Abstract
Instinctive reactions to danger are critical to the perpetuation of species and are observed throughout the animal kingdom. The scent of predators induces an instinctive fear response in mice that includes behavioral changes as well as a surge in blood stress hormones that mobilizes multiple body systems to escape impending danger1,2. How the olfactory system routes predator signals detected in the nose to achieve these effects is unknown. Here we identify a specific area of the olfactory cortex that induces stress hormone responses to volatile predator odors. Using monosynaptic and polysynaptic viral tracers, we found that multiple olfactory cortical areas transmit signals to hypothalamic CRH (corticotropin releasing hormone) neurons, which control stress hormone levels. However, only one minor cortical area, the amygdalo-piriform transition area (AmPir), contained neurons upstream of CRH neurons that were activated by volatile predator odors. Chemogenetic stimulation of AmPir activated CRH neurons and induced an increase in blood stress hormone, mimicking an instinctive fear response. Moreover, chemogenetic silencing of AmPir markedly reduced the stress hormone response to predator odors without affecting a fear behavior. These findings suggest that AmPir, a small area comprising <5% of the olfactory cortex, plays a key role in the hormonal component of the instinctive fear response to volatile predator scents.
Animals sense environmental dangers that challenge their survival and respond with behavioral and physiological changes to eliminate the challenge. The olfactory system detects chemical cues emitted from predators that signal danger and stimulate an instinctive fear response. In addition to characteristic behaviors, this response includes increases in blood stress hormones that are reminiscent of responses to fear and stress in humans and are controlled by CRH neurons in the paraventricular nucleus of the hypothalamus (PVN)1–3. The neural circuits that convey predator odor signals from the nose to CRH neurons are presently unknown.
Volatile predator cues are detected in the olfactory epithelium of the nose, which also detects common odorants, whereas fear-stimulating proteins are detected in the vomeronasal organ (VNO), an accessory olfactory structure4,5. Sensory signals travel from the nose through the olfactory bulb to the olfactory cortex (OC) and from the VNO through the accessory olfactory bulb to the vomeronasal amygdala (VA)6,7. The mouse nose has ~1000 different odorant receptors, each expressed by a unique subset of dispersed sensory neurons8–11. The olfactory bulb contains a semi-stereotyped map of receptor inputs in which individual glomeruli and projection neurons appear dedicated to one receptor12–14, but single bulb neurons can project widely in the OC15–17. The OC comprises multiple distinct areas whose respective functions are largely obscure6,7. Recent studies have linked an amygdala OC area to odor attraction and aversion18. Here, we investigated whether a specific OC area governs stress hormone responses to predator odors.
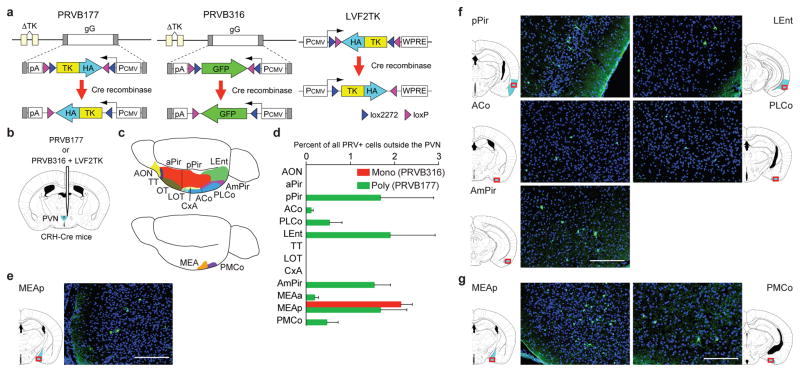
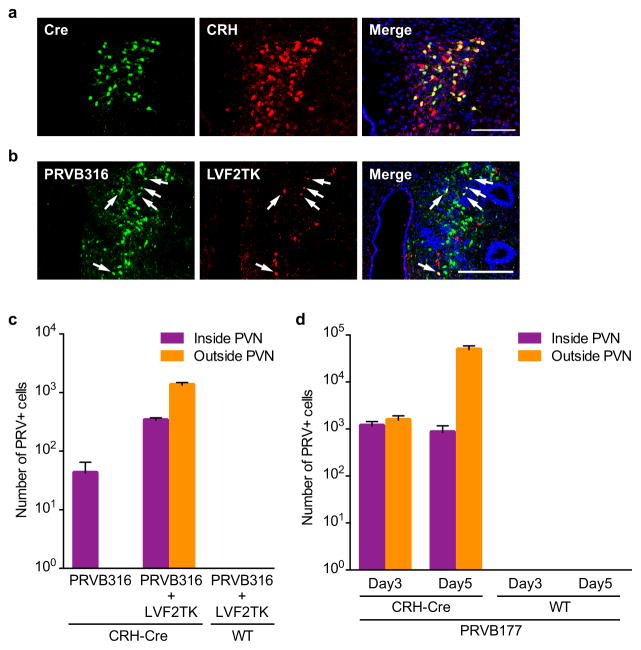
We first asked what OC areas can transmit signals to CRH neurons. We developed two Cre recombinase-dependent Bartha pseudorabies viruses (PRVs) that have nonfunctional thymidine kinase (TK) genes, but require TK to replicate and travel retrogradely across synapses19,20, and a lentivirus (LVF2TK) with Cre-dependent expression of hemagglutinin-tagged TK (HA-TK) (Fig. 1a). PRVB177 is a “polysynaptic PRV” with irreversible Cre-dependent expression of HA-TK that can cross multiple sequential synapses. PRVB316 is a “monosynaptic PRV” with Cre-dependent expression of GFP that can replicate in neurons coinfected with LVF2TK and spread across one synapse, but not further. We injected the viruses into the PVN of mice that express Cre in CRH neurons (CRH-ires-Cre (CRH-Cre) mice)21 (Fig. 1b) and then immunostained brain sections for (HA (PRVB177) or GFP (PRVB316)) to visualize virus-infected neurons.
Figure 1. Higher olfactory areas contain neurons upstream of CRH neurons.
a, Cre recombinase causes irreversible expression of HA-TK from PRVB177 and LVF2TK, and GFP from PRVB316.
b, Viruses were injected into the PVN (blue) of CRH-Cre mice.
c, The olfactory cortex (above) comprises multiple areas and the vomeronasal amygdala (below) two areas. See Methods for full names.
d, PRV+ neurons in olfactory areas four days after injection of PRVB316 (n=24) or PRVB177 (n=6). Error bars indicate SEM.
e–g, Photographs and diagrams of olfactory area (blue) sections30 (red rectangles) with neurons immunostained for PRVB316 (e) or PRVB177 (f, g) (green). Scale bars, 200 μm.
Preliminary experiments confirmed that Cre is expressed in PVN CRH neurons in CRH-Cre mice and that the viruses function as expected. In CRH-Cre mice crossed with Rosa-floxstop-GFP mice, 93.9±0.5% of GFP+ cells expressed CRH (n=4) (Extended Data Fig. 1a). Following PVN coinjection of PRVB316 and LVF2TK, some neurons expressed both viruses (Extended Data Fig. 1b). Neurons infected with PRVB316, but not LVF2TK, were subsequently detected outside the PVN, whereas none were detected outside the PVN when PRVB316 was injected without LVF2TK (Extended Data Fig. 1c). No infected neurons were seen in wildtype (WT) animals injected with the monosynaptic or polysynaptic PRV (Extended Data Fig. 1c, d).
Virus-infected neurons were detected in multiple brain areas following PRV infection of CRH neurons (Extended Data Fig. 2, 3). The monosynaptic PRV (PRVB316) infected 31 brain areas, including areas known to project axons to the PVN22 and areas linked to CRH neuron activation by certain non-olfactory stressors3.
In the olfactory system (Fig. 1c), PRVB316 infected only the posterior medial amygdala (MEAp), part of the VA (Fig. 1d, e), whereas, by day 4 after injection, PRVB177 infected five OC and both VA areas (Fig. 1d, f, g). Most brain areas infected with PRVB177 on day 3 were also infected by PRVB316 (Extended Data Fig. 2, 3), suggesting that OC areas infected on day 4 may transmit signals to CRH neurons via one intermediate relay.
We observed a regional bias in the locations of neurons upstream of CRH neurons in some olfactory areas, but not others. PRVB177+ neurons were randomly distributed along the anterior-posterior axis in most olfactory areas, but were concentrated in posterior regions of the posterior piriform cortex (pPir) and amydalo-piriform transition area (AmPir) and in the posteroventral quadrant of the MEA (MEApv) (Extended Data Fig. 4).
Dual labeling for PRVB177 and markers of excitatory glutamatergic neurons (Vglut1/2) or inhibitory GABAergic neurons (Gad1/2) indicated that some neurons upstream of CRH neurons in the MEA were GABAergic, but the majority of those labeled for the markers in higher olfactory areas were glutamatergic (Extended Data Fig. 5). The glutamatergic neurons could potentially activate either excitatory or inhibitory neurons directly presynaptic to CRH neurons in other brain areas, and thus either stimulate or inhibit CRH neurons.
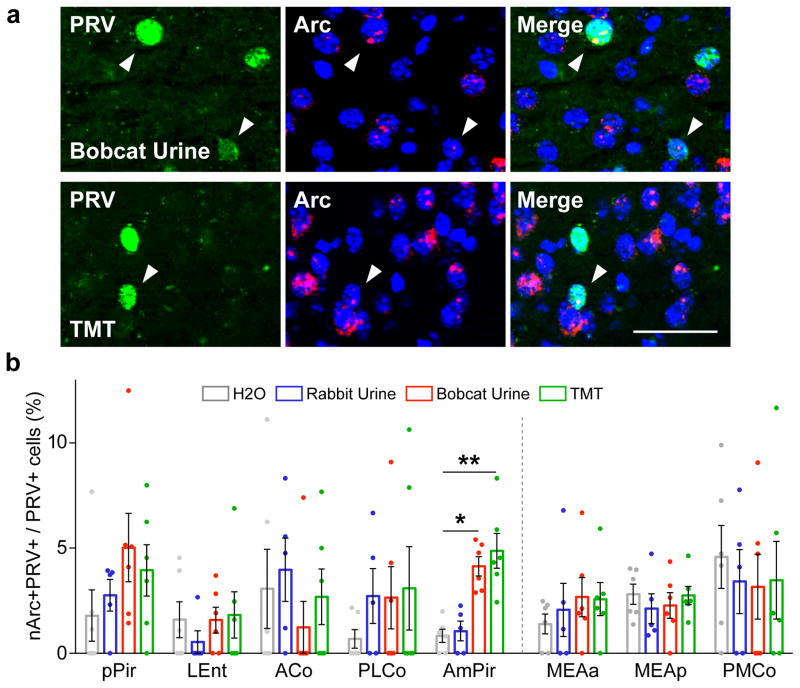
Neurons upstream of CRH neurons in different OC areas could conceivably transmit redundant predator odor signals to CRH neurons. To investigate this possibility, we used a neural activity marker, nuclear Arc mRNA (nArc)23 to identify PRVB177-infected neurons activated by the fox predator odorant, 2,5-dihydro-2,4,5- trimethylthiazoline (TMT)4,24 or bobcat (predator) urine. Preliminary experiments confirmed that TMT and bobcat urine, but not nonpredator (rabbit) urine, induce the activity marker c-Fos in PVN CRH neurons and increase blood levels of the stress hormones ACTH (adrenocorticotropic hormone) and corticosterone (Extended Data Fig. 6).
Surprisingly, both predator odors induced a significant increase in nArc in PRV+ neurons in only one OC area, AmPir (Fig. 2). The percentage of AmPir PRV+ neurons expressing nArc was increased 5.8-fold by TMT and 4.9-fold by bobcat urine, whereas rabbit urine had no significant effect. These results suggested that AmPir could play an important part in hormonal fear responses to volatile predator odors.
Figure 2. Predator odors activate neurons upstream of CRH neurons in AmPir.
a, AmPir neurons colabeled (arrowheads) for PRVB177 (green) and nArc (red) following exposure of mice to bobcat urine or TMT. Scale bar, 50 μm.
b, Percent of PRVB177+ neurons colabeled for nArc in areas of olfactory cortex (left) and vomeronasal amygdala (right) following exposure to TMT, bobcat urine, rabbit urine, or water. (n=5–6 per condition. Error bars indicate SEM. *p<0.05, **p<0.01. Kruskal-Wallis test with post-hoc Dunn’s test.)
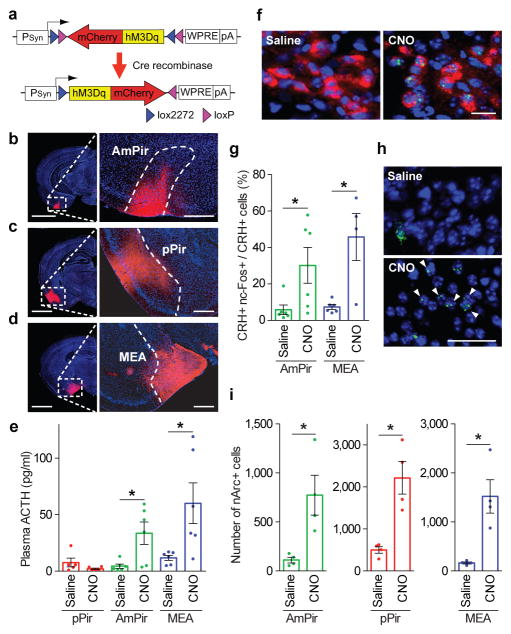
To explore this idea, we first asked if chemogenetic activation of AmPir induces stress hormone increases. We injected the adeno-associated virus (AAV), AAV-DIO-hM3Dq-mCherry25 bilaterally into the AmPir, pPir, or MEA of mice expressing Cre in projection neurons (Emx1-ires-Cre or Vglut2-ires-Cre mice)26,27 (Fig. 3a–d). This virus has Cre-dependent expression of mCherry fused to hM3Dq, a receptor that depolarizes neurons upon binding its pharmacologically inert ligand, clozapine-N-oxide (CNO)28. We then injected animals with CNO or saline and measured plasma ACTH.
Figure 3. Chemogenetic activation of AmPir induces stress hormone increase.
a–d, mCherry immunostaining (red) after injection of AAV DIO-hM3Dq-mCherry (a) into the AmPir (b), pPir (c), or MEA (d). Scale bars, 2 mm (left), 400 μm (right).
e, Plasma ACTH concentration following injection of CNO or saline into mice expressing hM3Dq in pPir, AmPir, or MEA. (n=5–6 per condition. Error bars indicate SEM. *p<0.05. Mann-Whitney U test.)
f, PVN neurons colabeled for CRH (red) and nuclear c-Fos (ncFos) (green) after injection of saline or CNO into mice expressing hM3Dq in AmPir. Scale bar, 20 μm
g, Percentage of PVN CRH+ neurons colabeled for nc-Fos in mice treated as in (e). (n=4–6 per condition. Error bars indicate SEM. *p<0.05. Mann-Whitney U test.)
h, AmPir neurons labeled for nArc (green) (arrowheads) following saline or CNO injection of mice expressing hM3Dq in AmPir. Scale bar, 50 μm.
i, Number of nArc+ neurons in AmPir, pPir, or MEA in mice treated as in (e). (n=4 per condition. Error bars indicate SEM. *p<0.05, Unpaired t-test.)
Chemogenetic activation of AmPir induced a striking (7.6-fold) increase in plasma ACTH (Fig. 3e). Activation of MEA, but not pPir, also increased plasma ACTH. Consistent with these results, activation of AmPir or MEA also induced c-Fos in PVN CRH neurons (Fig. 3f, g). Additional analyses showed that mCherry+ (AAV-infected) neurons were present in a large part of each targeted area (Extended Data Fig. 7) and that CNO induced nArc in neurons in each injected area (Fig. 3h, i). Those in AmPir could include neurons responsive to TMT or bobcat urine as well as neurons activated by other odorants. Due to the relatively small size of AmPir, infected neurons were also often seen in immediately adjacent olfactory areas (parts of the posterior PLCo and lateral PMCo). These results indicated that activation of neurons in and around AmPir induces a stress hormone response that mimics the instinctive fear response.
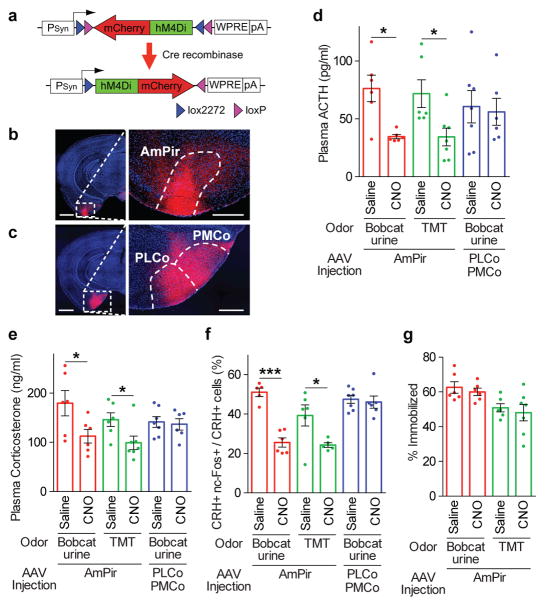
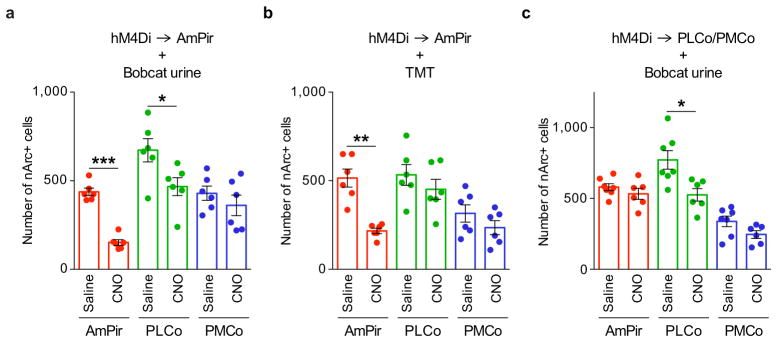
We next used chemogenetic silencing to ask whether the AmPir is required for stress hormone responses to predator odors. We injected the AmPir of Emx1-Cre mice bilaterally with AAV-DIO-hM4Di-mCherry25 (Fig. 4a, b and Extended Data Fig. 8a, b). This virus has Cre-dependent expression of hM4Di, which silences neurons upon binding to CNO28. We subsequently injected animals with CNO or saline, exposed to predator odors, and then determined plasma levels of stress hormones.
Figure 4. Chemogenetic silencing of AmPir inhibits stress hormone responses to predator odors.
a–c, mCherry immunostaining (red) after injection of AAV DIO-hM4Di-mCherry (a) into AmPir (b) or PLCo/PMCo (c). Scale bars, 1 mm (left), 400 μm (right).
d, e, Plasma ACTH (d) or corticosterone (e) concentration after mice expressing hM4Di in AmPir or PLCo/PMCo were injected with saline or CNO and then exposed to bobcat urine or TMT. (n=6–7 per condition. Error bars indicate SEM. *p<0.05. Mann-Whitney U test (d) or Unpaired t-test (e).)
f, Percentage of PVN CRH+ neurons expressing nc-Fos in animals treated as in (d, e). (n=5–7 per condition. Error bars indicate SEM. *p<0.05, ***p<0.001. Unpaired t-test.)
g, Animals expressing hM4Di in AmPir (d–f) were videotaped during exposure to bobcat urine and videos analyzed for the percentage of time spent immobile (% immobilized). (n=6–7 per condition. Error bars indicate SEM.)
Silencing of AmPir dramatically reduced stress hormone responses to predator odors (Fig. 4d). Plasma ACTH responses to TMT and bobcat urine were decreased by 52±11% and 55±02%, respectively (Fig. 4d). These decreases correlated with reductions of 58±03% and 65±04% in the percentages of AmPir neurons activated by TMT and bobcat urine, as assessed by nArc labeling (Extended Data Fig. 9a, b). AmPir silencing also decreased plasma corticosterone (Fig. 4e) and the percentage of c-Fos+ PVN CRH neurons observed following predator odor exposure (Fig. 4f). These results suggested that neurons in AmPir are required for full predator odor-induced increases in stress hormones.
To examine whether adjacent PLCo/PMCo regions were involved in the silencing effects observed for AmPir, we injected AAV-DIO-hM4Di-mCherry selectively into these regions, but not AmPir (Fig. 4c, Extended Data Fig. 8c). In sharp contrast to silencing of AmPir, silencing of these regions did not affect predator odor-induced increases in ACTH and corticosterone or CRH neuron activation (Fig. 4d–f, Extended Data Fig. 9c). These results clearly indicate that AmPir, but not adjacent regions of the PLCo or PMCo, is involved in predator odor-induced hormonal fear responses.
Interestingly, AmPir silencing did not affect the “freezing” behavior (immobilization) induced by bobcat urine or TMT, a typical fear behavior caused by predator odors (Fig. 4g). A recent study showed that inhibition of neurons in PLCo reduced freezing behavior to TMT18. Given the effects of AmPir silencing on stress hormone responses to predator odors, these findings suggest that different OC areas may be involved in the behavioral versus hormonal effects of predator odors, with AmPir and PLCo playing major roles in their hormonal and behavioral effects, respectively.
Instinctive fear responses to predator odors are observed in naïve animals, suggesting the involvement of stereotyped olfactory circuits. We have used a combination of monosynaptic and polysynaptic viral tracing, indicators of neuronal activation, and chemogenetic activation and silencing to investigate how the olfactory cortex translates volatile predator odor signals from the nose into a hormonal fear response. These studies implicate only one small area of the olfactory cortex in this response, the AmPir. They reveal that neurons in multiple olfactory cortical areas can transmit signals to CRH neurons that control stress hormones, but that fox and bobcat predator odors significantly activate only those in AmPir. Chemogenetic activation of AmPir induces stress hormone increases and its silencing causes a striking decrease in the stress hormone response to predator odors. Together, these findings suggest that AmPir plays a key role in generating hormonal fear responses to volatile predator cues that signal danger. The functions of neurons upstream of CRH neurons in other olfactory cortical areas are unknown. They might also contribute to stress hormone responses. However, given that rose oil can block stress hormone responses to a predator odor29, it is also conceivable that some transmit signals that suppress rather than activate hormonal responses associated with fear.
Methods
Mice
CRH-ires-Cre mice were generated previously21. C57BL/6J wild type mice, Emx1-ires-Cre and Vglut2-ires-Cre knock-in mice, and Rosa-floxstop-GFP reporter mice were purchased from the Jackson Laboratory. All procedures involving mice were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee.
We used both males and females in all experiments, with similar numbers where possible. The same number of each sex was used in experiments in Fig. 1 and Extended Data Figs 2, 3, 5 and 6.
No statistical methods were used to predetermine sample size. Animals were randomly chosen for experimental subjects. Animals were excluded from certain experiments. For Arc experiments in Fig. 2, animals were excluded if PRV+ cells were not found in all 5 OC and 3 VA areas analyzed. For chemogenetic activation or silencing of AmPir, animals were excluded if more than approximately 50% of mCherry+ cells were observed outside the AmPir.
Viral vectors
PRVs
PRVB177 and PRVB316 were constructed using homologous recombination between targeting vectors and genomic DNA of PRV TK-BaBlu, a thymidine kinase (TK)-deleted PRV Bartha strain derivative with a LacZ insertion into the gG locus20. For targeting vectors, a flexstop-flanked sequence31 encoding a PRV TK fused at its C-terminus to a hemagglutinin (HA) epitope tag (for PRVB177), or EGFP (for PRVB316), was first cloned with an inverse orientation into an EGFP-deleted pEGFP-N1 vector (Clontech). Next, NsiI fragments containing a CMV promoter, the flexstop-flanked coding sequence, and a SV40 polyadenylation signal, were cloned between gG locus sequences matching those 5′ and 3′ to the lacZ sequence in PRV TK-BaBlu to give the final targeting vectors. These vectors were then linearized and co-transfected with PRV TK-BaBlu genomic DNA into HEK 293T cells (ATCC). Recombinant virus clones were selected and confirmed following methods described previously32.
To propagate recombinant PRVs, PK15 cells (ATCC) were infected with the viruses using a multiplicity of infection (M.O.I.) = 0.1~0.01. After infected, cells showed a prominent cytopathic effect (~2 days). They were harvested by scraping, and the cell material was frozen using liquid nitrogen and then quickly thawed in a 37°C water bath. After three freeze-thaw cycles, cell debris was removed by centrifugation twice at 1000 × g for 5 minutes and the supernatant was then used for experiments. The titer of viral stocks was determined using standard plaque assays on PK15 cells19, with titers expressed in plaque-forming units (PFU).
Lentivirus
To generate LVF2TK, a flexstop-flanked sequence encoding TK-HA (see above) was cloned into the pLenti6.3 vector (Thermo Fisher). LVF2TK was produced using the ViraPower HiPerform Lentiviral Expression System (Thermo Fisher) according to the manufacturer’s instructions. Virus was concentrated using ultracentrifugation as described previously33. Viral titer was measured using the UltraRapid Lentiviral Titer Kit (System Biosciences) and titers described in infectious units (IFU).
AAVs
Serotype 8 AAVs with Cre recombinase-dependent flexstop cassettes that permit expression of mCherry-fused hM3Dq or mCherry-fused hM4Di under the control of the human synapsin promoter (AAV8-DIO-hM3Dq-mCherry or AAV8-DIO-hM4Di-mCherry) were purchased from the Vector Core at the University of North Carolina at Chapel Hill (the UNC vector core). Amount used is described in virus particles (VP).
Stereotaxic injection
Mice aged 2–6 months were used for injection. Viruses were injected into the brain using a Stereotaxic Alignment System (David Kopf Instruments) with an inhalation anesthesia of 2.5% Isoflurane. Virus suspensions (PRVs: 1–1.5 × 106 PFU (1 μl), LVF2TK: 1–1.5 × 106 IFU (1 μl), AAVs: 1–3 × 109 VP (200–330 nl)) were loaded into a 1-μl syringe, and injected at 100 nl per minute. The needle was inserted to the target locations based on a stereotaxic atlas30. After recovery, animals were singly housed with regular 12 h dark/light cycles, and food and water were provided ad libitum.
Odor exposure
Mice were exposed to a predator odor or distilled water within a modified polycarbonate vacuum-desiccator, as described previously34. A single mouse was placed on a platform in a chamber of 15 cm diameter with input and output ports, exposed to charcoal-filtered air for 16 h, and then to filtered air bubbled through 12 mM 2,5-dihydro-2,4,5-trimethylthiazoline (TMT) (Contech) diluted in water, water alone, bobcat urine (Maine outdoor solutions or Murray’s lures and trapping), or rabbit urine (Kishel’s quality animal scents and lures). For detection of Arc in PRV-infected cells, CRH-Cre mice injected with PRVB177 four days earlier were exposed to odors for 5 minutes. For analysis of c-Fos expression in CRH neurons, CRH-Cre mice crossed with Rosa-floxstop-GFP mice were exposed to odors for 10 minutes and then to clean air for 50 minutes. All odor exposures were performed between 9:00 and 11:00 AM.
Plasma ACTH and corticosterone assays
Following sacrifice of mice by cervical dislocation and decapitation, trunk blood was collected directly into blood collection tubes (Becton Dickinson) containing 50 μl aprotinin (Phoenix Pharmaceuticals). Plasma was obtained by centrifugation at 1600 × g for 15 minutes at 4°C, and stored at −80°C. Plasma ACTH concentrations were measured using the ACTH ELISA kit (MD Biosciences), according to the manufacturer’s instructions, with the following modifications: 1) 100 μl of the controls or blood plasma combined with 100 μl of phosphate buffered saline (PBS) (pH 7.4) were used in place of 200 μl plasma and 2) the results were assessed with the QuantaRed Enhanced Chemifluorescent HRP Substrate (Thermo Fisher). Plasma corticosterone concentrations were measured using the corticosterone ELISA kit (Abcam), according to the manufacturer’s instructions, with the following modifications: 1) Plasma was diluted 25 times instead of 100 times with buffer M and 2) the results were assessed with the QuantaRed Enhanced Chemifluorescent HRP Substrate. Fluorescence was measured with a CytoFluor4000 plate reader (Applied Biosystems).
Immunofluorescence
Animals were perfused transcardially with 4% paraformaldehyde (PFA). Their brains were then soaked in 4% PFA for 4 hours, in 30% sucrose for 48 hours, and then frozen in OCT (Sakura) and cut into 14–20 μm coronal sections using a cryostat. Brain sections were washed twice with PBS, permeabilized with 0.5% Triton X-100 in PBS for 5 minutes, washed twice with PBS, blocked with TNB (Perkin Elmer) for 1 hour at room temperature, and then incubated with primary antibodies diluted in TNB at 4°C overnight. Sections were then washed three times with TNT (0.1 M Tris pH 7.5, 150 mM NaCl, 0.05% Tween), incubated with the appropriate secondary antibodies and 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI, Sigma) for 1 hour at room temperature, and washed three times with TNT. Slides were coverslipped with Fluoromount-G (Southern Biotech). The following antibodies were used; (1) biotinylated mouse anti-HA (BioLegend, #901505, 1:300) followed by Alexa488-Streptavidin (Thermo Fisher, #S32354, 1:1000) for polysynaptic PRV tracing, (2) goat anti-GFP (Rockland, #600-101-215, 1:1000) and biotinylated anti-HA (BioLegend, 1:300) followed by Alexa488 donkey anti-goat IgG (Thermo Fisher, #A11055, 1:1000) and Alexa555-Streptavidin (Thermo Fisher, #S32355, 1:1000) for monosynaptic PRV tracing, (3) rabbit anti-GFP (Thermo Fisher, #A-11122, 1:500) and goat anti-c-Fos (Santa Cruz, #sc-52G, 1:300) followed by Alexa488 donkey anti-rabbit IgG (Thermo Fisher, #A21206, 1:1000) and Alexa555 donkey anti-goat IgG (Thermo Fisher, #A21432, 1:1000) for analysis of c-Fos expression in CRH neurons.
In situ hybridization
In situ hybridization was performed essentially as described previously11,35, with some experiments using additional steps for dual staining. Coding region fragments of Arc, Vglut1, Vglut2, Gad1, Gad2, and Crh, and the first intron sequence of c-Fos mRNA (for nuclear c-Fos staining) were isolated from mouse brain cDNA or mouse genomic cDNA using PCR, and cloned into the pCR4 Topo vector (Thermo Fisher). Digoxigenin (DIG) or Fluorescein (FLU) -labeled cRNA probes (riboprobes) were prepared using the DIG or FLU RNA Labeling Mix (Roche). Brains were frozen in OCT, and 16–20 μm coronal cryostat sections were hybridized to DIG- and/or FLU- labeled cRNA probes at 56°C for 13–16 hours.
nArc mRNA staining
After hybridization, sections were washed twice in 0.2× SSC at 63°C for 30 minutes, incubated with peroxidase (POD)-conjugated anti-DIG antibodies (Roche, #11207733910, 1:2000) at 37°C for 2 hours, and then treated using the TSA-plus FLU kit (Perkin Elmer). Sections were then coverslipped with Fluoromount-G with DAPI (Southern Biotech).
Costaining for nArc, Vglut1/2 or Gad1/2 mRNA and PRVB177 (HA)
After hybridization, sections were washed twice in 0.2× SSC at 63°C for 30 minutes, incubated with POD-conjugated anti-DIG antibodies (Roche, #11207733910, 1:2000) and biotinylated anti-HA antibodies (BioLegend, #901505, 1:300) at 37°C for 2 hours, and then treated using the TSA-plus Cy3 kit (Perkin Elmer). Sections were then incubated with 0.5 μg/ml DAPI and Alexa488 Streptavidin (Thermo Fisher, 1:1000) at room temperature for 1 hour, and were coverslipped with Fluoromount-G.
Costaining with Crh with nuclear c-Fos riboprobes
After hybridization, sections were washed twice in 0.2× SSC at 63°C for 30 minutes, blocked with Streptavidin/Biotin Blocking kit (Vector Laboratories), incubated with POD-conjugated anti-FLU antibodies (Roche, #11426346910, 1:300) and alkaline phosphatase (AP)-conjugated anti-DIG antibodies (Roche, #11093274910, 1:300) at room temperature for 2 hours, and then treated using the TSA-plus Biotin kit (Perkin Elmer). Sections were next incubated with 0.5 μg/ml DAPI and Alexa488 Streptavidin (Thermo Fisher, 1:1000) at room temperature for 30 minutes, incubated with HNPP Fluorescent Detection Set (Roche) at room temperature for 1 hour, and then coverslipped with Fluoromount-G.
Chemogenetic neuronal activation/silencing
Activation
AAV-DIO-hM3Dq-mCherry was injected into the pPir, AmPir or MEA of Emx1-ires-Cre or Vglut2-ires-Cre mice by stereotaxic injection (see above). At ≥ 2 weeks after injection, mice were intraperitoneally injected with clozapine-N-oxide (CNO, Sigma) (5.0 mg/kg body weight) or control saline. Twenty minutes later, trunk blood and brain were collected and the blood used for plasma ACTH or corticosterone assays (see above). Brains were fixed with 4% PFA for 4 hours, soaked in 30% sucrose for 24 hours, frozen in OCT, and cut into 20 μm coronal sections using a cryostat. To analyze expression of hM3Dq-mCherry, brain sections were immunostained with rabbit anti-RFP (Rockland, #600-410-379, 1:500) followed by Alexa555 donkey anti rabbit IgG (Thermo Fisher, #A31572, 1:1000) antibodies (see above).
Silencing
AAV-DIO-hM4Di-mCherry was injected into the AmPir of Emx1-ires-Cre mice by stereotaxic injection (see above). At ≥ 2 weeks after injection, mice were intraperitoneally injected with CNO (5.0 mg/kg body weight) or control saline. Forty minutes later, mice were exposed to bobcat urine or TMT for 10 minutes (see above). Animal behavior during exposure was recorded with a Canon PowerShot ELPH300HS camera placed above the chamber, and the duration that mice stayed immobile during the 10-minute odor exposure were scored by using Ethovision XT11 software (Noldus Information Technology). Trunk blood and brain were then collected. Blood was used for plasma ACTH or corticosterone assays (see above). Brains were treated and immunostained with rabbit-anti-RFP antibodies (see above).
Cell counting
Images were collected using an AxioCam camera and AxioImager.Z1 microscope with apotome device (Zeiss), or TissueFAXS system (Tissuegnostics). Brain structures were identified microscopically and in digital photos using a mouse brain atlas30.
To analyze the locations and number of PRV-infected cells in brain areas, every fifth section was analyzed, and numbers of PRV+ cells were multiplied by five to acquire approximate total number of cells per animal. Brain areas were judged to contain upstream neurons if they contained ≥ 10 labeled neurons in ≥ 50% of animals.
To analyze the location and number of virus-infected and Arc-expressing cells, three of five sections were analyzed. The number of PRV+ cells with or without Arc mRNA was blindly counted. The percentages of PRV+ cells with only nuclear Arc signals among all PRV+ cells lacking cytoplasmic Arc were calculated.
To analyze the activation of CRH neurons in chemogenetic experiments, the numbers of CRH+ neurons labeled for nuclear c-Fos mRNA were blindly counted.
To analyze the distribution of virus-infected cells, all sections through the regions of interest were analyzed. The section images were aligned from anterior to posterior, and the numbers of virus-infected cells were counted. The size of each brain was adjusted linearly so that each posterior piriform cortex had the same length (2.8 mm).
Statistical analysis
All data are shown as the mean±SEM. Data were tested with the Shapiro-Wilk test for normality. For data with a normal distribution, the unpaired t-test or one-way ANOVA with post-hoc Dunnett’s test was used to compare two groups or more than two groups, respectively, to analyze statistical significance. For t-tests, equality of variances was analyzed with the F-test and Welch’s correction was employed when variances of populations were significantly different. For data without a normal distribution, the Mann–Whitney U test or the Kruskal-Wallis test with post-hoc Dunn’s test was used to compare two groups or more than two groups, respectively. All tests were two-sided. Detailed information on the numbers of animals used, statistical analyses, and effect sizes is provided in Supplementary Table 1.
Extended Data
Extended Data Figure 1. Virus infection and spread in CRH-Cre and wildtype mice.
a, Photographs show a PVN section from CRH-Cre mice crossed with Rosa-floxstop-GFP mice that was costained with anti-GFP antibodies (green) (Cre) and a CRH riboprobe (red) (CRH). Colabeled cells (yellow) can be seen in the merged image, verifying the expression of Cre in CRH neurons. Of GFP+ (Cre+) neurons in PVN sections, 93.9±0.5% expressed CRH. (n=4) Scale bar = 100 μm.
b, Photographs show a section through the PVN of a CRH-Cre mouse previously injected with PRVB316 and LVF2TK. The section was costained with antibodies for Cre-dependent reporters of PRVB316 (GFP (green)) and LVF2TK (HA (red)). Some colabeled neurons are seen (arrows), verifying coinfection of some neurons by the two viruses. Scale bar = 200 μm.
c, The number of PRVB316+ cells inside or outside the PVN following PVN injection of PRVB316 alone (n=2) or PRVB316+LVF2TK (n=24) in CRH-Cre mice or PRVB316+LVF2TK in wildtype (WT) mice (n=3). Error bars indicate SEM. In CRH-Cre mice, PRVB316+ cells were seen both inside and outside the PVN following coinjection of PRVB316 and LVF2TK, but only inside the PVN when only PBVB316 was injected. In WT mice coinjected with the two viruses, PRV+ cells were not detected either inside or outside the PVN.
d, The number of PRV+ cells inside or outside PVN 3 or 5 days after PVN injection of PRVB177 in CRH-Cre or WT mice. (n=8 (CRH-Cre mice, Day3), n=6 (CRH-Cre mice, Day5), n=2 (WT mice, Day 3), n=2 (WT mice, Day 5). Error bars indicate SEM.). PRVB177+ neurons were seen both inside and outside the PVN of CRH-Cre mice on both days, but were not detected in WT mice at either location.
Extended Data Figure 2. Brain areas with neurons upstream of CRH neurons.
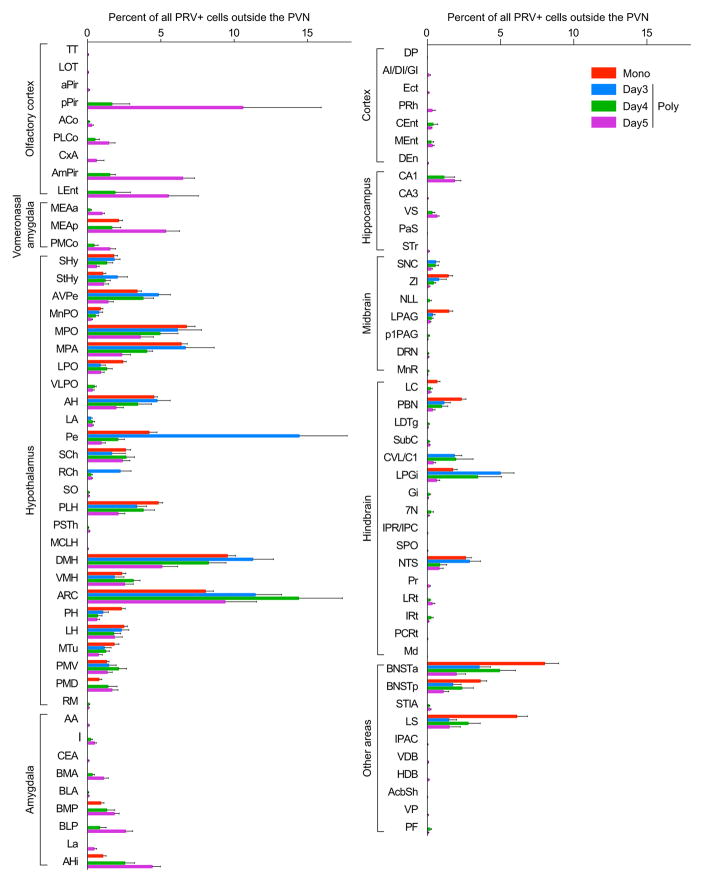
The percentage of all non-PVN PRV+ neurons found in individual brain areas four days after PVN injection of the monosynaptic PRV (PRVB316) (Mono) (n=24) or on day 3, 4, or 5 after PVN injection of the polysynaptic PRV (PRVB177) (Poly) (n=8, 6, and 6, for day 3, 4, and 5, respectively). Error bars indicate SEM. See Methods for full names of abbreviated brain areas.
Extended Data Figure 3. Brain areas containing neurons upstream of CRH neurons.
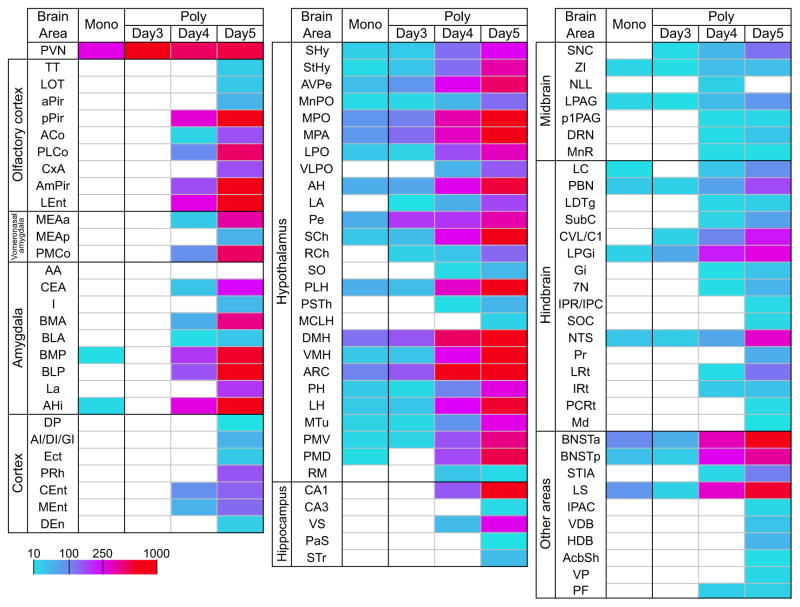
Colored boxes indicate the approximate number of infected neurons seen in individual brain areas (white indicates none) on day 4 after PVN injection of CRH-Cre mice with the monosynaptic PRV (PRVB316 (Mono)) (n=24) or on day 3, 4, or 5 after injection with the polysynaptic PRV (PRVB177 (Poly)) (n=8, 6, and 6, for day 3, 4, and 5, respectively). See Methods for full names of abbreviated brain areas.
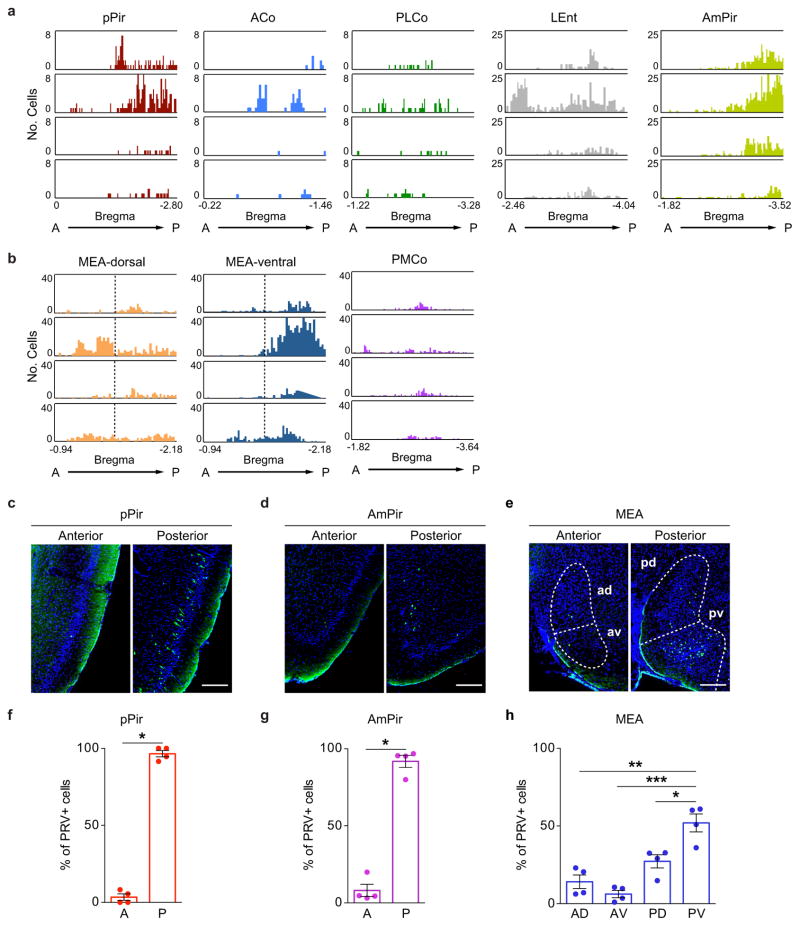
Extended Data Figure 4. Biased distribution of neurons upstream of CRH neurons in pPir, AmPir, and MEA.
a, b, Diagrams show the numbers of PRVB177-infected (HA+) neurons on day 4 after PVN injection in sequential 20 μm coronal sections along the anterior (A)-posterior (P) axis in different areas of the olfactory cortex (a) and vomeronasal amygdala (b). Data are shown in separate panels for four animals for each area. Infected neurons appear to be randomly distributed along the anterior-posterior axis in some areas, but not others.
c–e, Photographs of sections immunostained for PRVB177 (HA, green) on day 4 after PVN injection of PRVB177. Sections were counterstained with DAPI. More PRVB177+ neurons are seen in the posterior than anterior pPir (c) and AmPir (d). In the MEA (e), more are seen in the posteroventral (pv) quadrant than other quadrants (posterodorsal (pd), anterodorsal (ad), and anteroventral (av)). Scale bars = 200 μm.
f–h, The percentages of PRVB177-infected neurons in the anterior (A) versus posterior (P) halves of pPir (f) and AmPir (g) and in different quadrants of the MEA (h). (n=4. Error bars indicate SEM. *p<0.05, **p<0.01, ***p<0.001. Paired t-test (f, g) and paired one-way ANOVA with post-hoc Tukey’s test (h).) Infected neurons were concentrated in the posterior halves of pPir and AmPir and were more abundant in the posteroventral quadrant of the MEA than in other quadrants.
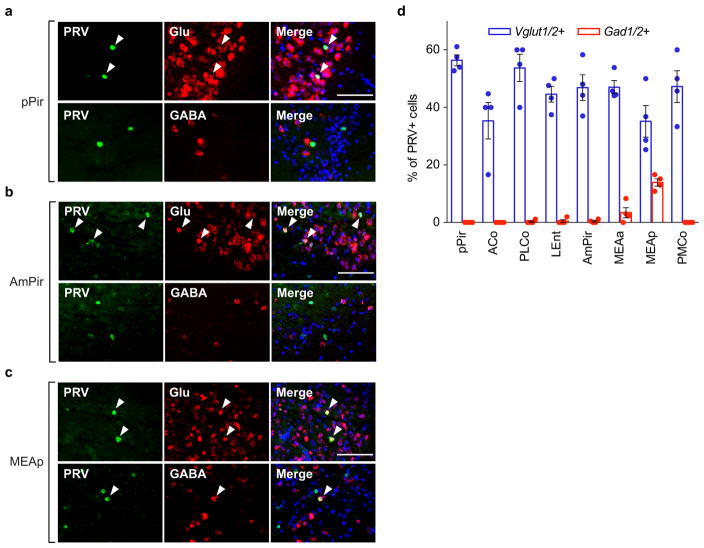
Extended Data Figure 5. Expression of glutamatergic and GABAergic markers in PRV-infected neurons.
a–c, Photographs of sections through the pPir (a), AmPir (b), or MEAp (c) four days after PVN injection of PRVB177. Sections were costained with antibodies against HA (PRVB177) (green) and riboprobes (red) for Vglut1 and Vglut2 (Vglut1/2), to identify glutamatergic neurons (Glu), or Gad1 and Gad2 (Gad1/2), to identify GABAergic neurons (GABA). Sections were counterstained with DAPI. Arrowheads indicate colabeled neurons. Numerous PRV-infected neurons were colabeled for Vglut1/2 in all three areas, but only the MEAp contained many colabeled for Gad1/2. Scale bars = 100 μm.
d, The percentage of PRVB177+ neurons colabeled for glutamatergic (Vglut1/2+) or GABAergic (Gad1/2+) markers in different areas of the olfactory cortex and vomeronasal amygdala. PRV+ neurons unlabeled by either probe could express other neurotransmitters or neuromodulators or reflect PRV effects on RNA transcription or degradation. (n=4 mice per condition. Error bars indicate SEM.) Most of the colabeled PRV+ cells in the olfactory areas were colabeled for the glutamatergic makers, but the MEA also contained some colabeled for the GABAergic markers.
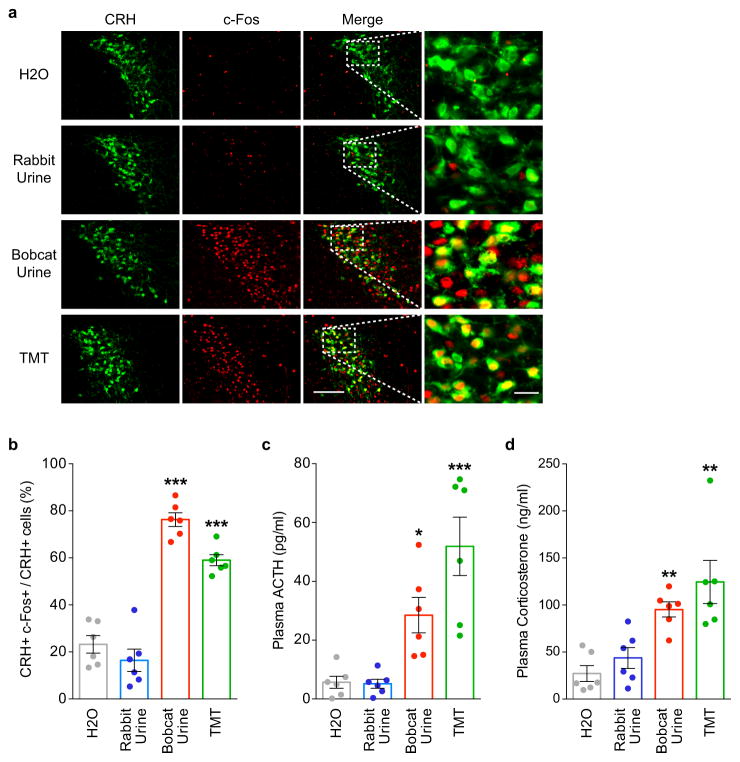
Extended Data Figure 6. TMT and bobcat urine activate CRH neurons and induce stress hormone increases.
a, Photographs of PVN sections from CRH-Cre mice crossed with Rosa-floxstop-GFP mice. Mice were exposed to water (H2O), rabbit urine, bobcat urine, or TMT and sections then costained with antibodies against GFP (green), to identify CRH neurons, and c-Fos (red), to detect activated neurons. Higher magnifications of boxed areas are shown at the right. Many CRH neurons (GFP+) expressed c-Fos after exposure to bobcat urine or TMT, but not rabbit urine. Scale bars = 100 μm (left) and 20 μm (right).
b, The percentage of PVN CRH (GFP+) neurons immunostained for c-Fos after odor exposure of CRH-Cre mice crossed with Rosa-floxstop-GFP mice. Exposure to bobcat urine or TMT, but not rabbit urine, increased the percentage of CRH neurons expressing c-Fos. (n=6 mice per condition. Error bars indicate SEM. ***p<0.001. One-way ANOVA with post-hoc Dunnett’s test.)
c, d, Plasma ACTH (c) or corticosterone (d) concentrations were measured in WT mice following odor exposure. Exposure of animals to TMT or bobcat urine increased blood levels of ACTH and corticosterone, whereas exposure to rabbit urine did not. (n=6 mice per condition. Error bars indicate SEM. *p<0.05, **p<0.01, ***p<0.001. One-way ANOVA with post-hoc Dunnett’s test (c) or Kruskal-Wallis test with post-hoc Dunn’s test (d).)
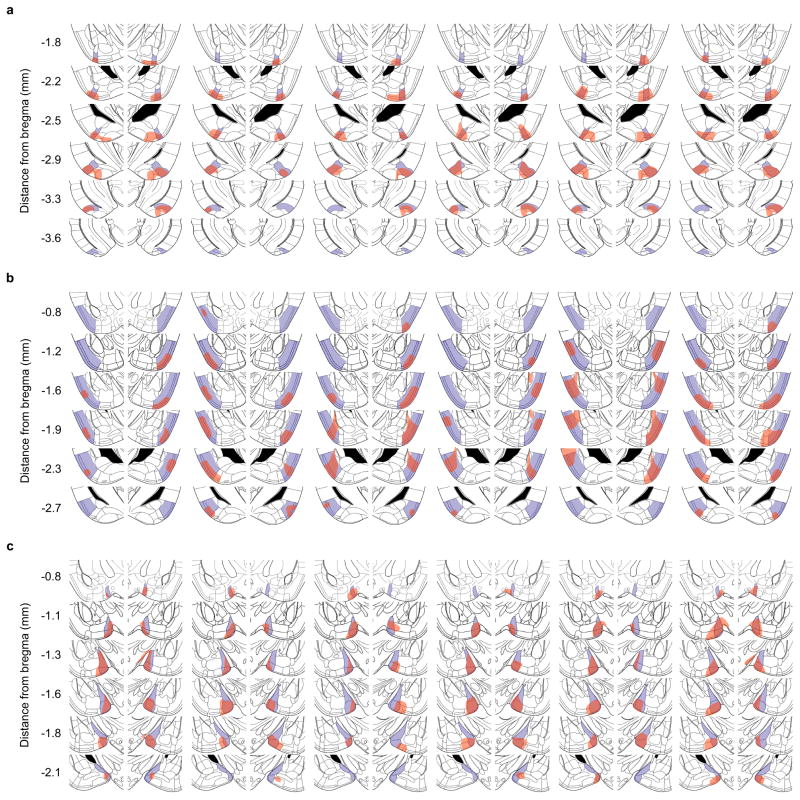
Extended Data Figure 7. Locations of infected neurons after injection of AAV DIO-hM3Dq-mCherry.
AAV DIO-hM3Dq-mCherry was injected into the AmPir (a), pPir (b), or MEA (c) and brain sections immunostained for mCherry to detect infected neurons. Shown are schematic sections from a mouse brain atlas30 that correspond to immunostained sections. The target area (AmPir, pPir, or MEA) is indicated in purple for each section. Regions that contained the nuclei of mCherry+ neurons are indicated in red. Schematic sections are shown for 6 animals per area that were injected with CNO to test the effects of activating the area.
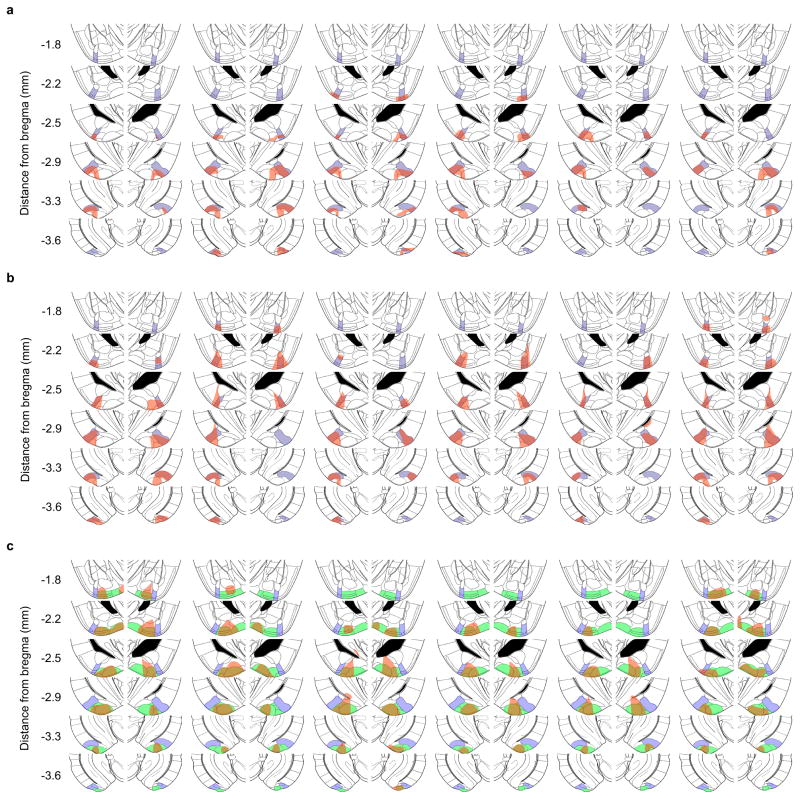
Extended Data Figure 8. Locations of infected neurons after injection of AAV DIO-hM4Di-mCherry.
AAV DIO-hM4Di-mCherry was injected into the AmPir (a, b) or PLCo/PMCo (c) and brain sections immunostained for mCherry to detect infected neurons. Shown are schematic sections from a mouse brain atlas30 that correspond to immunostained sections. The location of AmPir is indicated in purple (a–c), and the location of PLCo/PMCo is indicated in green (c). Regions that contained the nuclei of mCherry+ neurons are indicated in red in each section. Schematics are shown for 6 animals per area (per condition) injected with CNO and then exposed to bobcat urine (a, c) or TMT (b) to test the effects of silencing the AmPir or PLCo/PMCo.
Extended Data Figure 9. Effect of neuronal silencing on the activation of neurons by predator odors.
AAV DIO-hM4Di-mCherry was injected into the AmPir (a, b) or PLCo/PMCo (c). Animals were later injected with CNO or saline and exposed to bobcat urine (a, c) or TMT (b). Brain sections were hybridized with an Arc riboprobe and the number of cells labeled for nuclear Arc (nArc) determined for the AmPir, PLCo, and PMCo. Histograms show the number of cells with nArc in each area following injection with CNO or saline and exposure to predator odor. Based on the numbers of nArc+ neurons, silencing of AmPir neurons caused a significant decrease in AmPir and PLCo neurons activated by bobcat urine (a) and by AmPir (but not PLCo) neurons activated by TMT (b). Silencing of PLCo/PMCo (c) caused a significant decrease in neurons activated by bobcat urine in the PLCo, but not AmPir or PMCo. (n=6–7 per condition. Error bars indicate SEM. *p<0.05, **p<0.01, ***p<0.001. Unpaired t-test.)
Supplementary Material
Acknowledgments
We thank Lynn Enquist for providing the parental PRV, PRV TK-BaBlu and for helpful advice, and members of the Buck laboratory for discussions and comments. This work was supported by the Howard Hughes Medical Institute (HHMI) and by grants from the National Institutes of Health (National Institute on Deafness and Other Communication Disorders) (L.B.B.) and the Japan Society for the Promotion of Science (K.K.). L.B.B. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations for brain areas
Abbreviations used for brain areas are according to Franklin and Paxinos30, with a few minor modifications.
Olfactory areas
- ACo
Anterior cortical amygdala
- AON
Anterior olfactory nucleus
- AmPir
Amygdalo-piriform transition area
- CxA
Cortex-amygdala transition zone
- LEnt
Lateral entorhinal cortex
- LOT
Nucleus of lateral olfactory tract
- MEAa
Medial amygdala, anterior part
- MEAp
Medial amygdala, posterior part
- OT
Olfactory tubercle
- aPir
Piriform cortex, anterior part
- pPir
Piriform cortex, posterior part
- PLCo
Posterolateral cortical amygdala
- PMCo
Posteromedial cortical amygdala
- TT
Tenia tecta
Non-olfactory areas
- AA
Anterior amygdaloid area
- AcbSh
Accumbens nucleus, shell
- AH
Anterior hypothalamic area
- AHi
Amygdalohippocampal area
- AI
Agranular insular cortex
- ARC
Arcuate hypothalamic nucleus
- AVPe
Anteroventral periventricular nucleus
- BLA
Basolateral amygdala, anterior part
- BLP
Basolateral amygdaloid nucleus, posterior part
- BMA
Basomedial amygdala, anterior part
- BMP
Basomedial amygdaloid nucleus, posterior part
- BNSTa
Bed nucleus of the stria terminalis, anterior part
- BNSTp
Bed nucleus of the stria terminalis, posterior part
- CA1
Field CA1 of the hippocampus
- CA3
Field CA3 of the hippocampus
- CEA
Central amygdala CEnt, Caudomedial entothinal cortex
- CI
Caudal interstitial nucleus of the medial longitudinal
- CVL
Caudoventrolateral reticular nucleus
- DEn
Dorsal endopiriform claustrum
- DI
Dysgranular insular cortex
- DMH
Dorsomedial hypothalamic nucleus
- DP
Dorsal peduncular cortex
- DRN
Dorsal raphe nucleus
- Ect
Ectorhinal cortex
- 7N
Facial nucleu
- Gi
Gigantocellular reticular nucleus
- HDB
Nucleus of the horizontal limb of the diagonal band
- I
Intercalated nuclei of the amygdala
- IPAC
Interstitial nucleus of the posterior limb of the ant
- IPC
Interpeduncular nucleus, caudal subnucleus
- IPR
Interpeduncular nucleus, rostral subnucleus
- IRt
Intermediate reticular nucleus
- La
Lateral amygdala
- LA
Lateroanterior hypothalamic nucleus
- LC
Locus coeruleus
- LDTg
Laterodorsal tegmental nucleus
- LH
Lateral hypothalamic area
- LPAG
Lateral periaqueductal gray
- LPGi
Lateral paragigantocellular nucleus
- LPO
Lateral preoptic area
- LRt
Lateral reticular nucleus
- LS
Lateral septal nucleus
- MCLH
Magnocellular nucleus of the lateral hypothalamus
- Md
Medullary reticular nucleus
- MEnt
Medial entorhinal cortex
- MnPO
Median preoptic nucleus
- MnR
Median raphe nucleus
- MPA
Medial preoptic area
- MPO
Medial preoptic nucleus
- MTu
Medial tuberal nucleus
- NLL
Nucleus of the lateral lemniscus
- NTS
Nucleus of the solitary tract
- p1PAG
p1 periaqueductal gray
- PaS
Parasubiculum
- PBN
Parabrachial nucleus
- PCRt
Parvicellular reticular nucleus
- Pe
Periventricular nucleus of the hypothalamus
- PF
Parafascicular thalamic nucleus
- PH
Posterior hypothalamic nucleus
- PLH
Peduncular part of lateral hypothalamus
- PMD
Premammillary nucleus, dorsal part
- PMV
Premammillary nucleus, ventral part
- Pr
Prepositus nucleus
- PRh
Perirhinal cortex
- PSTh
Parasubthalamic nucleus
- PVN
Paraventricular nucleus of the hypothalamus
- RCh
Retrochiasmatic area
- RM
Retromammillary nucleus
- SCh
Suprachiasmatic nucleus
- SHy
Septohippocampal nucleus
- SNC
Substantia nigra
- SO
Supraoptic nucleus
- SOC
Subcommissural organ
- StHy
Striohypothalamic nucleus
- STIA
Bed nucleus of the stria terminalis, intraamygdaloid
- STr
Subiculum, transition area
- SubC
Subcoeruleus nucleus
- VDB
Vertical limb of the diagonal band
- VLPO
Ventrolateral preoptic nucleus
- VMH
Ventromedial hypothalamic nucleus
- VP
Ventral pallidum
- VS
Ventral subiculum
- ZI
Zona incerta
Footnotes
Online Content. Methods, along with any additional Extended Data display items are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author contributions. K.K. and L.B.B. conceived the project, Z.L. and K.K. designed and prepared the viruses, X.Y. performed experiments, D.P.O. and B.B.L. generated the CRH-Cre mice, K.K. developed methods and performed most of the experiments and data analysis, and K.K. and L.B.B. wrote the manuscript.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Apfelbach R, Blanchard CD, Blanchard RJ, Hayes RA, McGregor IS. The effects of predator odors in mammalian prey species: a review of field and laboratory studies. Neurosci Biobehav Rev. 2005;29:1123–1144. doi: 10.1016/j.neubiorev.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi LK. Olfactory systems and neural circuits that modulate predator odor fear. Front Behav Neurosci. 2014;8:72. doi: 10.3389/fnbeh.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci. 2009;10:397–409. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayakawa K, et al. Innate versus learned odour processing in the mouse olfactory bulb. Nature. 2007;450:503–508. doi: 10.1038/nature06281. [DOI] [PubMed] [Google Scholar]
- 5.Papes F, Logan DW, Stowers L. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell. 2010;141:692–703. doi: 10.1016/j.cell.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck LB, Bargmann C. In: Principles of Neuroscience. Kandel E, Schwartz J, Jessell T, Siegelbaum S, Hudspeth AJ, editors. McGraw-Hill; 2012. pp. 712–742. [Google Scholar]
- 7.Neville KR, Haberly LB. In: The synaptic organization of the brain. Shepherd GM, editor. Oxford University Press; 2004. pp. 415–454. [Google Scholar]
- 8.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 9.Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 10.Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 11.Hanchate NK, et al. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350:1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 13.Vassar R, et al. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 14.Mombaerts P, et al. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 15.Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472:213–216. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamichi K, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh S, et al. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 2011;472:217–220. doi: 10.1038/nature09945. [DOI] [PubMed] [Google Scholar]
- 18.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515:269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Card JP, Enquist LW. Transneuronal circuit analysis with pseudorabies viruses. Curr Protoc Neurosci. 2001;Chapter 1(Unit1 5) doi: 10.1002/0471142301.ns0105s09. [DOI] [PubMed] [Google Scholar]
- 20.DeFalco J, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- 21.Krashes MJ, et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 2014;507:238–242. doi: 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawchenko PE, Swanson LW. The organization of forebrain afferents to the paraventricular and supraoptic nuclei of the rat. J Comp Neurol. 1983;218:121–144. doi: 10.1002/cne.902180202. [DOI] [PubMed] [Google Scholar]
- 23.Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- 24.Vernet-Maury E. Trimethyl-thiazoline in fox feces: A natural alarming substance for the rat. Olfaction Taste. 1980:407. [Google Scholar]
- 25.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsukawa M, Imada M, Murakami T, Aizawa S, Sato T. Rose odor can innately counteract predator odor. Brain Res. 2011;1381:117–123. doi: 10.1016/j.brainres.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 30.Franklin K, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. 3. 2008. [Google Scholar]
- 31.Schnutgen F, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 32.Banfield BW, Bird GA. Construction and analysis of alphaherpesviruses expressing green fluorescent protein. Methods Mol Biol. 2009;515:227–238. doi: 10.1007/978-1-59745-559-6_15. [DOI] [PubMed] [Google Scholar]
- 33.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 34.Boehm U, Zou Z, Buck LB. Feedback loops link odor and pheromone signaling with reproduction. Cell. 2005;123:683–695. doi: 10.1016/j.cell.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 35.Liberles SD, Buck LB. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442:645–650. doi: 10.1038/nature05066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.