Abstract
Thioredoxin (Trx) is a major thiol-disulfide reductase that plays a role in many biological processes, including DNA replication and redox signaling. Although selenocysteine (Sec)-containing Trxs have been identified in certain bacteria, their enzymatic properties have not been characterized. In this study, we expressed a selenoprotein Trx from Treponema denticola, an oral spirochete, in Escherichia coli and characterized this selenoenzyme and its natural cysteine (Cys) homologue using E. coli Trx1 as a positive control. 75Se metabolic labeling and mutation analyses showed that the SECIS (Sec insertion sequence) of T. denticola selenoprotein Trx is functional in the E. coli Sec insertion system with specific selenium incorporation into the Sec residue. The selenoprotein Trx exhibited approximately 10-fold higher catalytic activity than the Sec-to-Cys version and natural Cys homologue and E. coli Trx1, suggesting that Sec confers higher catalytic activity on this thiol-disulfide reductase. Kinetic analysis also showed that the selenoprotein Trx had a 10-fold higher Km than Cys-containing homologues, suggesting that this selenoenzyme is adapted to work efficiently with high concentrations of substrate. Collectively, the results of this study support the hypothesis that selenium utilization in oxidoreductase systems is primarily due to the catalytic advantage provided by the rare amino acid, Sec.
Keywords: disulfide reductase, selenoprotein, thioredoxin, Treponema denticola
Introduction
Selenium is an essential trace element in mammals that is incorporated into proteins in the form of the rare amino acid selenocysteine (Sec) during protein translation [1]. Sec-containing proteins, which are known as selenoproteins, are found in all three kingdoms of life. Oxidoreductases, including formate dehydrogenase H, glutathione peroxidase, and methionine sulfoxide reductase, are among the most well characterized selenoproteins [2,3,4]. In selenoprotein forms of oxidoreductases, Sec replaces cysteine (Cys) residue in catalytic sites of their orthologue proteins. Sec is translationally inserted into protein using an UGA codon, which is normally a stop codon. A stem-loop structure known as the SECIS (Sec insertion sequence) element on the selenoprotein mRNA is necessary to recode the UGA codon for the Sec insertion [1,5]. SECIS elements are located immediately downstream of the Sec UGA codons in bacteria, whereas they are present in the 3′-untranslational regions in archaea and eukaryotes. A consensus bacterial SECIS structural model has been suggested in which a conserved G nucleotide in the apical loop of SECIS is critical to the function of SECIS [6].
The thioredoxin (Trx) system is a major disulfide reductase system that controls cellular redox status [7]. The Trx system is composed of NADPH, Trx reductase, and Trx, wherein electrons are transferred from NADPH to Trx via Trx reductase. Trxs, typically 12 kDa small thiol-disulfide oxidoreductases, were originally discovered as a hydrogen donor of Escherichia coli ribonucleotide reductase [8], and are now a well-known multifunctional protein involved in a variety of physiological processes, including DNA replication, transcription, antioxidant defense, and redox signaling [9]. Trxs involve two Cys residues in catalysis and contain a characteristic CGPC motif in the active site, in which the N-terminal Cys residue acts as a catalytic residue and the C-terminal Cys residue as a resolving Cys.
Sec-containing forms of Trx have been found in bacteria such as Treponema denticola [10], but have not been previously characterized. T. denticola is a Gram-negative, obligate anaerobic bacterium associated with periodontitis [11]. T. denticola growth is dependent on selenium in medium [12], and this organism contains at least seven selenoproteins, including Trx [10]. In this study, we expressed the T. denticola selenoprotein Trx in E. coli and characterized this selenoenzyme and its natural Cys homologue using E. coli Trx1 as a positive control.
Materials and Methods
Cloning of the T. denticola Sec- and Cys-containing Trxs
A coding region of the T. denticola selenoprotein Trx gene, trx1, was amplified by PCR using genomic DNA from T. denticola ATCC 33521 and forward (5′-GCGCCATATGATTATGGCAGTATTGG-3′) and reverse (5′-GCGCCTCGAGGATATGCTTTGAAACAAAG-3′) primers. The amplified DNA fragment digested with NdeI and XhoI was cloned into pET21b (Novagen). The resulting construct, designated pET21-Td-Trx1, encoded full-length selenoprotein Trx1 (tTrx1) with a C-terminal His-tag (LEHHHHHH). The Cys-containing homologue Trx gene, trx2, was also PCR-cloned into NdeI/XhoI sites of pET21b using forward (5′-GCGCCATATGATTGAATTGACAAAAG-3′) and reverse (5′-GCGCCTCGAGGATGTATTTTTTTACCATC-3′) primers. The resulting plasmid, pET21-Td-Trx2, encoded full-length Trx2 (tTrx2) with a C-terminal His-tag (LEHHHHHH). We also generated Sec-to-Cys version (tTrx1/U32C) and Trp mutant (tTrx1/U32W) of selenoprotein tTrx1 in which Sec32 was replaced with Cys and Trp, respectively, by site-directed mutagenesis.
Expression and purification of the Sec- and Cys-containing Trxs
To express tTrx1 in E. coli, the plasmid pET21-Td-Trx1 was introduced into BL21(DE3) cells also harboring a plasmid pSUABC [13], which encodes E. coli selA, selB, and selC. The transformed cells were grown in LB media containing 2 μM sodium selenite, 100 μg/ml ampicillin, and 25 μg/ml chloramphenicol at 37°C. IPTG (0.1 mM) was added when the culture reached an optical density at 600 nm of ~0.6–0.8, and the cells were cultured for an additional 4 h at 30°C. After centrifugation, the cell pellets were resuspended in extraction buffer (50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 10 mM imidazole, and 1 mM phenylmethylsulfonyl fluoride) and lysed by sonication. The supernatant of the lysate was then loaded onto a Talon-metal affinity resin (Clontech), washed with extraction buffer, and eluted with buffer containing 50 mM sodium phosphate (pH 7.0), 50 mM NaCl, and 150 mM imidazole. The eluted proteins were concentrated and dialyzed against 50 mM sodium phosphate (pH 7.5) and 50 mM NaCl. The typical yield of the purified selenoprotein Trx1 was 40–70 μg from 500 ml culture broth based on Western blot assays.
To express the T. denticola Cys-containing Trx forms (tTrx1/U32C or tTrx2) or Trp mutant tTrx1/U32W, E. coli BL21(DE3) cells transformed with the corresponding plasmids were cultivated in LB media containing 100 μg/ml ampicillin at 37°C until the optical density at 600 nm reached ~0.6–0.8, at which time 0.1 mM IPTG was added and the cells were cultured for an additional 4 h at 30°C. The proteins were purified as described above for selenoprotein tTrx1. We also expressed and purified E. coli Trx1 (eTrx1) by a Talon-metal affinity chromatography as previously described [14]. The samples were analyzed for purity by SDS–PAGE and found to consist almost exclusively of the ectopic protein.
75Se metabolic labeling
To verify expression of T. denticola selenoprotein Trx1 in E. coli, 75Se metabolic labeling was performed as previously described [15]. Briefly, E. coli BL21(DE3) cells transformed with an empty vector, pET21-Td-Trx1, or pET21-Td-Trx1/U32C were grown at 37°C in 5 ml LB media containing ampicillin until the optical density at 600 nm reached ~0.6. Next, 0.05 mCi of freshly neutralized [75Se]selenous acid and 1 mM IPTG were added to the cell culture, after which the cells were cultured at 37°C for 4 h. The harvested cells were then washed with phosphate-buffered saline and lysed. A total of 30 μg protein was separated by SDS-PAGE and transferred onto a PVDF membrane. Finally, the 75Se radioactivity pattern on the membrane was visualized using a PhosphorImager (GE Health Care).
Determination of protein concentration
Due to small amounts of purified recombinant selenoprotein tTrx1, the concentration of this selenoprotein was determined by Western blot analysis using anti-His antibodies, followed by quantification of the blot signals using the ImageJ (National Institute of Health) program. The tTrx1/U32C protein was used as an internal standard. Concentrations of purified Cys-containing Trxs and Trp mutant tTrx1/U32W were determined by the Bradford method using a BioRad protein assay reagent and bovine serum albumin as a standard.
Trx activity assay and analysis of kinetics
Trx activity was measured by the insulin disulfide reduction assay as described by Holmgren [16]. The reaction mixture (200 μl) contained 100 mM sodium phosphate (pH 7.0), 2 mM EDTA, 5–100 μM insulin (Sigma–Aldrich), and 0.8–2 μM Trx. The reaction was started by the addition of 0.5 mM dithiothreitol. Reduction of insulin was monitored as an increase in turbidity at 650 nm for 50 min due to insulin precipitation. Non-enzymatic reduction of insulin by dithiothreitol was measured as a negative control. Trx activity was defined as the increase in absorbance per min (ΔA/min) in the interval below 1.0 of optical density. Vmax and Km values were determined by non-linear regression using the Prism 5 (GraphPad) software.
Results
Sec- and Cys-containing Trxs from T. denticola
The T. denticola genome contains at least seven selenoprotein genes (six known and one predicted) [10]. The known selenoproteins identified include SelD, glutathione peroxidase, glycine reductase A, two glycine reductase Bs, and Trx. Sec-containing Trxs have also been found in other bacteria, including Geobacter metallireducens and Anaeromyxobacter dehalogenans, but are absent from eukaryotes [10]. In addition, the T. denticola genome harbors a separate Cys-containing Trx. We designated the T. denticola selenoprotein Trx as tTrx1 and the Cys-containing Trx as tTrx2. tTrx1 consists of 107 amino acids and contains the 32UPGC35 (U, Sec) catalytic motif (Figure 1). There are no other Cys residues in the sequence. tTrx2 protein has 105 amino acids, including the catalytic 29CVPC32 motif (Figure 1). tTrx1 protein shows 29.4% identity with tTrx2. Both selenoprotein tTrx1 and Cys-containing tTrx2 belong to the typical small Trx family. Multiple sequence alignment revealed differences between T. denticola Trxs and other known Trxs around the catalytic CxxC motif (Figure 1).
Figure 1. Multiple sequence alignment of Sec- and Cys-containing Trxs.
Catalytic Sec (U) or Cys residues are highlighted in red and resolving Cys residues in blue. GenBank accession numbers are as follows: T. denticola Trx1, 499259163; T. denticola Trx2, 488746655; E. coli Trx1, 388479466; Saccharomyces cerevisiae Trx1, 135747; Mus musculus Trx1, 6755911; Homo sapiens Trx1, 9280551.
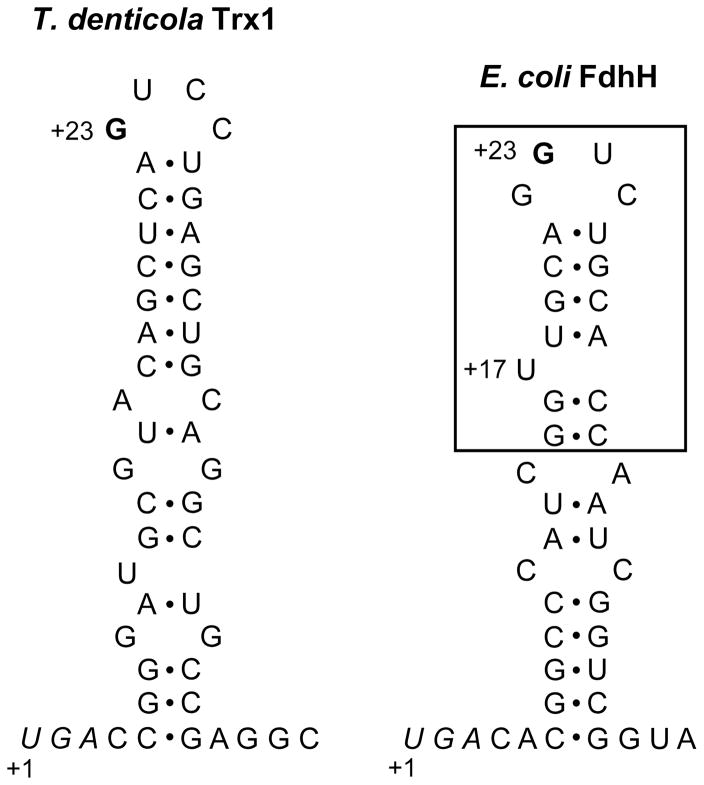
SECIS structure of T. denticola selenoprotein Trx and its functionality in E. coli
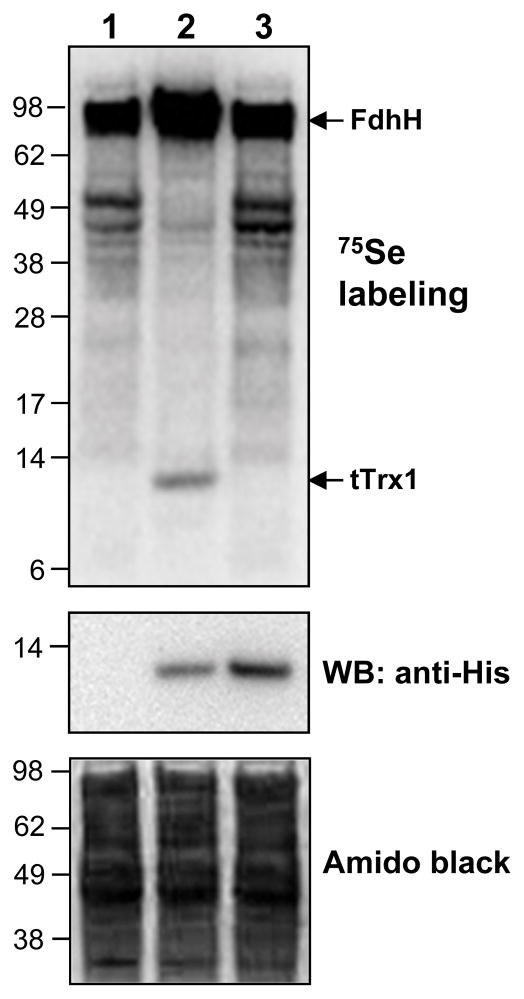
The SECIS element of tTrx1 had a conserved G nucleotide in the apical loop (Figure 2), which was consistent with the consensus bacterial SECIS structural model [6]. However, the tTrx1 SECIS element lacked a bulged U in the stem, which is present in the E. coli SECIS element and known to be important for the function of E. coli SECIS [17]. To determine whether the SECIS element of tTrx1 can be functional in E. coli, we metabolically labeled E. coli cells expressing tTrx1 with 75Se. Cells expressing a Sec-to-Cys mutant were also labeled to determine whether selenium incorporation is specific into Sec residue. As a result, a 12 kDa radioactive band corresponding in size to the calculated molecular mass of recombinant tTrx1 was detected (Figure 3, lane 2 of the upper panel). In contrast, no apparent Sec-to-Cys mutant protein band was observed (lane 3) in the Phosphor image, despite its higher expression level versus the native selenoprotein form (middle panel). We also confirmed the expression of recombinant selenoprotein tTrx1 with a C-terminal His tag by Western blot analysis using anti-His antibodies (middle panel). These results clearly demonstrated that selenoprotein tTrx1 can be expressed in E. coli with specific selenium incorporation into Sec. Therefore, our data indicate that the SECIS element tTrx1 is functional in the E. coli Sec insertion system.
Figure 2. Structures of SECIS elements of T. denticola Trx1 and E. coli formate dehydrogenase H.
The SECIS of T. denticola Trx1 contains a conserved G nucleotide (bold) in the apical loop, but lacks a bulged U, which is present in the minimal step-loop structure (boxed) in E. coli formate dehydrogenase H (FdhH) SECIS. Sec UGA codons are shown in italics and numbering of nucleotides is stated at these Sec codons. The SECIS structure of T. denticola Trx1 was predicted using RNAfold [24].
Figure 3. 75Se metabolic labeling.
E. coli BL21(DE3) cells containing an empty vector (lane 1), pET21-Td-Trx1 (lane 2), or pET21-Td-Trx1/U32C (lane 3) were metabolically labeled with 75Se. Proteins were separated by SDS–PAGE, transferred onto a PVDF membrane, and visualized with a PhosphorImager (upper panel). The same membrane was immunoblotted with anti-His antibody (middle panel) or stained with Amido black (lower panel).
Catalytic properties of Sec- and Cys- containing Trxs
To investigate catalytic properties of tTrx1 and tTrx2, we purified these proteins from E. coli cells. We also expressed and purified eTrx1 for comparison. We conducted an insulin disulfide reduction assay to measure Trx activity as described in the materials and methods. The Vmax value of tTrx1 was 11-fold higher than that of its Sec-to-Cys mutant (tTrx1/U32C) (Table 1). As expected, the Trp mutant tTrx1/U32W was completely inactive. These data show that Sec is the catalytic residue of tTrx1 and is critical to its catalysis. Although the Sec-to-Cys form (tTrx1/U32C) had a much lower activity than its original selenoprotein form, this protein showed activity comparable to eTrx1. Conversely, tTrx2 enzyme had activity similar to tTrx1/U32C. The Km value of selenoprotein tTrx1 was 10-fold higher than those of the Cys homologues, tTrx1/U32C, tTrx2, and eTrx1, suggesting that the tTrx1 selenoenzyme is adapted to work efficiently with high concentrations of substrate.
Table 1.
Kinetic parameters of T. denticola Trx forms
| Protein | Vmax (ΔA/min/mg) | Km (Insulin) (μM) |
|---|---|---|
| tTrx1 | 49 ± 4 | 1240 ± 230 |
| tTrx1/U32C | 4.2 ± 0.6 | 40 ± 13 |
| tTrx1/U32W | ND | - |
| tTrx2 | 2.8 ± 0.8 | 35 ± 18 |
| eTrx1 | 7.0 ± 1.4 | 46 ± 20 |
Trx activity was measured by an insulin disulfide reduction assay and defined as the increase in absorbance at 650 nm per min. Vmax and Km values were determined by non-linear regression using the Prism 5 program. tTrx1, wild-type selenoprotein T. denticola Trx1; tTrx1/U32C, Sec-to-Cys mutant of tTrx1; tTrx1/U32W, Sec-to-Trp mutant of tTrx1; tTrx2, Cys-containing T. denticola Trx2; eTrx1, E. coli Trx1. ND, activity not detectable.
Discussion
Here, the first characterization of a selenoprotein Trx is presented. Sec residue, which is chemically characterized by a lower pKa, is normally more reactive than Cys residue. Sec-containing oxidoreductases that have been characterized to date, including Trx reductase and methionine sulfoxide reductase, exhibit higher catalytic activities (at least 10-fold) than their Sec-to-Cys mutants or natural Cys-containing counterparts [18,19,20]. The higher catalytic activity provided by Sec residue is thought to be a primary reason for selenium utilization in oxidoreductase systems. Consistent with this assumption, our data show that Sec confers a higher catalytic activity on the selenoprotein Trx.
The catalytic activity of selenoprotein tTrx1 was 7 to 17-fold higher than those of its Sec-to-Cys version and natural Cys-containing tTrx2 and eTrx1. However, this selenoprotein tTrx1 activity might have been underestimated because, to the best of our knowledge, no recombinant selenoproteins have contained 100% Sec insertion when expressed in E. coli. It has been reported that Trp insertion through opal suppression at the UGA codon always occurs during translation of recombinant selenoproteins in E. coli cells [15,21,22].
We previously characterized a natural selenoprotein glutaredoxin (Grx) from Clostridium oremlandii [15]. Grx is another major cellular thiol-disulfide oxidoreductase that constitutes the glutathione redox system. Sec also provides a catalytic advantage with the C. oremlandii selenoprotein Grx (200-fold higher activity than Sec-to-Cys mutant). In addition to the higher catalytic activity provided by Sec, our previous study suggested that selenium utilization by Grx was due to substrate specificity. C. oremlandii selenoprotein Grx efficiently reduces selenoprotein methionine sulfoxide reductase A of this species, whereas Cys-containing Grxs from C. oremlandii and E. coli poorly reduce this selenoenzyme. Among the known selenoproteins identified in T. denticola, glycine reductase A would be a strong candidate substrate of selenoprotein Trx since Trx is a well-known reductant of glycine reductase A [23]. Therefore, it would be interesting to determine if Sec-containing tTrx1 specifically reduces selenenylsulfide (Se–S) bonds in oxidized glycine reductase A from T. denticola relative to the reduction ability of Cys-containing tTrx2.
Interestingly, the results of the present study indicate that the selenoprotein tTrx1 expression in E. coli regulates the expression of E. coli host selenoproteins. 75Se metabolic labeling data showed that a radioactive protein band of 80 kDa is significantly elevated by the expression of selenoprotein tTrx1, whereas all other radioactive protein bands were diminished (Figure 3). The expression of Sec-to-Cys mutant (tTrx1/U32C) did not affect the expression of E. coli host selenoproteins. The 80 kDa protein band corresponds well in size to E. coli formate dehydrogenase H. These data suggest that expression of Sec-containing Trx in E. coli may modulate specific Sec incorporation into host selenoproteins.
In summary, we report for the first time the catalytic properties of Sec-containing Trx. The selenoprotein Trx exhibited approximately 10-fold higher catalytic activity versus Cys-containing Trx forms. Our study supports the hypothesis that Sec utilization in oxidoreductase systems is primarily due to the higher catalytic activity provided by this rare amino acid.
Acknowledgments
The authors thank Dr. Bong-Kyu Choi (Seoul National University, Korea) for kindly providing T. denticola genomic DNA. This work was supported by the 2015 Yeungnam University Research Grant.
References
- 1.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. Embo J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 5.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Gladyshev VN. An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics. 2005;21:2580–2589. doi: 10.1093/bioinformatics/bti400. [DOI] [PubMed] [Google Scholar]
- 7.Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 8.Laurent TC, Moore EC, Reichard P. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J Biol Chem. 1964;239:3436–3444. [PubMed] [Google Scholar]
- 9.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun HK, Kang YM, Lee HR, Lee SH, Choi BK. Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect Immun. 2008;76:2428–2438. doi: 10.1128/IAI.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rother M, Bock A, Wyss C. Selenium-dependent growth of Treponema denticola: evidence for a clostridial-type glycine reductase. Arch Microbiol. 2001;177:113–116. doi: 10.1007/s002030100351. [DOI] [PubMed] [Google Scholar]
- 13.Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 14.Kim HY. Glutaredoxin serves as a reductant for methionine sulfoxide reductases with or without resolving cysteine. Acta Biochim Biophys Sin (Shanghai) 2012;44:623–627. doi: 10.1093/abbs/gms038. [DOI] [PubMed] [Google Scholar]
- 15.Kim MJ, Lee BC, Jeong J, Lee KJ, Hwang KY, Gladyshev VN, Kim HY. Tandem use of selenocysteine: adaptation of a selenoprotein glutaredoxin for reduction of selenoprotein methionine sulfoxide reductase. Mol Microbiol. 2011;79:1194–1203. doi: 10.1111/j.1365-2958.2010.07500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 17.Liu Z, Reches M, Groisman I, Engelberg-Kulka H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SR, Bar-Noy S, Kwon J, Levine RL, Stadtman TC, Rhee SG. Mammalian thioredoxin reductase: oxidation of the C-terminal cysteine/selenocysteine active site forms a thioselenide, and replacement of selenium with sulfur markedly reduces catalytic activity. Proc Natl Acad Sci U S A. 2000;97:2521–2526. doi: 10.1073/pnas.050579797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HY, Fomenko DE, Yoon YE, Gladyshev VN. Catalytic advantages provided by selenocysteine in methionine-S-sulfoxide reductases. Biochemistry. 2006;45:13697–13704. doi: 10.1021/bi0611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su D, Li Y, Gladyshev VN. Selenocysteine insertion directed by the 3′-UTR SECIS element in Escherichia coli. Nucleic Acids Res. 2005;33:2486–2492. doi: 10.1093/nar/gki547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandman KE, Noren CJ. The efficiency of Escherichia coli selenocysteine insertion is influenced by the immediate downstream nucleotide. Nucleic Acids Res. 2000;28:755–761. doi: 10.1093/nar/28.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreesen JR. Glycine reductase mechanism. Curr Opin Chem Biol. 2004;8:454–461. doi: 10.1016/j.cbpa.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proc Biol Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]