Abstract
Chromatin, primarily a complex of DNA and histone proteins, is the physiological form of the genome. Chromatin is generally repressive for transcription and other information transactions that occur on DNA. A wealth of post-translational modifications on canonical histones and histone variants encode regulatory information to recruit or repel effector proteins on chromatin, promoting and further repressing transcription and thereby form the basis of epigenetic information. During metazoan oogenesis, large quantities of histone proteins are synthesized and stored in preparation for the rapid early cell cycles of development and to elicit maternal control of chromatin assembly pathways. Oocyte and egg cell-free extracts of the frog Xenopus laevis are a compelling model system for the study of chromatin assembly and transcription precisely because they exist in an extreme state primed for rapid chromatin assembly or for transcriptional activity. We show that chromatin assembly rates are slower in X. laevis oocyte than in egg extracts, while conversely only oocyte extracts transcribe template plasmids. We demonstrate that rapid chromatin assembly in egg extracts represses RNA Polymerase II dependent transcription, while pre-binding of TATA-Binding Protein (TBP) to a template plasmid promotes transcription. Our experimental evidence presented here supports a model in which chromatin assembly and transcription are in competition and that the onset of zygotic genomic activation may be in part due to stable transcriptional complex assembly.
INTRODUCTION
Chromatin is the complex of DNA and histones that is the biological form of the eukaryotic genome (Shechter and Allis, 2007). Chromatin is generally repressive for transcription of RNA from the genome. During early metazoan development, chromatin assembly and transcriptional activation and repression are simultaneously acting to establish the embryonic epigenome and transcriptome, building a program essential for development and life of the organism. Chromatin and transcription are interrelated, especially during these important regulatory transitions that occur during early metazoan embryogenesis. The organization of DNA into nucleosomes is critical as it not only allows the long stretch of DNA to be housed inside the nucleus but as it also serves as the fundamental regulatory center for gene expression (Bannister and Kouzarides, 2011, Das and Tyler, 2013).
Early development in animals is primarily controlled by maternal RNAs and proteins, including histones and histone chaperones that form chromatin. New zygotic control over the maternal program occurs at various stages depending on the species at a time point variously called the mid-blastula transition (MBT), maternal-to-zygotic transition (MZT), and zygotic genome activation (O'Farrell et al., 2004, Schier, 2007, Tadros and Lipshitz, 2009). The MBT is typically the period of dramatic cell cycle changes as well as the beginning of differentiation. Since new zygotic transcription occurs during this time, a reasonable hypothesis is that the dramatic chromatin and transcriptional changes are critical for subsequent differentiation. We previously showed that substantial and elaborate changes in abundance of different core and linker histones and discrete classes of histone modifications occur during vertebrate MBT (Nicklay et al., 2009, Shechter et al., 2009a, Shechter et al., 2009b, Wang et al., 2014). Others have shown differences in chromatin assembly ability and rates in different oocyte and embryo contexts (Almouzni et al., 1990, Almouzni et al., 1991, Dimitrov et al., 1993, Hair et al., 1998, Prioleau et al., 1995, Prioleau et al., 1994, Wolffe et al., 1993). A still unanswered question is the direct relationship between the distinct chromatin states and the transcriptional competence of that chromatin.
Xenopus frogs are commonly used vertebrate model systems for studying development, cell cycle, cancer, chromatin, and many other biological phenomena. In particular, studies in Xenopus cell free extracts and embryos have established robust biological pathways regulating chromatin assembly and transcription, however many questions about how maternal chromatin modifications and deposition machinery function to control gene expression remain. In this issue (Shechter, co-submitted), we provide a short review and overview of the literature on chromatin and transcription in early development and the significance of the cross-talk between these biological events. Here, we present new experiments testing how chromatin assembly and transcription are coincident and correlated with each other.
RESULTS AND DISCUSSION
Utility of Xenopus extracts for studying transcription
Xenopus oocyte and egg extracts are prepared from dissected ovaries, prior to germinal vesicle breakdown and arrested in meiosis I, or laid eggs that have entered the somatic cell cycle (Figure 1). Importantly, there is no active transcription in extracts from Xenopus eggs through the mid-blastula transition around developmental stage 8 (Figure 1A) (Maller et al., 2001, Newport and Kirschner, 1982), yet any DNA can be duplicated in the extract (Figure 1B). We have used multiple reporter plasmids in the assay presented here as any plasmid with a strong viral promoter (for RNA Polymerase II) will induce transcription in these conditions (Toyoda and Wolffe, 1992). We mainly used the plasmid pG5ML (An et al., 2004, An and Roeder, 2004) and the commercial pC1-CMV-EYFP plasmid.
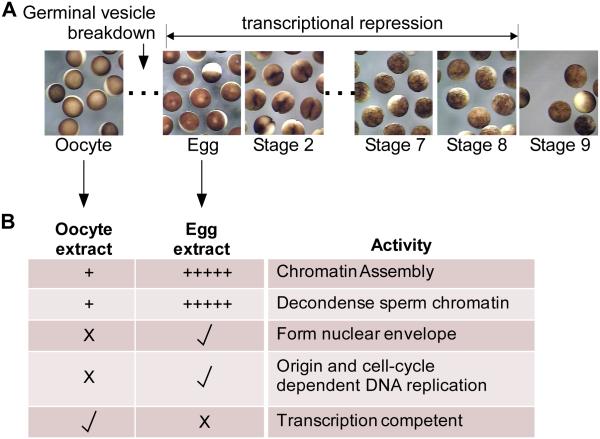
Figure 1. Xenopus laevis early embryology and extract functionality.
A. Stereomicrographs of Xenopus laevis stage IV oocytes, eggs (stage 1), and Stage 2 through Stage 10 embryos as indicated. Timing of germinal vesicle breakdown (GVBD) and the onset of transcriptional repression is indicated.
B. Cell-free oocyte extract is prepared from a pool of dissected oocytes (stages II-VI, primarily the later stages as the early stage oocytes are lost), while cell-free egg extract is prepared from laid eggs. The table shows various activities present in the extracts (+ = modest activity; +++++ = high activity; X = no activity; check = activity).
Chromatin Assembly in Oocyte and Egg Extracts
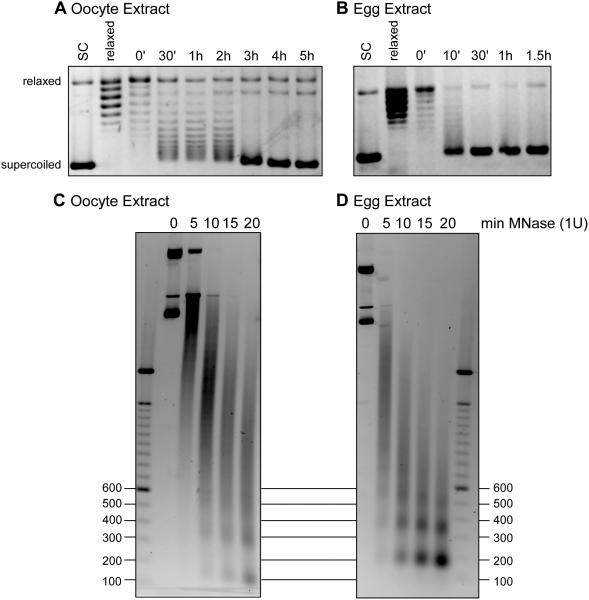
To clearly establish the difference in chromatin assembly and assembly rates, we first prepared Xenopus laevis oocyte and egg extracts as we had previously described (Figure 1B) (Banaszynski et al., 2010). We incubated oocyte and egg extract with supercoiled plasmid DNA as well as Topoisomerase I relaxed plasmid DNA. Products were stopped in SDS-containing solution and DNA was phenol-chloroform isolated at time points up to 5 hours (oocyte extract) or 1.5 hours (egg extract) and run on an agarose gel in the absence of ethidium bromide. Stained gels are shown in Figures 2A,B, with increased supercoiled DNA indicative of chromatin assembly. The rate of assembly was substantially faster in egg extract compared to oocyte extract.
Figure 2. Plasmid chromatin assembly in oocyte and egg extracts.
A. pG5ML plasmid was relaxed by Topoisomerase I and incubated in oocyte extract for up to 5 hours as indicated. Reactions were stopped at indicated time, deproteinized and the DNA was run on an agarose gel and stained with EtBr. Supercoiled and relaxed positions are indicated.
B. pG5ML plasmid was relaxed by Topoisomerase I and incubated in egg extract for up to 1.5 hours as indicated. Reactions were stopped at indicated time, deproteinized and the DNA was run on an agarose gel and stained with EtBr. Supercoiled and relaxed positions are indicated
C. Plasmid DNA was incubated in oocyte extract for 2.5 hours and reactions were treated with 1U of micrococcal nuclease (MNase) for 0, 5, 10, 15, or 20 minutes. Reactions were deproteinized and the DNA was run on an agarose gel and stained with EtBr. Marker positions are indicated in basepair length. Nucleosomes are 146bp to 200bp fragments.
D. Plasmid DNA was incubated in egg extract for 2.5 hours and reactions were treated with 1U of micrococcal nuclease (MNase) for 0, 5, 10, 15, or 20 minutes. Reactions were deproteinized and the DNA was run on an agarose gel and stained with EtBr. Marker positions are indicated in basepair length. Nucleosomes are 146bp to 200bp fragments.
To confirm that the supercoiled DNA product represented bona fide nucleosome assembly, we subjected the 1 hour time point of the oocyte and egg extract chromatin assembly reactions to micrococcal nuclease (MNase) digestion. MNase specifically cleaves chromatin between nucleosomes and reveals the size and regularity of assembled chromatin by producing a ladder of 146-200bp DNA molecules, the typical size of nucleosome fragments. As shown in Figure 2C, oocyte extract assembled chromatin digested by 1 unit of MNase resulted in a smear of DNA and a few regularly spaced but smaller fragments. This suggests either poor and incomplete assembly or irregular assembly. In contrast, chromatin assembled on a plasmid in egg extract exhibited a clear ladder of nucleosomal sized DNA fragments, indicating robust assembly (Figure 2D).
Our conclusions from these initial experiments are that egg extract is more efficient at assembling chromatin than is oocyte extract. This is likely due to differences in regulation of histone chaperones, as we previously observed for Nucleoplasmin (Onikubo et al., 2015), differences in post-translational modifications of core and linker histones (Nicklay et al., 2009, Shechter et al., 2009b), or some other modulation of remodeling factors, such as ISWI (Kikyo et al., 2000). As with most work from Xenopus extracts due to the heterogeneity of the population of frogs, while there were batch-to-batch variations, egg extract was routinely more efficient than oocyte extract at assembling chromatin (data not shown).
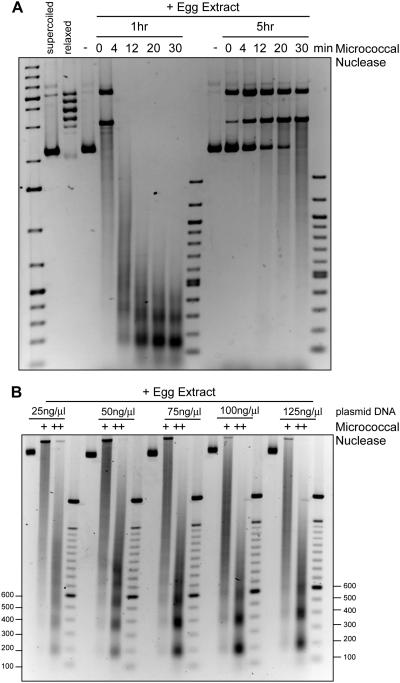
The time course of chromatin assembly in egg extract revealed apparently complete supercoiling by 1 hour. To further probe chromatin assembly rate and density, we probed assembled plasmid DNA in egg extract at 1 and 5 hours with micrococcal nuclease. As shown in Figure 3A, chromatin assembled at 1 hour showed a repeating nucleosomal ladder, consistent with our experiment in Figure 2D. However, chromatin assembled for 5 hours was largely resistant to micrococcal nuclease, consistent with a large assembly of proteins on the chromatin, perhaps indicative of the artificial nature of this experimental setup. In embryos, cell divisions occur at 90 minutes after fertilization and then roughly every 45 minutes, so chromatin assembly would not persist for 5 hours. Alternatively, there are large quantities of linker histones and HMG proteins in the egg and robust assembly of these proteins onto chromatin may preclude MNase digestion.
Figure 3. Plasmid chromatin assembly in egg extract is concentration and time dependent.
A. pG5ML plasmid was relaxed by Topoisomerase I and incubated in egg extract for 1 or 5 hours, as indicated. Reactions were treated with 1U of MNase for 0, 4, 12, 20, or 30 minutes and the DNA was run on an agarose gel and stained with EtBr. Nucleosomal sized fragments were readily apparent in the 1 hour incubation but not the 5 hour incubation.
B. pG5ML plasmid was relaxed by Topoisomerase I and incubated in egg extract for 1 hour at 25, 50, 75, 100, or 125 ng DNA / μl of egg extract, as indicated. Reactions were treated with 1 or 2 units of MNase and DNA was run on an agarose gel and stained with EtBr.
Hair et al. (1998) showed that titration of plasmid DNA into egg extract revealed a limited ability to supercoil DNA above 25 ng DNA per μl of egg extract, likely due to titration of histones from the extract. This observation of limiting histones in eggs was recently shown to be responsible for setting the DNA-to-cytoplasm ratio in embryos, partly responsible for regulating timing of the MBT (Amodeo et al., 2015). To test the extent of limited histones in micrococcal nuclease digestion, we titrated the concentration of DNA from 25 to 125 ng/μl of egg extract (Figure 3B). We observed maximal laddering of nucleosomes at 50 ng/μl of egg extract, but clear nucleosomal formation at the highest concentration. This suggests that even though histones are limiting in the egg, nucleosomal formation still occurs but at reduced density.
Transcription in oocyte and egg extracts
To measure transcription in oocyte and egg extracts, we spiked extracts with 32P-α-ATP and a plasmid with a CMV promoter and TATA box (pCMV-EYFP), incubated the extract, isolated total RNA with Trizol, and counted the resulting alcohol precipitated RNA in a scintillation counter. As shown in Figure 4A, oocyte extract incubated with the plasmid exhibited robust incorporation of the radiolabeled ribonucleotide and this incorporation was blocked upon the addition of α-amanitin, a highly potent and specific RNA Polymerase II inhibitor (Wieland and Faulstich, 1978), consistent with Pol II dependent transcription off of the plasmid. In contrast, the same plasmid incubated in egg extract was not transcribed.
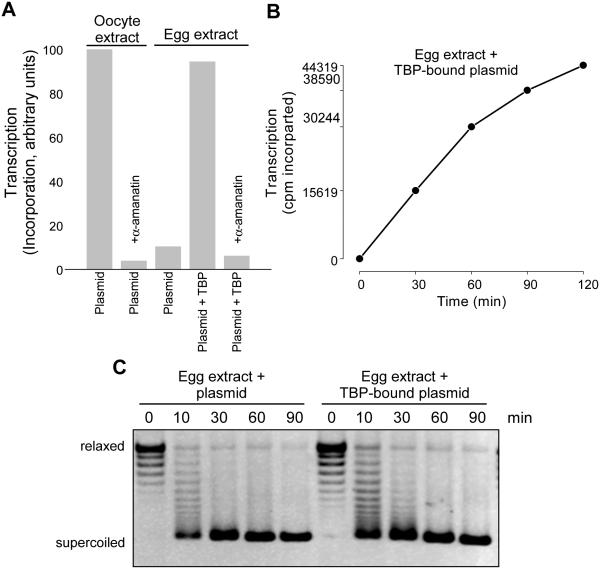
Figure 4. Transcription in oocyte and egg extracts.
A. pCMV-EYFP plasmid was incubated in oocyte or egg extracts in the presence of 32P-α-ATP α-amanitin was added or the plasmid pre-incubated with TBP protein, as indicated. The reaction products were stopped and RNA extracted with TRIzol. Precipitated RNA was directly counted in a scintillation counter and resulting transcription radioactivity was scaled to the oocyte plasmid control.
B. pCMV-EYFP plasmid was incubated with TBP protein and then in egg extract in the presence of 32P-α-ATP and the reactions stopped and RNA extracted at 0, 30, 60, 90, and 120 minutes after addition. Resulting radioactive incorporation is plotted (cpm) demonstrating transcription over time.
C. Topoisomerase-I relaxed pCMV-EYFP plasmid or plasmid pre-bound with TBP was incubated in egg extracts and stopped at indicated time. The reactions were deproteinized and the DNA was run on an agarose gel and stained with EtBr. Supercoiled and relaxed positions are indicated.
We confirmed the earlier studies that suggested that pre-incubating the plasmid template with TATA binding protein (TBP) stimulated transcription (Almouzni and Wolffe, 1995), which we also demonstrated was blocked by α-amanitin confirming the specific Pol II transcription (Figure 4A). Plasmid incubated with exogenous TBP at the same time as egg extract was not transcribed (data not shown). We then performed a time course of incorporation into RNA on TBP-bound plasmid, showing a linear increase in incorporation (Figure 4B).
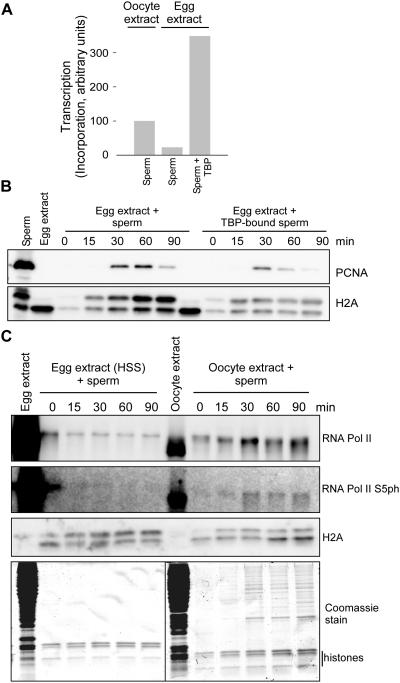
These observations were all consistent with a potential model in which chromatin assembly was competitive with transcriptional complex assembly. Therefore we tested supercoiling on a plasmid in egg extract or pre-bound with TBP and then incubated in extract and observed a modest reduction in the rate of supercoiling (Figure 5A), consistent with a reduction in chromatin assembly as a mechanism of promoting transcription. To test this model with a different approach, we incubated Xenopus sperm chromatin in oocyte or egg extract and measured transcription (Figure 5B). Sperm pre-incubated with TBP was transcribed while sperm in the absence of TBP was not transcribed. To measure chromatin assembly on sperm pronuclei, we isolated chromatin at time points after incubation in egg extract and immunoblotted for PCNA as an indication of DNA replication and histone H2A as an indication of histone deposition and chromatin assembly. As shown in Figure 4E, we observed reduced PCNA and H2A abundance on the TBP-bound sperm chromatin as opposed to the sperm alone, consistent with our hypothesis. To independently confirm that the observed incorporation off of the template was indeed due to RNA Pol II, we isolated chromatin assembled in egg or oocyte extract and immunoblotted for Pol II and Pol II CTD Ser5 phosphorylation (S5ph), an indication of transcriptional initiation. We only observed S5ph in chromatin from oocyte extract, consistent with the observed transcriptional readout (Figure 5C).
Figure 5. Sperm Chromatin transcription in oocyte and egg extracts.
A. Sperm chromatin was incubated in oocyte or egg extracts in the presence of 32P-α-ATP. Sperm was pre-incubated with TBP protein as indicated. The reaction products were stopped and RNA extracted with TRIzol. Precipitated RNA was directly counted in a scintillation counter and resulting transcription radioactivity was scaled to the oocyte sperm control.
B. Sperm chromatin was incubated in egg extract either without or pre-incubated with TBP protein as indicated. The reaction product chromatin was isolated after 0, 15, 30, 60 or 90 minutes through a sucrose cushion and pelleted proteins were immunoblotted for PCNA and histone H2A.
C. Sperm chromatin was incubated in oocyte or egg extracts. The reaction product chromatin was isolated after 0, 15, 30, 60 or 90 minutes through a sucrose cushion and pelleted proteins were immunoblotted for RNA Pol II, Pol II S5ph, and H2A as indicated. A total Coomassie stained gel of the isolated products is also shown at the bottom.
Implications for the regulation of chromatin assembly and transcription: a race between chromatin assembly and transcription?
Here we demonstrated that Xenopus oocyte and egg extracts are compelling models for the crosstalk between chromatin assembly and transcription as they represent an extreme state not found in somatic cells. Since there are dramatic shifts between transcriptional activity and transcriptional quiescence, concomitant with altered chromatin assembly rates, the developing embryos and as recapitulated in the cell free extracts, these experimental systems can be directly probed for regulatory mechanisms. We confirmed earlier reports that simple pre-incubation of TBP protein to a plasmid containing a TATA-box promoter induced RNA Pol II dependent transcription in egg extracts. Furthermore, we showed that TBP modestly altered the rate of chromatin assembly and histone deposition.
In conclusion, we showed experiments probing the relative roles of chromatin assembly in the regulation of transcription in a cell free system. Since we can add and remove components from cell free extracts to probe chromatin assembly directly (Onikubo et al., 2015), future studies can directly test how chromatin assembly competes with basal and specific transcriptional output. In particular, studies of transcriptional regulation by abundant histone modifications and modifying activities present in the egg, such as extensive arginine methylation and serine phosphorylation (Wang et al., 2014, Wilczek et al., 2011), may provide novel insights not testable in any other model system. The utility of cell free extracts will continue to be important for new discovery.
MATERIALS AND METHODS
Chemicals and Antibodies
Chemicals and reagents were obtained from Sigma (St. Louis, MO), RPI (Illinois) or Fisher Scientific (Pittsburgh, PA). Antibodies were from Millipore/Upstate.
Animals
Frogs were handled in an ethical manner according to our animal use protocols 20151003, 20121005 and 20110603 approved by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (IACUC). The following procedures were performed in strict accordance with the protocol and every effort was made to minimize suffering. Xenopus frogs were primed with 50 U PMSG 3-5 days before injection with Human Chorionic Gonadotropin (Sigma) for egg laying or before dissection for oocyte collection. Frogs were anesthetized with 0.2 % MS-222 (Thermo Fisher Scientific, Bridgewater, NJ) pH 7.0 and sacrificed according to IACUC-approved protocols.
Egg and Oocyte preparation
Extracts were prepared as described (Banaszynski et al., 2010, Wang et al., 2014). Briefly, High-speed supernatant clarified egg extract (HSS) was prepared from low speed supernatant by spinning in an SW-55 rotor at 55,000 rpm × 45 min. The clarified middle layer was removed and respun for 30 min, and glycerol was added to 5%, aliquoted, and flash frozen. Xenopus oocyte extracts were prepared from freshly dissected ovaries by disrupting the follicular layer as described above. The defolliculated oocytes were then washed extensively with 1× MMB containing 200 mm sucrose and 1 mm DTT, and the later staged oocytes settled to the bottom (the less dense stage I and II oocytes were mostly lost during the preparation). The oocytes were settled in 13 × 51-mm Beckman ultracentrifuge tubes, and excess buffer was removed. They were then spin-crushed at 35,000 rpm (150,000 × g) for 40 min in an SW-55 rotor. The middle layer was removed with a pipette or a syringe punctured through the side wall and respun for 30 min. The middle layer was again removed, glycerol was added to 5% final, and the extract was aliquoted and flash frozen in liquid nitrogen.
Plasmid Chromatin assembly reactions
25 μl egg or oocyte extract was incubated with 1 μl relaxed-pG5ML vector (625 ng/μl) and 1 μl 50X energy mix at 23°C for the indicated time. Then 5 μl of product was transferred into 200 μl of chromatin assembly stop buffer (10 mM Tris pH 8.0, 20 mM EDTA, 0.5% SDS) with 2 μl of RNase A (10 mg/ml) and incubated at 37°C for 30 min. The sample was digested with 5 μl 20mg/ml Proteinase K (final 50 μg/ml) at 56 °C for 1hr. Plasmid DNA were then purified through Phenol/Chloroform exraction and ethanol precipitated in the presence of NaOAC and 1 μl GlycoBlue (15 mg/ml). Finally, the pellets were dissolved in 12 ul of DNA loading buffer containing RNase A (final 10 ng/ml) and loaded on 0.8 % agarose gel run at 25 V overnight in the cold room. The gel was stained afterwards with ethidium bromide (EtBr).
Micrococcal nuclease digestion of assembled chromatin
After the assembly reaction in extract, 11 μl of the product was transferred into 90 ul of MNase Reaction buffer (10 mM Hepes pH8.0, 50 mM KCl, 5 mM MgCl2, 3 mM CaCl2, 1mM DTT, 0.1 % NP40 and 8% Glycerol) containing 1 unit of MNase and incubated at room temperature for the indicated length of time. 110 ul Stop Buffer was added to stop the reaction. 8 μl RNase A (10 mg/ml) was added to each sample and incubated at 37 °C overnight. 5 μl of Protenase K (20 mg/ml) was added to each sample and incubate at 56 °C for 1 hr. Plasmid DNA was phenol-chloroform extracted and ethanol precipitated as above. The final products were loaded on a 2% agarose gel and run at 100 V for 60 min at room temperature.
Sperm Chromatin Isolation
2,000 demembranated sperm chromatin per μl were incubated in 80 μl of extract. Samples were flash frozen, suspended in 800 μl of ELB-CIB buffer (10 mM Hepes, pH 7.8, 250 mM sucrose, 2.5 mM MgCl2, 50 mM KCl, 1 mM DTT, 1 mM EDTA, 1 mM spermidine, 1 mM spermine, 0.1% Triton X-100, 10 mM sodium butyrate, 1× phosphatase inhibitors, and 1× protease inhibitors) and chromatin isolated via centrifugation at 4000 rpm for 5 min through a 0.3 ml sucrose cushion of ELB-CIB with 0.5 M sucrose underlayered in the tube. The pellet was washed once with ELB-CIB plus 250 mM KCl.
Immunoblotting
Gels were transferred to polyvinylidene difluoride membrane (Millipore) using 1× NuPAGE transfer buffer (Invitrogen) plus 20% methanol and 0.1 % SDS. Membranes were stained using Direct Blue 71 stain to ensure proper transfer; any membranes with inadequate or uneven transfer were discarded. Membranes were blocked in 2% skim milk or 3% BSA and blotted with antibodies. Secondary horseradish peroxidase-coupled antibodies were applied and then visualized using ECL Advance, with images captured using the ImageQuant LAS 4000 digital system (GE, Pittsburgh). Images were incrementally exposed until CCD saturation, and the exposure before saturation was used for analysis. Images were aligned with molecular weight markers by simultaneous capture of the lit membrane and subsequently cropped, and levels were adjusted for contrast in Adobe Photoshop; no exposed bands were eliminated upon adjustment.
ACKNOWLEDGMENTS
This work was supported by startup funds from the Albert Einstein College of Medicine, The American Cancer Society – Robbie Sue Mudd Kidney Cancer Research Scholar Grant (124891-RSG-13-396-01-DMC) and NIH R01GM108646-01A1 (all to D.S.).
Footnotes
AUTHOR CONTRIBUTIONS
WW and DS planned experiments, performed analyses and wrote the manuscript. Conflict of interest: None declared.
REFERENCES
- ALMOUZNI G, MÉCHALI M, WOLFFE A. Competition between transcription complex assembly and chromatin assembly on replicating DNA. Embo J. 1990;9:573–82. doi: 10.1002/j.1460-2075.1990.tb08145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALMOUZNI G, MÉCHALI M, WOLFFE A. Transcription complex disruption caused by a transition in chromatin structure. Mol Cell Biol. 1991;11:655–65. doi: 10.1128/mcb.11.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALMOUZNI G, WOLFFE A. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. Embo J. 1995;14:1752–65. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMODEO AA, JUKAM D, STRAIGHT AF, SKOTHEIM JM. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci U S A. 2015;112:E1086–95. doi: 10.1073/pnas.1413990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AN W, KIM J, ROEDER RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–48. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- AN W, ROEDER RG. Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 2004;377:460–74. doi: 10.1016/S0076-6879(03)77030-X. [DOI] [PubMed] [Google Scholar]
- BANASZYNSKI LA, ALLIS CD, SHECHTER D. Analysis of histones and chromatin in Xenopus laevis egg and oocyte extracts. Methods. 2010;51:3–10. doi: 10.1016/j.ymeth.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANNISTER AJ, KOUZARIDES T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAS C, TYLER JK. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta. 2013;1819:332–42. doi: 10.1016/j.bbagrm.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIMITROV S, ALMOUZNI G, DASSO M, WOLFFE A. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev Biol. 1993;160:214–27. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- HAIR A, PRIOLEAU MN, VASSETZKY Y, MECHALI M. Control of gene expression in Xenopus early development. Dev Genet. 1998;22:122–31. doi: 10.1002/(SICI)1520-6408(1998)22:2<122::AID-DVG2>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- KIKYO N, WADE PA, GUSCHIN D, GE H, WOLFFE AP. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science. 2000;289:2360–2. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- MALLER J, GROSS S, SCHWAB M, FINKIELSTEIN C, TAIEB F, QIAN Y. Cell cycle transitions in early Xenopus development. Novartis Found Symp. 2001;237:58–73. doi: 10.1002/0470846666.ch6. discussion 73-8. [DOI] [PubMed] [Google Scholar]
- NEWPORT J, KIRSCHNER M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–96. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- NICKLAY JJ, SHECHTER D, CHITTA RK, GARCIA BA, SHABANOWITZ J, ALLIS CD, HUNT DF. Analysis of histones in Xenopus laevis. II. mass spectrometry reveals an index of cell type-specific modifications on H3 and H4. J Biol Chem. 2009;284:1075–85. doi: 10.1074/jbc.M807274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'FARRELL PH, STUMPFF J, SU TT. Embryonic cleavage cycles: how is a mouse like a fly? Curr Biol. 2004;14:R35–45. doi: 10.1016/j.cub.2003.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ONIKUBO T, NICKLAY JJ, XING L, WARREN C, ANSON B, WANG WL, BURGOS ES, RUFF SE, SHABANOWITZ J, CHENG RH, et al. Developmentally Regulated Post-translational Modification of Nucleoplasmin Controls Histone Sequestration and Deposition. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRIOLEAU MN, BUCKLE RS, MECHALI M. Programming of a repressed but committed chromatin structure during early development. Embo J. 1995;14:5073–84. doi: 10.1002/j.1460-2075.1995.tb00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRIOLEAU MN, HUET J, SENTENAC A, MECHALI M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–49. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- SCHIER AF. The maternal-zygotic transition: death and birth of RNAs. Science. 2007;316:406–7. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- SHECHTER D, ALLIS CD. A lasting marriage: histones and DNA tie a knot that is here to stay. Nature Reviews Genetics. 2007:S23. DNA Technologies Supplement. [Google Scholar]
- SHECHTER D, CHITTA RK, XIAO A, SHABANOWITZ J, HUNT DF, ALLIS CD. A distinct H2A.X isoform is enriched in Xenopus laevis eggs and early embryos and is phosphorylated in the absence of a checkpoint. Proc Natl Acad Sci U S A. 2009a;106:749–754. doi: 10.1073/pnas.0812207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHECHTER D, NICKLAY JJ, CHITTA RK, SHABANOWITZ J, HUNT DF, ALLIS CD. Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. J Biol Chem. 2009b;284:1064–74. doi: 10.1074/jbc.M807273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TADROS W, LIPSHITZ HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–42. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- TOYODA T, WOLFFE AP. In vitro transcription by RNA polymerase II in extracts of Xenopus oocytes, eggs, and somatic cells. Anal Biochem. 1992;203:340–7. doi: 10.1016/0003-2697(92)90322-x. [DOI] [PubMed] [Google Scholar]
- WANG W-L, ANDERSON L, NICKLAY J, CHEN H, GAMBLE M, SHABANOWITZ J, HUNT D, SHECHTER D. Phosphorylation and arginine methylation mark histone H2A prior to deposition during Xenopus laevis development. Epigenetics & Chromatin. 2014;7:22. doi: 10.1186/1756-8935-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIELAND T, FAULSTICH H. Amatoxins, phallotoxins, phallolysin, and antamanide: the biologically active components of poisonous Amanita mushrooms. CRC Crit Rev Biochem. 1978;5:185–260. doi: 10.3109/10409237809149870. [DOI] [PubMed] [Google Scholar]
- WILCZEK C, CHITTA R, WOO E, SHABANOWITZ J, CHAIT BT, HUNT DF, SHECHTER D. Protein Arginine Methyltransferase Prmt5-Mep50 Methylates Histones H2A and H4 and the Histone Chaperone Nucleoplasmin in Xenopus laevis Eggs. Journal of Biological Chemistry. 2011;286:42221–42231. doi: 10.1074/jbc.M111.303677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLFFE A, ALMOUZNI G, URA K, PRUSS D, HAYES J. Transcription factor access to DNA in the nucleosome. Cold Spring Harb Symp Quant Biol. 1993;58:225–35. doi: 10.1101/sqb.1993.058.01.027. [DOI] [PubMed] [Google Scholar]