage is the possibility to use the same optic fiber for delivering high-intensity light pulses for optogenetic stimulation27 and for the Ca2+ readout.
Here, we investigated the similarity of optogenetic and sensory activation of brain networks in terms of (1) evoked primary neuronal response and its relation to the BOLD response, and (2) numbers and density of primarily activated neurons upon optogenetic stimulation. For this purpose we have implemented the combination of simultaneous optogenetic stimulation and Ca2+ recordings during BOLD fMRI measurements.
Materials and methods
All experiments were carried out according to the German Tierschutzgesetz, approved by the Landesamt für Natur und Umweltschutz Nordrhein-Westfalen, Germany (A787.54.04.2010.A274), and are reported according to the ARRIVE guidelines. We performed experiments on 27 female Fisher rats with a body weight between 160 and 180 g. Twenty-two rats obtained fMRI and Ca2+ recordings, out of which four rats received additional LFP recordings. Three rats were subjected to Ca2+ recordings only, and two additional rats were used for histology only.
Recording setup
We used a custom-built setup for optical fiber-based Ca2+ recordings and optogenetic stimulation. The light for excitation of the fluorescent Ca2+ indicator, Oregon Green 488 BAPTA-1 (OGB-1, Invitrogen, Life Technologies, Carlsbad, CA) and for stimulation of ChR2 was delivered by a blue 20 mW solid-state laser at 488 nm (Sapphire, Coherent, Dieburg, Germany). An acousto-optic modulator (AOM 3080-125, Crystal Technology, Palo Alto, CA) was used for rapid control of laser intensity. The laser beam was coupled into a 200 µm optic multimode fiber using a fiber collimator (Thorlabs, Grünberg, Germany). The emitted fluorescent light was guided back through the fiber, separated from the excitation light with a dichroic mirror and detected by an avalanche photodiode (LCSA500-01, Lasercomponents GmbH, Olching, Germany). The signal was sampled at 2 kHz using a multifunction data acquisition board (PCI 6259, National Instruments, Austin, TX) and a custom-written LabView (National Instruments) script.
Stereotactic viral injections
Stereotactic injections were conducted under isoflurane (Forene, Abbott, Wiesbaden, Germany) anesthesia in 18 animals. Rats were placed on a warming pad (37 ℃) and fixed in a stereotactic frame. A small craniotomy was made with a dental drill (Ultimate XL-F, NSK, Trier Germany, and VS1/4HP/005, Meisinger, Neuss, Germany) with the aid of a dissecting microscope. Solutions were delivered by a glass micropipette connected to a 10 ml syringe by manual pressure.
Animals were pretreated with the analgesic Metacam (1 mg/kg s.c.). For transduction of ChR2, two adeno-associated virus (AAV) preparations, serotype 2, were mixed at a ratio of one volume AAV-CAG-Cre and four volumes AAV-EF1A-DIO-hChR2(H134R)-YFP/mCherry. 0.5 µl of the viral solution was slowly injected via a small craniotomy into primary somatosensory cortex, frontlimb region (S1FL; AP 0 mm, ML + 3.5 mm, DV 0.8 mm) according to stereotactic coordinates (Paxinos Watson Rat brain atlas28). To avoid reopening of the craniotomy for subsequent Ca2+ indicator injections, we conducted an angular injection in 12/18 animals (55 ° vertical from medial; AP 0 mm, ML + 2.5 mm, DV 1.2 mm). The scalp incision was closed with a suture. Optical stimulation and recordings were carried out after a minimum of 14 days after viral injections (cluster size of BOLD responses was not influenced by time after virus injection).
Administration of fluorescent Ca2+ indicator
OGB-1 was prepared as previously described29 and injected at a concentration of 1 mM in S1FL, at AP 0 mm, ML + 3.5 mm under surgical depth of anesthesia at DV 700, 500, and 300 µm (approximately 0.3 µl each).
Optical fiber implantation
After removing the cladding, the fiber delivering blue light for both optogenetic stimulation and Ca2+ recordings was inserted via the craniotomy into the stained region, at a depth of 300 µm in S1FL and glued to the skull with UV glue (Polytec, PT GmbH, Waldbrunn, Germany).
Electrical recordings
Local field potential (LFP) recordings were performed using an EXT-02F/2 amplifier (npi Electronic, Tamm, Germany). Pipettes with a tip resistance of 1–2 mOhm filled with phosphate buffered saline (Sigma) were inserted through a second craniotomy lateral to the fiber insertion site at a depth of 300 µm. The final position of the tip was aimed to target the same region as the optical fiber. Signals were filtered at 300 Hz (low pass) without high pass filter, digitized at 2 kHz, and acquired together with the optical signals using the same acquisition program.
Fluorescence microscopy and cell quantification
After experiments, rats were transcardially perfused with 4% paraformaldehyde (PFA) under deep isoflurane anesthesia. Brains were excised, fixed overnight (4% PFA), and transferred to 30% sucrose solution. For characterization of opsin expression, coronal sections were prepared using a vibratome (Leica, Wetzlar, Germany). For the identification of neuronal somata, in two animals, slices were stained with green fluorescent Nissl stain (Neurotrace, Molecular Probes, Life Technologies, Carlsbad, CA). Briefly, to permeabilize cells, slices were incubated with 0.1% Triton X-100 (Invitrogen, Life Technologies, Carlsbad, CA) in PBS. Thereafter, slices were stained with Neurotrace (1:300) for 60 min and washed in PBS, followed by treatment with 0.1% Triton X-100 in PBS overnight. Slices were mounted using antiquenching Vectashield (Vector Laboratories, Burlingame, CA).
Quantification of ChR2-expressing cells and estimation of light-activated cells
To determine the total number of ChR2-expressing cells in cortex, slices (100 µm thickness) of two animals were imaged with a confocal microscope (SP8, Leica, Mannheim, Germany) and a 20x (HCX PL APO dry, Leica) objective, with a numerical aperture of 0.70. Using ImageJ30 software we quantified the total number of ChR2-expressing cells by counting all cells with a smooth, membrane-bound fluorescence in image stacks covering the entire area of opsin expression.
To derive densities of ChR2-expressing neurons, Nissl-stained coronal slices with a slice thickness of 70 µm of two additional animals were used, covering more than 800 µm in anterio-posterior direction. From these, four slices of each animal in the center of the expression area were used for stereological assessment. Morphometric measurements were performed on a stereology workstation consisting of a light microscope (Olympus BX51, Japan, equipped with a 20X NA = 0.75 UPlanSApo Olympus objective), a motorized specimen stage, CCD video color camera, and stereology software (StereoInvestigator, Microbrightfield, Williston, USA) as described previously.31 The region of interest (ROI) in the cortex was determined using the optical dissector probe of the StereoInvestigator software. The region of homogenous virus expression was outlined manually (ROI). Cells were distinguished by both color and morphological distribution of fluorescence: First, all cells exhibiting green cytosolic fluorescence were defined as neurons positive for Neurotrace. Second, all cells showing homogenous membrane-bound red fluorescence were defined as neurons functionally expressing ChR2-mCherry. Both sets overlapped; all red fluorescent cells exhibited also green fluorescence, whereas only a subset of green fluorescent cells additionally exhibited red fluorescence. Counting frames of 100 µm × 100 µm were randomly placed by the software within the ROI, sampling a 3D ROI by an unbiased counting procedure. In these frames, all ChR2-mCherry-expressing neurons and all Neurotrace-stained neurons were counted. Considering the total volume of the expression area per slide, defined by the slice thickness and the ROI, the average number of cells in the randomly placed counting frames was used to extrapolate the cells per mm3 within a virus expressing area (cells/mm3).
Calculating light transmission through cortical tissue
To calculate the tissue penetration depth of light in one dimension (z), we consider an optical fiber with radius r = 0.1 mm and an aperture of NA = 0.48, running along the z-coordinate, and having a perfectly planar and circle shaped ending at z=0 in brain tissue with refractive index n = 1.36.32 We assume monochromatic light of intensity I0 in the fiber and homogeneous tissue with isotropic scattering. The same assumptions and geometry were used previously by Aravanis et al.32 to model light penetration by a combination of losses from scattering and absorption, described by a transmission factor T(z) using the Kubelka–Munk model, and decreasing light intensity due to geometrical spread, described by a geometry factor G(z).
We follow a similar approach and assume that the light intensity in tissue I(z) can be described as
| (1) |
Using the transmission factor according to the Kubelka–Munk model for diffuse scattering media
| (2) |
where S is a damping constant (S=10.3 mm−1 in rat gray matter, as measured by Aravanis et al.32).
This model assumed a conical spread of light from the fiber tip, which is characterized by an angle of divergence, given by . Along z, the increasing cross sectional area of the cone can be calculated from , which can be expressed in terms of . With the definition of
| (3) |
the geometry factor for conical light spread can be expressed as
| (4) |
And consequently
| (5) |
However, recent experimental measurements have shown that for blue light, propagation deviates from a conical spread.33 In particular, the lateral illumination is largely underestimated. To account more accurately for this volume, we assume that geometrical spread is dominated by scattering. The initial light intensity through the fiber cross section propagates over a half sphere with radius R from the center of the fiber. The geometry factor becomes
| (6) |
and the light intensity
| (7) |
fMRI procedures
MRI was performed on a 9.4 T small animal imaging system with a 0.7 T/m gradient system (Biospec 94/20, Bruker Biospin GmbH, Ettlingen, Germany) equipped with a RF surface coil with fiber lead-through. Animals were mounted on a heated MRI cradle and supplied with a mixture of 80% air and 20% oxygen. The skull was covered with a 1–2 mm thick layer of dental alginate (Weiton, Johannes Weithas dental-Kunststoffe, Lütjenburg, Germany) or 1% agar, to reduce susceptibility artifacts at the bone–air interface. The fiber was guided through the RF surface coil placed on the animal's head. Local shimming was applied (shimming volume 0.6–1.6 cm³, Mapshim, Bruker). For anatomical images, a T2-weighted 2D RARE sequence, TR/TE 2000/12.7 ms, RARE factor 8, 256 matrix, 110 ×100 µm2 spatial resolution and slice thickness 1.2 mm, 6–9 contiguous slices was used. For BOLD fMRI measurements, T2*-weighted images were acquired with a single-shot gradient echo EPI (Echo Planar Imaging) sequence with TR = 1 s and TE = 18 ms. The spatial resolution of the MR images was 350 × 325 µm2 and slice thickness 1.2 mm, 6–9 contiguous slices.
In vivo BOLD fMRI and Ca2+ recordings
For sensory stimulation, two needle electrodes were subcutaneously inserted into the left forepaw (between digits 2 and 4) and connected to a constant current stimulator (DS5, Digitimer, Welwyn Garden City, UK). Stimulation pulses were of constant duration (1 ms) and strength (1 mA), if not stated otherwise. We used a block paradigm with a pulse train of 10 s length at 9 Hz (if not stated otherwise), followed by 20 s baseline, for 10 min, resulting in 20 stimulations per experiment.
In four animals, subjected to fMRI and consecutive simultaneous Ca2+ and LFP recordings, the stimulation frequency of electrical pulse trains of 10 s duration was varied between 3, 5, 7, 9, and 12 Hz.
In three animals we performed single-pulse electrical stimulations (10 ms duration, one pulse every 10 s) at different stimulation strengths of 0.5, 1.0, 1.5 mA and acquired Ca2+ recordings. For optogenetic stimulation, we used light pulses of 10 ms duration at 9 Hz with a light intensity of 80 mW/mm2 at the tip of the fiber, below the threshold of heat-induced artifacts (unpublished data). For optogenetic stimulation during Ca2+ recordings, high-intensity light pulses were switched on during continuous illumination (1.3 mW/mm2) for fluorescence excitation. Functional data acquisition was synchronized with the stimulation series.
When animals were transferred to the scanner, sedation was induced by a subcutaneous bolus injection of 0.04 mg/kg medetomidine (Domitor 1 mg/ml, Pfizer, Orion Pharma, Espoo, Finland), followed by continuous subcutaneous infusion of 0.05 mg/kg/h medetomidine. Isoflurane inhalation was reduced stepwise while monitoring the respiration rate and discontinued within the first 5 min after bolus injection. As described previously,34 we observed a drop of heart rate, respiration rate, and mean arterial blood pressure directly after starting the medetomidine infusion. In a subset of experiments we performed transcutaneous pCO2 measurements and found that elevated CO2 levels after starting medetomidine infusion declined to a stable value during 30 min. Experiments were performed 30 min after change of anesthesia. Optic fiber recordings were performed at least 1 h after dye injection to allow for the cleavage of OGB-1 ester. Respiration rate and body core temperature were routinely monitored during experiments. The duration of functional experiments did not exceed 2 h and was terminated earlier if animal physiology did not remain stable. The temporal sequence of stimulation paradigms was varied between animals.
fMRI analysis
fMRI data from individual animals were analyzed with SPM8 (Statistical Parametric Mapping) (Functional Imaging Laboratory, Wellcome Trust Centre for Neuroimaging, UK London). The initial five scans were discarded from each imaging series to remove signal variations at the beginning of data acquisition. Remaining images were motion corrected and spatially filtered with a Gaussian kernel of 0.5 mm. BOLD time courses were obtained by summing image intensities over a ROI placed inside the target region, over all 20 stimulations of one experiment. The area under the curve for the BOLD response was determined from average time courses over all experiments with sensory or optogenetic stimulation, respectively, by integrating the percent signal change deviations of the time courses from the baseline. For assessment of the size of activated clusters and graphical representation of the activation maps, the canonical hemodynamic response function implemented in SPM was used to model the BOLD response to stimulation. Statistical significance level for activation maps was set at p < 0.05, corrected for multiple comparisons (family wise error (FWE)). Resulting t-score maps were superimposed to the mean EPI images and average cluster size ± standard error of the mean (SEM) was reported. For the analysis of the frequency dependence of the BOLD response, spatially filtered data were subjected to a t-test at a significance level of p < 0.001 in ImageJ. The BOLD response (%) was superimposed to a smoothed representative EPI image set and average cluster size ± SEM was reported.
Analysis of Ca2+ and LFP recordings
Analysis of optical and LFP recordings was performed with Igor (WaveMetrics, Portland, OR). All Ca2+ traces represent relative changes in fluorescence (Δf/f). Amplitudes were determined as the intensity difference (Δf) from the baseline and the highest intensity value of the transient.
Because of the same excitation wavelength for Ca2+ indicator and ChR2, the detector was saturated by the high fluorescence emission of the indicator during the high-intensity optic stimulation pulses, as in previous studies.4,24 We used low light intensities (1.3 mW/mm2) for constant excitation of OGB-1 versus high intensities (80 mW/mm2) for pulsed excitation of ChR2. For analysis of Ca2+ signal following optogenetic stimulation, the signal resulting from detector saturation was removed using two different approaches. After the high-intensity light pulse the saturation decayed. This decay could be fitted with a monoexponential function (see suppl. Figure 1) with a time constant of 0.9954 ±0.0038 ms (R2 = 0.9998). In the initial approach, the stimulus artifact was removed for the first 25 ms after onset of the stimulation pulse (10 ms). In the second approach, the detector recovery curve (monoexponential signal decay after saturation) was subtracted from the raw data (Suppl. Figure 1). With this correction, already 5–6 ms after the end of the optical pulse, the Ca2+ signal could be identified without a residual signal contribution from the detector recovery.
Data were tested for normal distribution using Lilliefors test. If normal distribution could be assumed (p > 0.05), the parametric two-tailed Student's t-test was employed to compare means. A p-value below 0.05 was considered significant.
Results
Implementing a novel setup that allows for simultaneous optical recordings and optogenetic stimulation during BOLD fMRI
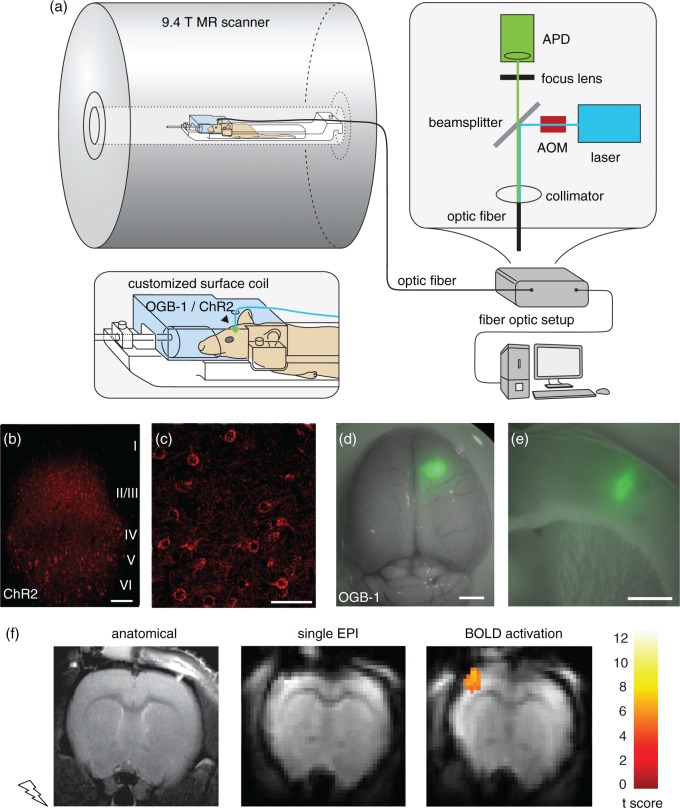
We established a setup combining in vivo optical Ca2+ recordings, optogenetic neuronal control, and BOLD fMRI on a 9.4 T small animal MR scanner (Figure 1(a)). We used a custom-built optical setup providing simultaneous excitation of both the Ca2+-sensitive fluorescent indicator OGB-1 and the optogenetic actuator ChR24 through one 200 µm diameter multimode optical fiber.
Figure 1.
Setup for simultaneous optical recordings and optogenetic stimulation during BOLD fMRI. (a) Scheme of experimental setup. Optic fiber-based stimulation/recording setup is placed outside of the scanner cabin. Light intensities of the blue laser are modulated with high temporal precision by use of an acousto-optic-modulator (AOM), emitted light is recorded by an avalanche photodiode (APD). The customized surface coil has a lead-through for the optical fiber. Electric stimulation, laser excitation, fMRI data acquisition, and optical recordings are synchronized with a customized LabView program. (b, c) Confocal images from fixed sections of rat brain injected with ChR2-mCherry AAV in somatosensory cortex. (b) mCherry expression in cortical layers II/III, V, and VI becomes evident. Note that layer IV does not exhibit fluorescence; scale bar = 100 µm. (c) High-resolution imaging reveals membrane-bound localization of ChR2-mCherry fusion protein; scale bar = 50 µm. (d, e) Photomicrographs of rat brain injected with Ca2+ indicator OGB-1. (d) Whole rat brain, overlay of transmitted light image with green fluorescence channel; scale bar = 2 mm. Spatially confined area of OGB-1 staining is visible in right somatosensory cortex. (e) Coronal brain slice at the level of the somatosensory cortex of OGB-1 fluorescence; scale bar = 1.5 mm. Staining is restricted to the cortex, with a diameter of approximately 600 µm. (f) Anatomical RARE (left) and functional EPI MR images (center) of one animal, and BOLD activation map showing response upon forepaw stimulation (10 s at 9 Hz, 1 mA, 1 ms) overlayed on mean EPI image (right). The color scale represents the t-score of the BOLD analysis. (g) Averaged BOLD time course of experiment shown in (f). (h) Ca2+ responses from same experiment as (f, g). (i) Primary Ca2+ responses induced by forepaw stimulation at 3 Hz, 10 s pulse train.
For experiments combining BOLD fMRI with Ca2+ recordings and ChR2 stimulation, we injected ChR2 encoding AAVs stereotactically in rat somatosensory cortex (S1FL) of 18 rats. Confocal imaging of histological slices revealed strong, yet sparse expression of ChR2-mCherry in neurons of layer II/III, layer V, and layer VI (Figure 1(b)). At the cellular level, membrane-bound expression with no apparent cytotoxicity was observed (Figure 1(c)). For Ca2+ recordings, we stereotactically injected OGB-1 into the same region of rat somatosensory cortex, resulting in a circumscribed stained column with a mean diameter of approximately 600 µm (Figure 1(d) and (e)). Subsequently, the optical fiber was implanted dorsal to the stained area. With this setup, optical Ca2+ detection could be performed without any distortions by radio frequency, static or dynamic magnetic fields of the MR scanner. Slight MR signal intensity disturbances at the site of fiber implantation were observed in 15 of 18 animals, caused by fiber or dye injection in anatomical T2-weighted and corresponding EPI MR images.
Forepaw stimulation resulted in a positive BOLD response in contralateral primary somatosensory cortex (37 ± 13 voxels, mean cluster size ± SEM) in 11 of 18 animals (Figure 1(f)). Without correction for the FWE, 14 of 18 animals showed activation. The BOLD time course showed a delayed rise in signal after the stimulation onset, peaking approx. 5 s after stimulation onset and returning to baseline approx. 5 s after the end of the stimulation (Figure 1(g)). In simultaneous Ca2+ recordings, sensory stimulation resulted in Ca2+ transients with short rise times and uniform latencies. At 9 Hz, Ca2+ transients showed initial adaptation in terms of a decrease in amplitude, followed by a rebound of transients in this animal (Figure 1(h)). In contrast, for 3 Hz stimulation frequency (Figure 1(i)) nearly every pulse evoked a Ca2+ response.
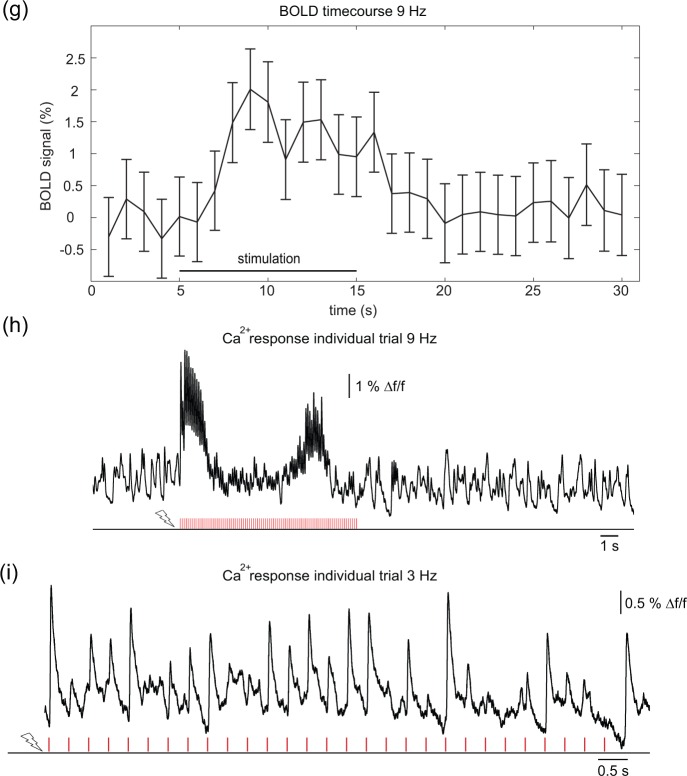
Differential response of suprathreshold Ca2+ and BOLD signals upon increasing frequencies of sensory stimulation
To probe the stimulation frequency dependence of fMRI and Ca2+ recordings, electrical pulse trains of 10 s duration were varied between 3, 5, 7, 9, and 12 Hz (Figure 2). With higher frequency, a more pronounced BOLD signal was observed (Figure 2(a) and (b)) while Ca2+ responses showed adaptation, evident on the level of individual pulse trains (Figure 2(c)), as well as in averages of pulse trains over an entire experiment (Figure 2(d)). No robust BOLD response was detected at 3 Hz. Mean cluster sizes within S1FL for the different stimulation frequencies were 14 ± 3 for 5 Hz, 11 ± 3 for 7 Hz, 20 ± 4 for 9 Hz, and 12 ± 4 at 12 Hz. The time course of the BOLD response was determined in S1FL and averaged over all experiments. At 7 Hz, a weak BOLD signal change of approx. 0.4% was discernible. At 9 Hz the highest signal change of approx. 0.6% could be detected (Figure 2(b)). Ca2+ recordings in S1FL showed a primary neuronal response following nearly each stimulation pulse at 3 and at 5 Hz stimulation. At higher frequencies a faster decrease of signal amplitudes indicated earlier adaptation. A plot of average BOLD versus Ca2+ responses shows that different frequencies elicit different relations of BOLD and Ca2+ response. An increase in frequency is not leading to a related increase in BOLD and Ca2+ (Suppl. Figure 2). LFP recordings, reflecting both supra- and subthreshold activity, showed less prominent adaptation at higher frequencies (Suppl. Figure 3).
Figure 2.
Impact of varying stimulation frequencies on BOLD activation and Ca2+ responses. Rats were subjected to forepaw stimulation at different frequencies with a pulse train duration of 10 s. (a) Exemplary BOLD maps from one animal. Weak activation was observed at 5 and 7 Hz, strong activation at 12 Hz, and strongest activation at 9 Hz. Color bar represents % signal change. (b) Averaged BOLD time courses in S1FL upon stimulation (four animals, eight experiments). Ten-second stimulation period is indicated on top (from 5 to 15 s). (c) Exemplary single-trial Ca2+ responses upon stimulation are shown only for the stimulation period (5–15 s) as indicated in (b). The number of evoked responses decreased at high frequencies (9 and 12 Hz). (d) Averaged Ca2+ responses of stimulation periods of entire trials (one animal). 3D graphs show normalized amplitude (vertical axis), number of pulses within the stimulation sequence (horizontal axis), and interstimulus intervals (time axis). Response amplitudes were similar for all stimulation pulses within a stimulation sequence at low frequencies (3 and 5 Hz). At 7, 9, and 12 Hz increasing adaptation became apparent. Note the different lengths of time axes due to the decrease of the interstimulus interval with higher stimulation frequency.
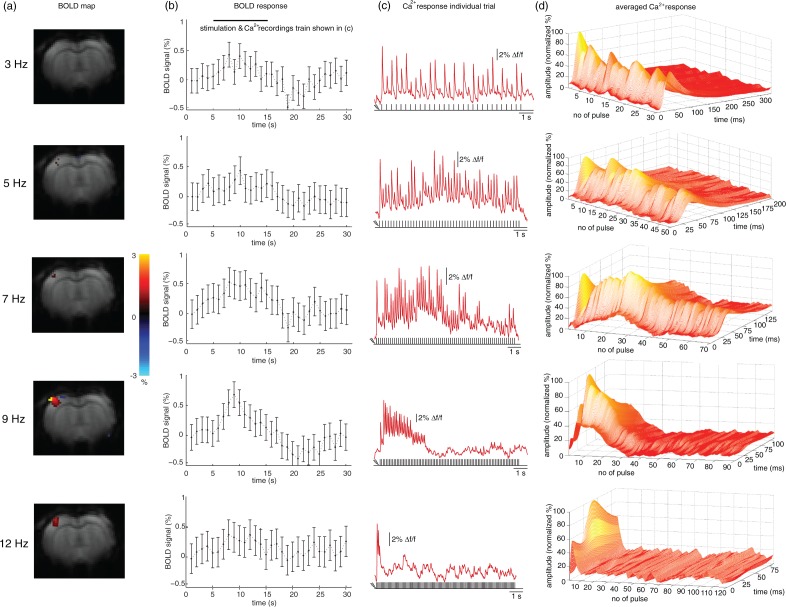
Assessing the density of ChR2-expressing neurons
To estimate the number of ChR2-expressing cells activated by light from the optic fiber, we quantified the total number of ChR2-expressing neurons in cortex in confocal images of two animals. The entire area of strong, membrane-bound expression spanned 800–1000 µm in anterior–posterior and 950–1200 µm in medial–lateral direction of the neocortex. A total number of 810 and 835 ChR2-expressing neurons was counted (Figure 1(b) and (c)).
In two additional animals with cortical ChR2 injections, we evaluated the proportion of ChR2-expressing neurons in relation to the total number of neurons in the local network. We conducted a semiautomatized cell counting within the area of strong, homogenous ChR2-mCherry expression (Figure 3(a)) using Nissl-stained coronal slices at the level of S1. In dorso-ventral direction, we detected homogenous ChR2 expression in layers V/VI and layers II/III. Within layer IV, almost no ChR2-expressing neurons could be found.
Figure 3.
Assessment of the density of ChR2-expressing neurons and their optogenetically activated subset. (a) Fluorescence micrograph, used for cell quantifications, displaying all neurons in green (Neurotrace staining), and the subset of ChR2-mcherry-expressing neurons in red. Coronal section at the level of layer V. Scale bar = 100 µm. (b) Schematic of a 100 µm3 tissue volume illustrating the average number of neurons and portion of transduced neurons in somatosensory cortex. (c) Scheme of distribution of ChR2-expressing neurons in cortex and illumination with blue light emitted from optical fiber. (d) Logarithmic plot of light attenuation calculated with Aravanis model (squares) and spherical model (circles). Vertical scale is also valid for (c).
All neurons exhibiting smooth, membrane-bound mCherry fluorescence were defined as ChR2-positive cells. For assessing the total number of neurons, all cells exhibiting somata with delineated green fluorescence were counted. These quantifications resulted in average cell densities of 32,300 ± 3400 (animal 1) and 30,600 ± 7700 (animal 2) neurons per mm3, similar to densities found in the rat barrel cortex using an alternative approach.35 The density of ChR2-expressing neurons ranged at 2200 ± 500 (animal 1) and 1800 ±300 neurons (animal 2) per mm3. Consequently, 6% (animal 2) and 7% (animal 1) of the local neuronal population expressed ChR2 and could potentially be activated by a light pulse (for a schematic illustration see Figure 3(b) and (c)).
Estimating the number of optogenetically activated neurons
The number of activated cells depends on the density of neurons expressing the opsin and on the light intensity sufficient for ChR2 activation reaching those cells. We assumed a threshold to drive a ChR2-mediated action potential at 1 mW/mm2,23,32,36,37 and estimated the volume of tissue illuminated with above-threshold light power density from two different models of light propagation through brain tissue (Figure 3(d) and Suppl. Figure 4). The Aravanis model, equation (5)32 assumes light spreads in a perfect conical shape. However, it largely underestimates lateral illumination and overestimates penetration, as recently shown by light propagation measurements.33 To account for these shortcomings, we used a second model (spherical model, equation (7)) that assumes strong scattering leading to a spherical propagation, most likely overestimating lateral spread and underestimating penetration. Using the experimental light intensity I0 = 80 mW/mm2, above threshold volumes of 0.098 and 0.062 mm3 were obtained for Aravanis and spherical model, respectively. Assuming that no ChR2 was expressed in layer IV and using the above calculated homogeneous cell densities in layer II/III and V/VI (Figure 3(c) and (d)), the number of typically activated cells, ranged between 189 and 155 neurons for the Aravanis model, and between 115 and 94 neurons for the spherical model, for animal 1 and animal 2, respectively.
Spatial and temporal characteristics of optogenetically evoked BOLD
Functional data were acquired using GE-EPI and animals were subjected to alternating optogenetic and forepaw stimulations. Light pulses of 10 ms duration at 9 Hz with a light intensity of 80 mW/mm2 at the tip of the fiber resulted in a local BOLD activation at the site of virus injection in 16 of 18 rats with a cluster size of 40 ± 8 voxels (17 of 18 uncorrected). Optogenetically induced activation areas overlapped with sensory-evoked activation areas observed in 11 animals (Figure 4 and Suppl. Figure 5).
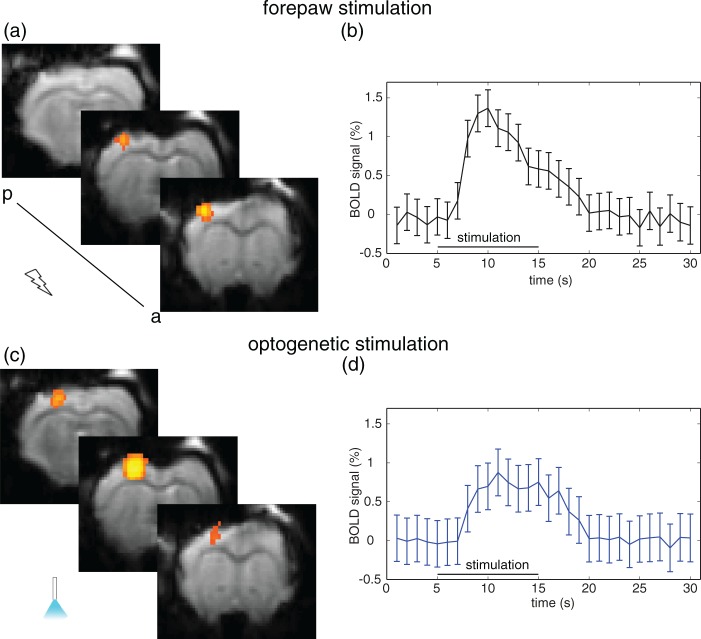
Figure 4.
BOLD response upon sensory and optogenetic stimulation. (a) Representative activation map upon electric forepaw stimulation (1 mA), and (b) averaged time course (11 animals) showing the BOLD response in S1FL. Peak BOLD amplitude was 1.3%, and the area under the curve was 10.2 a.u. (c) Representative activation map upon optogenetic stimulation, and (d) averaged time course (11 animals) showing the BOLD response in the optogenetically activated region. Peak BOLD amplitude was 0.7%, the area under the curve was 7.3 a.u.
Temporal dynamics of BOLD responses were similar for sensory and optogenetic stimulations (Figure 4). Both time courses, averaged over all activated voxels in 11 animals, showed an increase of 3 s after the start of the stimulation and reached a maximum after 5–6 s that decayed to baseline level after 20 s. BOLD amplitude was approx. 0.7% for optogenetic and 1.3% for forepaw stimulation, decaying over 10 s. The area under the curve was 10.2 ± 0.9 a.u. for sensory and 7.3 ± 1.1 a.u. for optogenetic stimulation.
Assessment of secondary recruitment of optogenetic stimulation
We assessed the temporal dynamics and magnitude of the recruitment of the neuronal network, using optical Ca2+ recordings as region-specific readout of neuronal spiking activity. To this end, we first subjected animals to forepaw stimulation pulses of varying strength, yet well below pain threshold (Figure 5(a)). For all stimulation strengths we found identical temporal dynamics, yet a significant increase in peak amplitude for higher stimulation strength. This shows that Ca2+ recordings can serve as a quantitative readout of neuronal population activity.
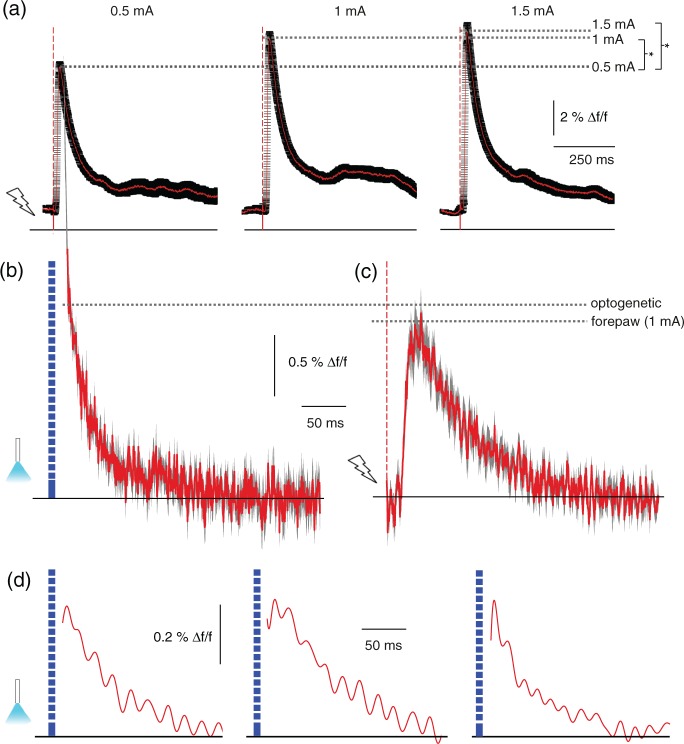
Figure 5.
Cortical optogenetic stimulation did not result in a widespread secondary recruitment of the neuronal network. (a) Primary Ca2+ responses induced by forepaw stimulation, single pulses at different stimulation strength. Mean (red line) ± SEM (black line), significant increase of peak amplitudes comparing 0.5 to 1 mA (p < 0.001, 21 versus 18 transients) and comparing 0.5 to 1.5 mA (p < 0.001, 18 versus 23 transients). (b) Average primary responses upon optogenetic stimulation with a pulse duration of 10 ms in a different animal expressing ChR2 in somatosensory cortex, mean (red line) ± SEM (black line), 11 transients. Ca2+ transients were recorded after single stimulation pulses without preceding fluorescence excitation to exclude influence of constant illumination. Note that the stimulation artifact has been removed. (c) Average responses of same animal upon 1 ms, 1 mA electric forepaw stimulation, identical scaling as in (b), mean (red line) ± SEM (black line), 11 transients. Importantly, fluorescence intensity changes can only be compared within the same experimental subject. (d) Single Ca2+ transients from three different animals upon optogenetic stimulation. Exponential detector recovery was removed and data were low-pass filtered. Data are shown starting at 6 ms after the end of the optic pulse.
For assessment of the response to optogenetic activation, we used low light intensity for constant excitation of OGB-1 and high intensity for pulsed excitation of ChR2.4 We could not record neuronal activity during the short stimulation pulse. Due to the same excitation wavelength for Ca2+ indicator and ChR2, the detector was saturated by the high fluorescence emission of the indicator during the high-intensity optic stimulation pulses (stimulation artifact, see Suppl. Figure 1), as in previous studies.4,24 Despite the stimulation artifact, we could estimate the amplitude of neuronal responses by subsequently stimulating with either electric forepaw pulses or combined electric and optic pulses (Suppl. Figure 6). Similar signal amplitudes for both paradigms were observed. Yet, latency and rise times could not be resolved. We compared averaged primary neuronal Ca2+ responses to either optogenetic or 1 mA sensory stimulation and found similar amplitudes and a monotonous signal decay (Figure 5(b) and (c)). After subtracting the detector saturation from the data, single responses could be resolved starting from 6 ms after end of the stimulation pulse (Suppl. Figure 1). Monotonic signal decay was observed throughout (Figure 5(d)), indicating a primary neural response as origin of the detected signal.
Discussion
In this study, we simultaneously recorded BOLD fMRI and Ca2+ transients upon optogenetic and forepaw stimulation. This novel methodological combination affords both to manipulate neuronal networks and to detect specifically neuronal spiking activity during simultaneous brain-wide functional imaging by BOLD fMRI. We employed Ca2+ recordings for a spatially confined optical readout of neuronal spiking activity, to assess the cortical representation of sensory and local optogenetic stimulation. We found a similar scale of local neuronal network recruitment upon both stimulation paradigms. To explore the extent and dynamics of optogenetic network recruitment, we assessed the relative portion of optogenetically activated neurons in the volume with suprathreshold light intensities. By using Ca2+ recordings as a specific, local and unperturbed fast spiking readout of neuronal activity, we were able to resolve primary neuronal response alongside brain-wide BOLD fMRI.
The application of optogenetics has great potential in advancing causal therapeutic strategies in neurological disorders, aiming at rebalancing neuronal circuitry.5,6 It has been shown previously that ChR2 can drive neuronal spiking using Poisson stimulus trains, which resemble endogenous stimulus patterns.3 Yet, on a network level, to emulate sensory inputs, optogenetic stimulation should recruit a similar proportion of the local network. Based on a combination of our experimental cell quantifications and the estimation of illuminated volume, we conclude that upon a light stimulus we recruit about 100–200 neurons, representing approximately 7% of the local neuronal population, within the area of activation. This percentage is in the same range as typically recruited by sensory stimulation.1 Yet, the complex interplay of cortical layers cannot be mimicked by local cortical optogenetic stimulation; we cannot recapitulate the temporal succession of the activation of the individual network components, even more given that no significant expression of ChR2 in layer IV could be found.4
Based on previous experiments that combined confocal image analysis with single-cell electrophysiology and titrated down the number of transduced cells,4 we assume that we indeed functionally activate those cell numbers determined by our quantification methods, provided that they are situated within the area of activation. However, the exact determination of activated neurons remains challenging, particularly in the in vivo situation as the spatial distribution of light in heterogeneous brain tissue can only be estimated.32,33 The volume of tissue illuminated with above threshold light was calculated with two models. The Aravanis model of conical light distribution, which is commonly accepted and routinely used in optogenetic studies,7,15 provides an estimate of maximum penetration depth and minimum lateral spread. The spherical model provides maximum lateral light spread and underestimates penetration depth. Therefore, our calculations define the limits of illuminated volume. The scope of optogenetic network activation will furthermore critically depend on virus titer, serotype, promoter, injected volume, and cytoarchitecture of the targeted regions. Nevertheless, our findings may serve as an order-of-magnitude estimate of network recruitment for future studies using comparable experimental protocols.
Direct cortical optogenetic stimulation predominantly activated those cortical layers with ChR2-expressing cells, layer II/III, V, and VI. Yet, the spatial and temporal characteristics of the BOLD response were similar to those observed upon sensory stimulation, in agreement with a recent comprehensive study of the neurovascular response upon optogenetic and sensory stimulation.18 We found that BOLD activity extended over up to three image slices, representing more than the illuminated area. However, the vascular recruitment may exceed the area of primary neuronal activation and BOLD signal will be detected from a larger volume. This effect may be more pronounced for optogenetic activation, as a recent study has measured higher oxygen demand upon optogenetic versus sensory stimulation.38
As a complementary functional readout we used optic fiber-based Ca2+ recordings to assess spatially confined spiking activity.39 Ca2+ imaging using single-cell loading of neurons with OGB-1 is capable of resolving Ca2+ dynamics of individual spines upon synaptic input.40 However, bolus loading of neuronal populations results in a predominant contribution of the increased somatic Ca2+ concentration to the population signal.24,41 High amplitude and fast changes in somatic Ca2+ levels are mainly driven by action potential-coupled influx of Ca2+ through voltage-gated Ca2+ channels.41
It has to be noted that the Ca2+ indicator OGB-1 is also taken up by astrocytes.41 However, even though astrocytes exhibit high-amplitude Ca2+ transients, we can exclude a significant contribution of astrocytic activity. The temporal profile of astrocytic Ca2+ transients differs significantly from neuronal responses, exhibiting slow rise times and durations in the range of up to 30 s,24,41 in contrast to durations of less than 100 ms of neuronal responses. Therefore, Ca2+ recordings may be interpreted as a selective readout of neuronal population activity.
We probed the relationship of Ca2+ recordings and BOLD fMRI. We employed sensory stimulations at different frequencies to differentiate between spiking responses and synaptic activity, as in cortex the spiking response adapts at higher frequencies by the postsynaptic mechanism of synaptic depression.42 The interrelation of sensory stimulation parameters on BOLD fMRI has been studied previously,43–45 showing a differential impact of stimulus frequency and duration on CBF, CBV, and BOLD, and reporting drastically lower BOLD amplitudes at low stimulation frequencies below 5 Hz,25,46–48 in agreement with our observations. While the BOLD response persisted during the stimulation trains for higher stimulation frequencies, the Ca2+ signal, reflecting spiking activity of a local neuronal population, already adapted within the first 1–4 s. LFP measurements, reflecting both spiking activity and subthreshold potentials, showed a less pronounced adaptation with increasing frequencies. This suggests a different adaptation behavior of spiking and subthreshold activity on local population level. Among other signal components, BOLD integrates both spiking as well as subthreshold potentials, and their relative contributions to the signal may vary.21,49,50 Consequently, BOLD and Ca2+ recordings provide complementary readouts of neural network activity.
Our results, showing similar BOLD time courses in response to optogenetic and sensory stimulation, support recent findings that network activation is comparable in both regimens.18 The BOLD amplitude after both stimulations scales with the stimulation strength.11,18 Yet, a similar BOLD time course does not exclude that upon optogenetic stimulation an imminent secondary recruitment of the network might dominate the network response.23 Our data provide a measure of suprathreshold neuronal network activity alongside BOLD upon optogenetic stimulation. Even though we could not record during the stimulus pulse due to the spectral overlap of the excitation wavelengths of ChR2 and OGB-1, we could reduce the downtime to 16 ms after stimulation onset. We can exclude that we miss a significant proportion of the transient, as the time of spike initiation upon ChR2 activation ranges at 7 ms3, and additional time is required for Ca2+ influx and binding to OGB-1. From the absence of a secondary response peak in the Ca2+ recordings, we conclude that the imminent local network activation is dominated by the primarily activated ChR2-expressing neurons. In future studies, the control of opsin expression by promoters of immediate early genes such as cfos51 might allow a direct recapitulation of the spatial patterns of network recruitment evoked by sensory stimulation.
Ultimately, optogenetics may not only serve as a basic science tool, but may allow for a causal assessment of therapeutic strategies in the preclinical setting, aiming at rebalancing aberrant circuit function.6 Here, we provide an order-of-magnitude estimate on the scope and temporal dynamics of local optogenetic network activation, mitigating concerns about a nonphysiological activation in ofMRI.23
Conclusions
The methodological approach that we have established here allows for a comprehensive assessment of the spatiotemporal dynamics of optogenetically and sensory evoked activity of local brain networks. A causal assessment of the contribution of individual network components to BOLD fluctuations alongside optical recordings of neuronal population activity becomes feasible, opening the door for investigating functional networks of the brain in unprecedented detail and comprehensiveness.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Amanda Tose and Zeke Barger for conducting confocal imaging, Michaela Moisch, for excellent technical assistance.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Bavarian State Ministry of Sciences, Research and the Arts (“ForNeuroCell”); the DFG (SFB 1080); the Focus Program translational Neuroscience (ftn); the Interdisciplinary Center for Clinical Research Münster (Fa3/01613 and PIX); and the Excellence Cluster Cells in Motion (DFG EXEC 1003, FF-2013-17).
Authors' contributions
FS, LW, MS, CF, and AS are equally contributing first/last authors. AS and CF designed the study. FS built the head coil and assembled the laser setup. FS and LW conducted the fMRI experiments. FS, LW, CF, CZ, and AS established the fMRI procedures; AS and LW established optogenetics. PHP and CFo implemented, conducted, and analyzed the LFP recordings. MS performed cell quantifications, GP conducted cell stainings. ERJ, FS, CF conducted the light propagation calculations; LW, FS, AS, and MS conducted and MS, FS, PHP, and AS analyzed optical fiber experiments. AS, CF, FS, LW, ERJ, and MS wrote the manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary material
Supplementary material for this paper can be found at http://jcbfm.sagepub.com/content/by/supplemental-data
References
- 1.Kuhlman SJ, Olivas ND, Tring E, et al. A disinhibitory microcircuit initiates critical-period plasticity in the visual cortex. Nature 2013; 501: 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olshausen BA, Field DJ. Sparse coding of sensory inputs. Curr Opin Neurobiol 2004; 14: 481–487. [DOI] [PubMed] [Google Scholar]
- 3.Boyden ES, Zhang F, Bamberg E, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 2005; 8: 1263–1268. [DOI] [PubMed] [Google Scholar]
- 4.Stroh A, Adelsberger H, Groh A, et al. Making waves: initiation and propagation of corticothalamic ca(2+) waves in vivo. Neuron 2013; 77: 1136–1150. [DOI] [PubMed] [Google Scholar]
- 5.Gradinaru V, Mogri M, Thompson KR, et al. Optical deconstruction of parkinsonian neural circuitry. Science 2009; 324: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 2011; 477: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JH, Durand R, Gradinaru V, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature 2010; 465: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai M, Kahn I, Knoblich U, et al. Mapping brain networks in awake mice using combined optical neural control and fMRI. J Neurophysiol 2011; 105: 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez AL, Fukuda M, Crowley JC, et al. Neural and hemodynamic responses elicited by forelimb- and photo-stimulation in channelrhodopsin-2 mice: insights into the hemodynamic point spread function. Cereb Cortex 2014; 24: 2908–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie IN, Wells JA, Southern P, et al. fMRI response to blue light delivery in the naive brain: implications for combined optogenetic fMRI studies. NeuroImage 2013; 66: 634–641. [DOI] [PubMed] [Google Scholar]
- 11.Kahn I, Desai M, Knoblich U, et al. Characterization of the functional MRI response temporal linearity via optical control of neocortical pyramidal neurons. J Neurosci 2011; 31: 15086–15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn I, Knoblich U, Desai M, et al. Optogenetic drive of neocortical pyramidal neurons generates fMRI signals that are correlated with spiking activity. Brain Res 2013; 1511: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerits A, Farivar R, Rosen BR, et al. Optogenetically induced behavioral and functional network changes in primates. Curr Biol 2012; 22: 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honjoh T, Ji ZG, Yokoyama Y, et al. Optogenetic patterning of whisker-barrel cortical system in transgenic rat expressing channelrhodopsin-2. PLoS One 2014; 9: e93706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitz AJ, Fang Z, Lee HJ, et al. Optogenetic fMRI reveals distinct, frequency-dependent networks recruited by dorsal and intermediate hippocampus stimulations. NeuroImage 2015; 107: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Z, Watson GD, Alloway KD, et al. Mapping the functional network of medial prefrontal cortex by combining optogenetics and fMRI in awake rats. NeuroImage 2015; 117: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takata N, Yoshida K, Komaki Y, et al. Optogenetic activation of CA1 pyramidal neurons at the dorsal and ventral hippocampus evokes distinct brain-wide responses revealed by mouse fMRI. PLoS one 2015; 10: e0121417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iordanova B, Vazquez AL, Poplawsky AJ, et al. Neural and hemodynamic responses to optogenetic and sensory stimulation in the rat somatosensory cortex. J Cereb Blood Flow Metab 2015; 35: 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SG, Ogawa S. Biophysical and physiological origins of blood oxygenation level-dependent fMRI signals. J Cereb Blood Flow Metab 2012; 32: 1188–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogawa S, Lee TM, Kay AR, et al. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci USA 1990; 87: 9868–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logothetis NK, Pauls J, Augath M, et al. Neurophysiological investigation of the basis of the fMRI signal. Nature 2001; 412: 150–157. [DOI] [PubMed] [Google Scholar]
- 22.Li CY, Poo MM, Dan Y. Burst spiking of a single cortical neuron modifies global brain state. Science 2009; 324: 643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logothetis NK. Bold claims for optogenetics. Nature 2010; 468: E3–E4. [DOI] [PubMed] [Google Scholar]
- 24.Grienberger C, Adelsberger H, Stroh A, et al. Sound-evoked network calcium transients in mouse auditory cortex in vivo. J Physiol 2012; 590: 899–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz K, Sydekum E, Krueppel R, et al. Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex. Nat Methods 2012; 9: 597–602. [DOI] [PubMed] [Google Scholar]
- 26.Rochefort NL, Garaschuk O, Milos RI, et al. Sparsification of neuronal activity in the visual cortex at eye-opening. Proc Natl Acad Sci USA 2009; 106: 15049–15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adelsberger H, Grienberger C, Stroh A, et al. In vivo calcium recordings and channelrhodopsin-2 activation through an optical fiber. Cold Spring Harbor Protocols 2014; 2014 pdb prot084145. [DOI] [PubMed] [Google Scholar]
- 28.Paxinos G, Watson C. The rat brain in stereotaxic coordinates, Amsterdam: Academic Press, 2013. . [Google Scholar]
- 29.Garaschuk O, Milos RI, Konnerth A. Targeted bulk-loading of fluorescent indicators for two-photon brain imaging in vivo. Nat Protoc 2006; 1: 380–386. [DOI] [PubMed] [Google Scholar]
- 30.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogt J, Paul F, Aktas O, et al. Lower motor neuron loss in multiple sclerosis and experimental autoimmune encephalomyelitis. Ann Neurol 2009; 66: 310–322. [DOI] [PubMed] [Google Scholar]
- 32.Aravanis AM, Wang LP, Zhang F, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng 2007; 4: S143–S156. [DOI] [PubMed] [Google Scholar]
- 33.Yizhar O, Fenno LE, Davidson TJ, et al. Optogenetics in neural systems. Neuron 2011; 71: 9–34. [DOI] [PubMed] [Google Scholar]
- 34.Weber R, Ramos-Cabrer P, Wiedermann D, et al. A fully noninvasive and robust experimental protocol for longitudinal fMRI studies in the rat. NeuroImage 2006; 29: 1303–1310. [DOI] [PubMed] [Google Scholar]
- 35.Meyer HS, Egger R, Guest JM, et al. Cellular organization of cortical barrel columns is whisker-specific. Proc Natl Acad Sci USA 2013; 110: 19113–19118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bi A, Cui J, Ma YP, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron 2006; 50: 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishizuka T, Kakuda M, Araki R, et al. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res 2006; 54: 85–94. [DOI] [PubMed] [Google Scholar]
- 38.Parpaleix A, Goulam Houssen Y, Charpak S. Imaging local neuronal activity by monitoring PO(2) transients in capillaries. Nat Med 2013; 19: 241–246. [DOI] [PubMed] [Google Scholar]
- 39.Adelsberger H, Garaschuk O, Konnerth A. Cortical calcium waves in resting newborn mice. Nat Neurosci 2005; 8: 988–990. [DOI] [PubMed] [Google Scholar]
- 40.Jia H, Rochefort NL, Chen X, et al. Dendritic organization of sensory input to cortical neurons in vivo. Nature 2010; 464: 1307–1312. [DOI] [PubMed] [Google Scholar]
- 41.Grienberger C, Konnerth A. Imaging calcium in neurons. Neuron 2012; 73: 862–885. [DOI] [PubMed] [Google Scholar]
- 42.Puccini GD, Sanchez-Vives MV, Compte A. Selective detection of abrupt input changes by integration of spike-frequency adaptation and synaptic depression in a computational network model. J Physiol 2006; 100: 1–15. [DOI] [PubMed] [Google Scholar]
- 43.Hirano Y, Stefanovic B, Silva AC. Spatiotemporal evolution of the functional magnetic resonance imaging response to ultrashort stimuli. J Neurosci 2011; 31: 1440–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goloshevsky AG, Silva AC, Dodd SJ, et al. BOLD fMRI and somatosensory evoked potentials are well correlated over a broad range of frequency content of somatosensory stimulation of the rat forepaw. Brain Res 2008; 1195: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masamoto K, Kim T, Fukuda M, et al. Relationship between neural, vascular, and BOLD signals in isoflurane-anesthetized rat somatosensory cortex. Cereb Cortex 2007; 17: 942–950. [DOI] [PubMed] [Google Scholar]
- 46.Zhao F, Zhao T, Zhou L, et al. BOLD study of stimulation-induced neural activity and resting-state connectivity in medetomidine-sedated rat. Neuroimage 2008; 39: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masamoto K, Fukuda M, Vazquez A, et al. Dose-dependent effect of isoflurane on neurovascular coupling in rat cerebral cortex. Eur J Neurosci 2009; 30: 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukuda M, Vazquez AL, Zong X, et al. Effects of the alpha(2)-adrenergic receptor agonist dexmedetomidine on neural, vascular and BOLD fMRI responses in the somatosensory cortex. Eur J Neurosci 2013; 37: 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lima B, Cardoso MM, Sirotin YB, et al. Stimulus-related neuroimaging in task-engaged subjects is best predicted by concurrent spiking. J Neurosci 2014; 34: 13878–13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirotin YB, Das A. Anticipatory haemodynamic signals in sensory cortex not predicted by local neuronal activity. Nature 2009; 457: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Ramirez S, Pang PT, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 2012; 484: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.