Summary
Forming distinct representations and memories of multiple contexts and episodes is thought to be a crucial function of the hippocampal-entorhinal cortical network. The hippocampal dentate gyrus (DG) and CA3 are known to contribute to these functions but the role of the entorhinal cortex (EC) is poorly understood. Here, we show that Ocean cells, excitatory stellate neurons in the medial EC layer II projecting into DG and CA3, rapidly form a distinct representation of a novel context and drive context-specific activation of downstream CA3 cells as well as context-specific fear memory. In contrast, Island cells, excitatory pyramidal neurons in the medial EC layer II projecting into CA1, are indifferent to context-specific encoding or memory. On the other hand, Ocean cells are dispensable for temporal association learning, for which Island cells are crucial. Together, the two excitatory medial EC layer II inputs to the hippocampus have complementary roles in episodic memory.
Introduction
The hippocampal (HPC)-entorhinal cortex (EC) network plays a crucial role in episodic memory (Eichenbaum, 2000; Scoville and Milner, 1957; Tulving, 2002). It contributes to the formation of distinct memories of similar episodes by generating separate representations of the spatial and temporal relationships comprising events in an episode. Earlier theoretical work on this topic had suggested that the EC layer II (ECII)→dentate gyrus (DG)→CA3 pathway is crucial for forming discriminatory representations of similar spaces or contexts (pattern separation) based on the greater number of granule cells in DG (DG-GCs), their relatively sparse activity, and the limited redundancy of DG→CA3 connections (Bakker et al., 2008; Leutgeb et al., 2007; Marr, 1971; O'Reilly and McClelland, 1994; Treves and Rolls, 1994). These theories were supported by subsequent experimental data on the physiological response of CA3 pyramidal cells (PCs) to switching between a pair of similar contexts (Leutgeb et al., 2007; Leutgeb et al., 2004; McHugh et al., 2007; Wintzer et al., 2014), and on the behavioral performance of mice lacking functional N-methyl-D-aspartate (NMDA) receptors in the DG-GCs (McHugh et al., 2007). However, more recent studies conducted on mice in which adult neurogenesis (Altman and Das, 1965; Eriksson et al., 1998; Schlessinger et al., 1975; Seki and Arai, 1993) and/or the DG→CA3 inputs were blocked indicated that mossy fiber (MF) input from the overwhelming majority of DG-GCs onto CA3-PCs is dispensable for the discrimination of a similar pair of contexts (Nakashiba et al., 2012). Rather, the minority population of DG-GCs, the young GCs generated during adult neurogenesis, play the crucial role in the discrimination of similar contexts (Clelland et al., 2009; Creer et al., 2010; Nakashiba et al., 2012; Sahay et al., 2011; Scobie et al., 2009).
These previous studies have all focused on the role of HPC cells (both DG-GCs and CA3 PCs) and their circuits as the substrates for pattern separation, but have not investigated the potential role of ECII cells in the discrimination of more different contexts. However, it is possible that EC cells are sensitive to contextual differences and may respond to various contexts in different ways. Such context-specific activity of EC cells could drive discriminatory encoding of contexts in the downstream HPC sub-regions and thereby contribute to context-specific memory. Recent studies have revealed novel ways to dissect ECII excitatory neurons and function. The two major populations - Ocean and Island cells - differ in molecular markers, anatomical features, and projection targets (Kitamura et al., 2014; Ray et al., 2014; Varga et al., 2010). Whereas Ocean cells project to the DG-GCs and CA3-PCs, Island cells project to CA1 and predominantly to interneurons (Kitamura et al., 2014; Ray et al., 2014; Varga et al., 2010).
Here, we investigated the potential role of medial ECII (MECII) cells in context discrimination using in vivo Ca2+ imaging to study the activation of Ocean and Island cells as mice freely explored two alternating different contexts. We also examined the effects of optogenetic manipulations of Ocean or Island cell activity on contextual representations in the HPC sub-regions and on the formation of context-specific fear memory.
Results
Specific labeling of Ocean cells and Island cells by GCaMP6f
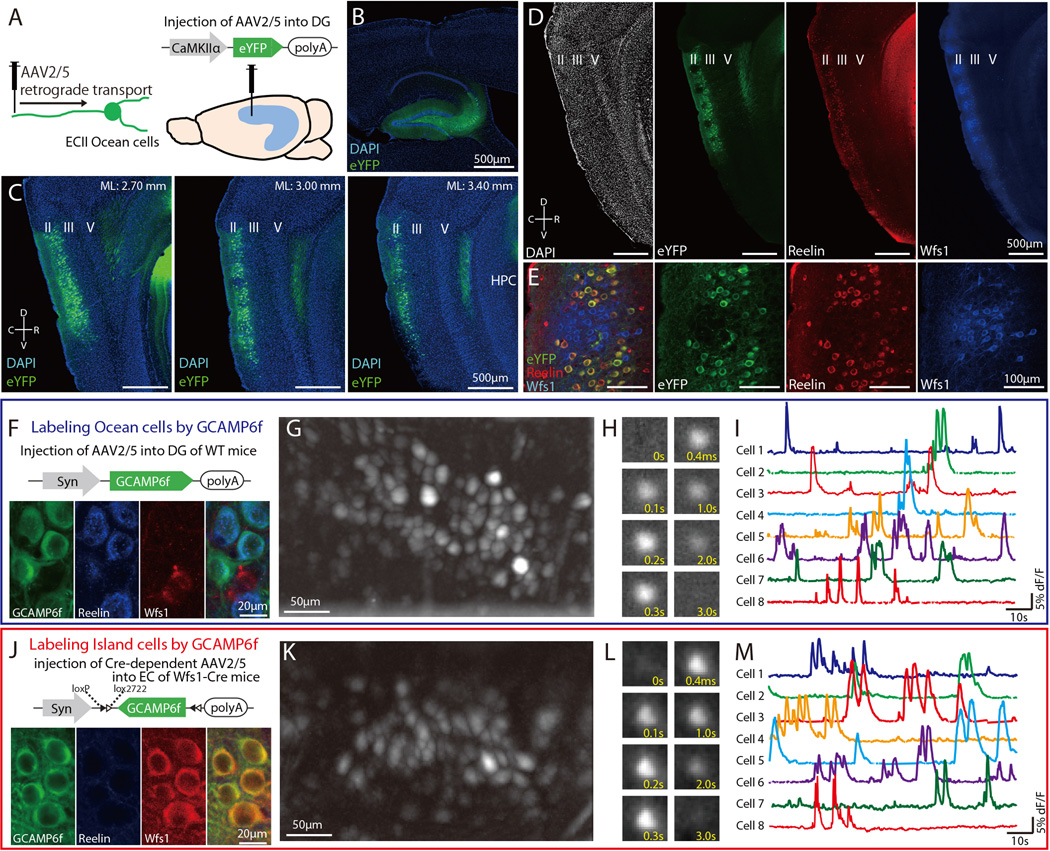
We first sought a means to specifically label the DG-projecting stellate Ocean cells in MECII. As a test, we injected AAV2/5-CaMKIIα-eYFP into the dorsal DG of C57BL/6 mice (Figures 1A and 1B). The AAV was retrogradely transported to the somata of Ocean cells through their axons in DG (Figures 1A and 1C). eYFP expression in MEC was restricted to Reelin+ Ocean cells and absent in Island cells, which are marked by Wolfram syndrome 1 (Wfs1) expression. This test confirmed that AAV2/5 injected into the dorsal DG can enable gene expression specifically in MEC Ocean cells (Figures 1D and 1E).
Fig. 1. Specific-labeling of Ocean cells and Island cells by GCaMP6f.
(A) Injection of AAV2/5-CaMKIIα-eYFP in DG to retrogradely label Ocean cells from their axons. (B) Injection site of AAV2/5 in DG. (C) Parasagittal sections of MEC visualized with eYFP-labeled cell bodies (green) and stained with DAPI (blue). (D) Parasagittal sections of MEC visualized with eYFP-labeled cell bodies (green) and immunostained with anti-Reelin (red) and anti-Wfs1 (blue). (E) Magnification image from (D). Reelin+ cells never overlap with Wfs1+ cells in MECII. (F) Labeling method of Ocean cells by GCaMP6f. Injection of AAV2/5-Syn-GCaMP6f in DG and implantation of microendoscope into MEC. Parasagittal sections of MEC visualized with GCaMP6f-labeled cell bodies (green) and immunostained with anti-Reelin (blue) and anti-Wfs1 (blue) (J) Labeling method of Island cells by GCaMP6f. Injection of AAV2/5-Syn-DIO-GCaMP6f in MEC and implantation of microendoscope into MEC. Parasagittal sections of MEC visualized with GCaMP6f-labeled cell bodies (green) and immunostained with anti-Reelin (blue) and anti-Wfs1 (red) (G, K) Stacked image acquired through the microendoscope over 20 min of imaging in MEC of an Ocean-GCaMP6f mouse (G) and an Island-GCaMP6f mouse (K) as they explored multiple open fields. (H, L) Time-lapse image sequence of GCaMP6f fluorescence in an individual Ocean cell (H) and Island cell (L) (I, M) Relative fluorescence changes (ΔF/F) for 8 Ocean cells (I) and 8 Island cells (M). D, dorsal; V, ventral; R, rostral; C, caudal.
For Ca2+ imaging studies, we injected AAV2/5-Syn-GCaMP6f into dorsal DG of C57BL/6 mice, to specifically express the GCaMP6f Ca2+ indicator in MECII Ocean cells (Figure 1F) (Chen et al., 2013). As expected, expression of GCaMP6f in MEC of C57BL/6 mice (henceforth termed Ocean GCaMP6f mice) was restricted to Reelin+ Ocean cells, with no expression in Wfs1+ Island cells (Figure 1F, Figure S1A) (Sun et al., 2015). 85.3% of Reelin+ cells in dorsal MECII were also GCaMP6f+, demonstrating efficient transport of the AAV2/5 virus from DG to Ocean cells (256 GCaMP6f+ cells out of 300 Reelin+ cells from 6 mice).
For Ca2+ imaging studies of MECII Island cells, we injected AAV2/5-Syn-DIO-GCaMP6f into the superficial layers of MEC of Wfs1-Cre transgenic mice (Island GCaMP6f mice; Figure 1J; Kitamura et al., 2014). Expression of GCaMP6f in MEC was restricted to Wfs1+ Island cells, with no expression in Reelin+ Ocean cells (Figure 1J, Figure S1B). Consistent with our previous study (Kitamura et al., 2014; Sun et al., 2015), a high proportion of Wfs1+ cells in dorsal MECII expressed GCaMP6f (91%, 222 GCaMP6f+ cells out of 244 Wfs1+ cells from 5 mice). For both cohorts of mice, we implanted a microendoscope (Ziv et al., 2013) into dorsal MEC and imaged Ca2+ signals using a miniaturized, head-mounted fluorescence microscope as the mice freely explored an open field. In both Ocean GCaMP6f and Island GCaMP6f mice, somatic Ca2+ transients were apparent in many individual neurons (Figures 1G, 1K) across the population (about 40 cells per mouse; Figures 1H, 1I, 1L and 1M).
Ocean cells, but not Island cells, exhibit context-specific Ca2+ activity
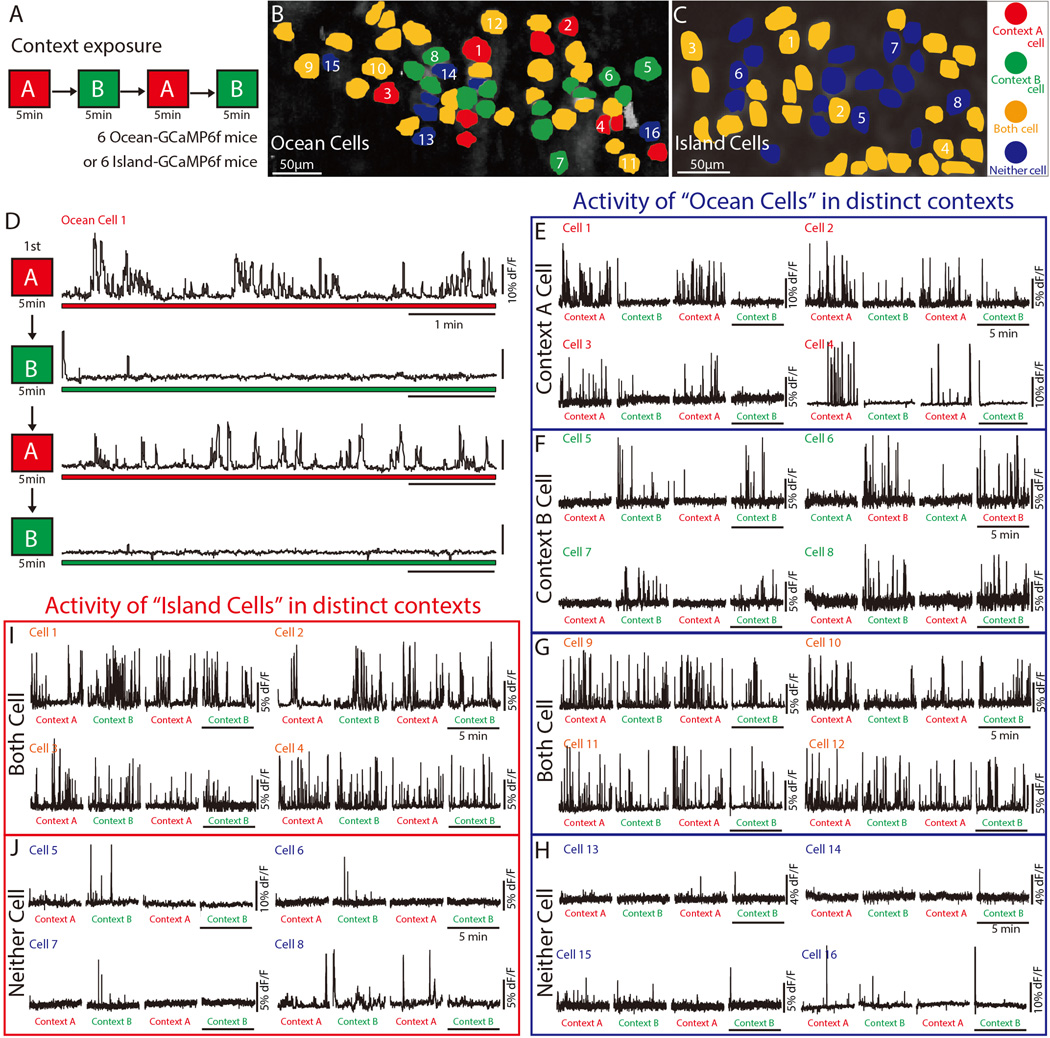
To understand the potential role of Ocean cells and Island cells in context discrimination, we monitored Ca2+ activity as Ocean and Island mice were exposed to two different novel contexts - A and B (See Methods, Figure S2) - in succession in the following order: A→B→A→B (Figure 2A). We monitored Ca2+ activity in ~40 cells per session per mouse (Figure 2B and 2C), yielding >210 cells from 6 Ocean mice. One sub-population of Ocean cells exhibited robust Ca2+ activity preferentially in Context A, but far less so in Context B (Figure 2D and 2E). Conversely, another sub-population of Ocean cells had higher Ca2+ activity in Context B, but not in Context A (Figure 2F). These abrupt changes in activity occurred when the context was shifted. The altered state of activity was stable for the entirety of the 5 min context exposure. A third and larger subpopulation of Ocean cells showed equally robust Ca2+ activity in both Context A and Context B (Figure 2G). In a fourth sub-population, Ca2+ activity was low in both contexts (Figure 2H). Ocean cells that were only active during the first Context A exposure but not in the second Context A exposure (and vice versa for Context B) were rare (Table 1). There was no evidence of context-specific cells in Island cells of Island mice; the majority of Island cells were active in both contexts and the remaining cells were relatively inactive in both contexts (Figure 2I and 2J).
Fig. 2. Ocean cells, but not Island cells, exhibit context-specific Ca2+ activity.
(A) Experimental schedule showing sequential exposure to two contexts (each exposure is 5 min): A → B → A → B. (B and C) Distribution of different types of cell responses in MEC of an Ocean mouse (B) and an Island mouse (C) observed through microendoscopy during exposures to both contexts. Response to Context A (red cells), Context B (green cells), both contexts (yellow cells), or neither context (blue cells). Cell responses were found by picking small regions of interest (ROIs) (~ 1/3 of cell body size) at the center of the cell bodies. (D) Example of Context A-specific Ca2+ activity in a cell from an Ocean mouse that explored both Context A and B. (E–H) Ca2+ activity in an Ocean mouse that explored both contexts. Four examples are shown for each different type of Ocean cell response: Context A-specific Ca2+ activity (E), Context B-specific Ca2+ activity (F), Ca2+ activity in both contexts (G), and Ca2+ activity in neither context (H). (I–J) Ca2+ activity in an Island mouse that explored both contexts: 4 example Island cells that were active in both contexts (I), and 4 example Island cells that were active in neither context (J).
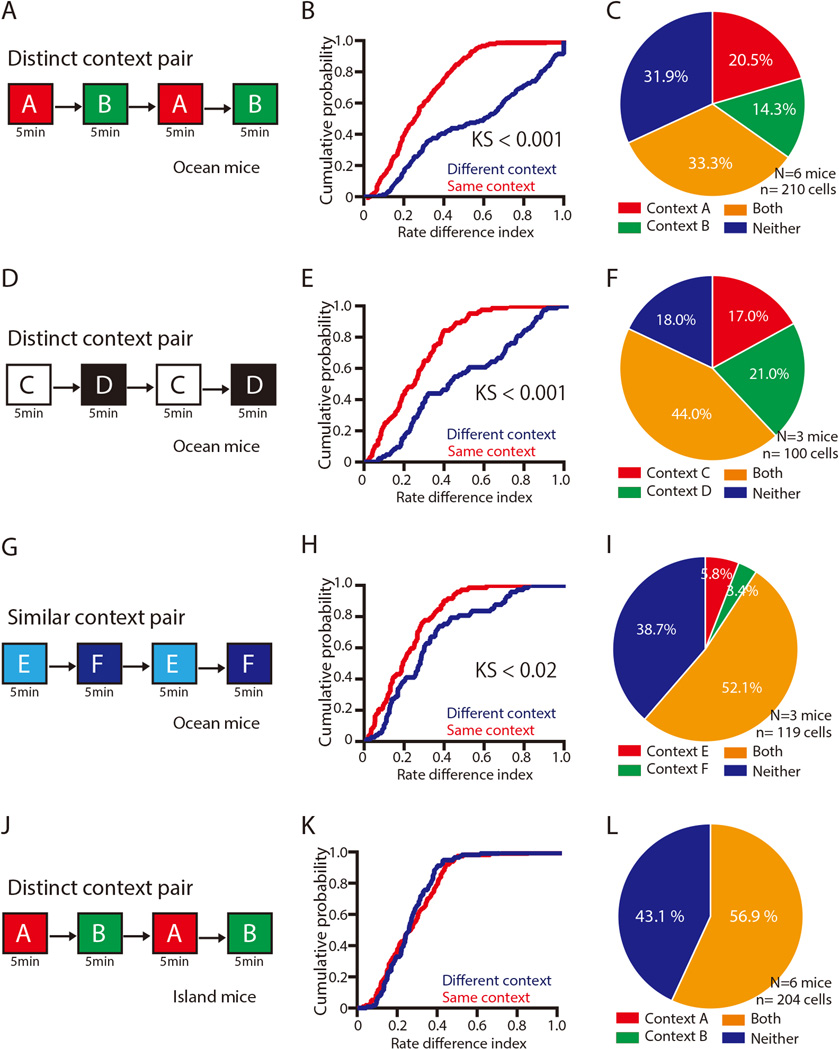
We calculated the rate difference index of Ca2+ activity between the two contexts for Ocean and Island cells by comparing the average activity rates of each cell during exploration of the two contexts (Figure 3A), using the normalized changes in mean activity rates in the two contexts (See Methods for the definition of rate difference index). Figure 3B shows the cumulative probability histograms for the rate difference indices of Ocean cells. The rate difference index of Ocean cells showed a significant shift of the cumulative histogram to the right when exposures between different contexts were compared with multiple exposures to the same context (Kolmogorov–Smirnov (KS) test, P< 0.001), indicating that a greater proportion of Ocean cells undergoes a change in Ca2+ event rates when the contexts are different (Figure 3B). Using the 99th percentile of the rate difference distribution as a threshold for defining context-specificity (see Methods), we found that 20.5% of Ocean cells were Context A-specific, and that 14.3% of Ocean cells were Context B-specific (threshold = 0.6) (Figure 3C). We examined another pair of distinct contexts (Context C and Context D) (Figure 3D, Figure S2), and obtained similar results showing context-specific cells in Contexts C versus D (KS test, P< 0.001) (Figure 3D–F).
Fig. 3. Proportions of various types of Ocean cells and Island cells in different context pairs.
(A) Experimental schedule showing sequential exposure to two distinct contexts (each exposure is 5 min): A → B → A → B. (B) Cumulative probability of the rate difference indices in Ocean cells from exposure to two distinct contexts versus same contexts. (C) Proportion of Ocean cells showing response to Context A (red), Context B (green), both contexts (yellow), or neither context (blue) (D) Experimental schedule showing sequential exposure to two distinct contexts (each exposure is 5 min): C→ D → C → D. (E) Cumulative probability of the rate difference indices in Ocean cells from exposure to two distinct contexts versus same contexts (F) Proportion of Ocean cells showing response to Context C (red), Context D (green), both contexts (yellow), or neither context (blue) (G) Experimental schedule showing sequential exposure to two similar contexts (each exposure is 5 min): E → F → E → F. (H) Cumulative probability of the rate difference indices in Ocean cells from exposure to two distinct contexts versus same contexts. (I) Proportion of Ocean cells showing response to Context E (red), Context F (green), both contexts (yellow), or neither context (blue) (J) Experimental schedule showing sequential exposure to two distinct contexts (each exposure is 5 min): A → B → A → B. (K) Cumulative probability of the rate difference indices in Island cells from exposure to two distinct contexts versus same contexts. (F) Proportion of Island cells showing response to Context A (red), Context B (green), both contexts (yellow), or neither contexts (blue). N indicates number of animals. n indicates number of cells.
To determine whether Ocean cells can discriminate between more similar context pairs, we monitored the Ca2+ activity of Ocean cells in another group of Ocean mice that we exposed to a pair of similar contexts (Context E and Context F; Figure 3G–I, Figure S2). The rate difference index showed a significant shift of the cumulative histogram to the right when exposures between Context E and Context F were compared with multiple exposures to the same context (KS test, P< 0.02) (Figure 3H). However, we found that a much smaller percentage of cells were specific for Context E and for Context F, than in groups of mice that were exposed to more distinct context pairs (Context A vs B, and Context C vs D) (P < 0.001, P < 0.001, respectively, by Fisher’s exact test) (Figure 3I), indicating that Ocean cells may be less sensitive in the discrimination of the similar context pair. In contrast, none of the Island cells showed a context-specific response when the animals were exposed to distinct Contexts A and B (Context-specific cells; 0%, 0 out of 204 neurons, Figure 3J–L), even though a large proportion of them (57%) were active in both contexts.
The rapid context-specific firing of some Ocean cells could be driven at least in part by the loops of activity going from EC to HPC and back to EC (Amaral and Witter, 1989; Witter et al., 2000). We tested this hypothesis by injecting muscimol, a γ-aminobutyric acid subtype A receptor agonist, into dorsal CA1 (Figure S3). This treatment induced a significant reduction of multiunit CA1 activity. Nevertheless, we observed a similar percentage of context-specific Ocean cells in these mice (26% in Figure S3) as in non-muscimol-injected Ocean mice (35% n Figure 3C). Latencies of the first rise of Ca2+ activity of these cells were also unaltered (Figure S3 and Figure 4). These results suggest that the contribution of the EC→HPC→EC loops in the context-specific firing of Ocean cells is minimal, if any.
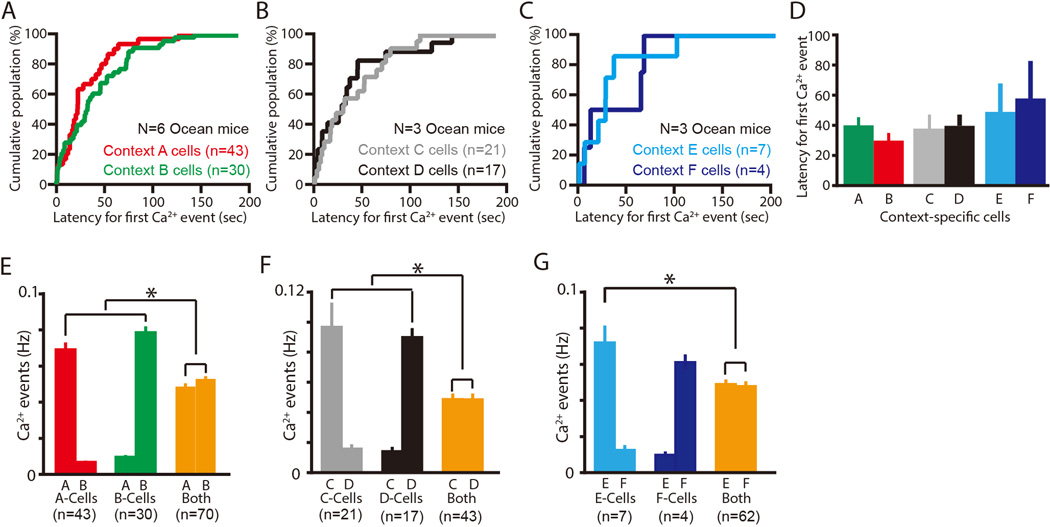
Fig. 4. Ca2+ event latencies and frequencies in context-specific Ocean cells.
(A–C) Cumulative population of all the latencies to the first Ca2+ event in each context-specific Ocean cell in their respective preferred context: Context A and B cells (A), Context C and D cells (B), and Context E and F cells (C). (D) Average of all the latencies to the first Ca2+ event in each context-specific cell active in their respective preferred contexts. (E) Average Ca2+ event frequency in each specific context (top row of horizontal axis label) for each type of Ocean cell (Context A cells, Context B cells, and Both). (F) Average Ca2+ event frequency in each specific context (top row of horizontal axis label) for each type of Ocean cell (Context C cells, Context D cells, and Both). (G) Average Ca2+ event frequency in each specific context (top row of horizontal axis label) for each type of Ocean cell (Context E cells, Context F cells, and Both). Data are represented as mean±SEM. *P< 0.05. N indicates number of animals. n indicates number of cells.
Response latency of context-specific Ocean cells and the effects of context specificity on their Ca2+ events
To further characterize context-specific Ocean cells, we measured the latency to the first Ca2+ event in each cell after exposure to their preferred context (Figure 4A–4D). The latency varied between 10–150 s with a mean of 40–60 s (Figure 4D). No significant effect of the similarity or dissimilarity of the context pairs was observed. However, interestingly, the average frequencies of Ca2+ activity of context-specific Ocean cells were significantly greater than in those cells that responded to both contexts (Context A vs both; P < 0.05, Context B vs both; P < 0.05, Context C vs both; P < 0.05, Context D vs both; P < 0.05, Context E vs both; P < 0.05, one-way ANOVA followed by Scheffe's test) (Figure 4E–4G). These results indicate that context-specific cells use rate coding to discriminate between distinct context pairs.
Ocean cells, but not Island cells, drive context exposure-dependent activation of DG cells and CA3 cells
Next, to investigate the role of Ocean cells in driving the activities of hippocampal DG and CA3 cells induced by exposure to a novel context (Context A), we optogenetically inhibiting Ocean cell activity using eArchT (Ocean-eArchT mouse, Figure 5A–5C, 5G). For comparison, we performed an analogous set of experiments by specifically inhibiting Island cell activity (Figure 5D–5F, 5G). Although we have previously shown that Island cells project to CA1 and not DG or CA3, we cannot rule out that Island cells indirectly influence Ocean cell activity (and downstream DG and CA3 activity) via the CA1 → EC Layer V pathway, which projects to superficial EC (Amaral and Witter, 1989; Witter et al., 2000). Consistent with our previous findings (Liu et al., 2012; Sarinana et al., 2014), context exposure increased expression of c-Fos, an immediate early gene, in DG and CA3 in C57BL/6 mice, compared to homecage exposure (DG; t10 = −9.93, P < 0.001, CA3; t10 = −11.3, P < 0.001) (Figure 5H).
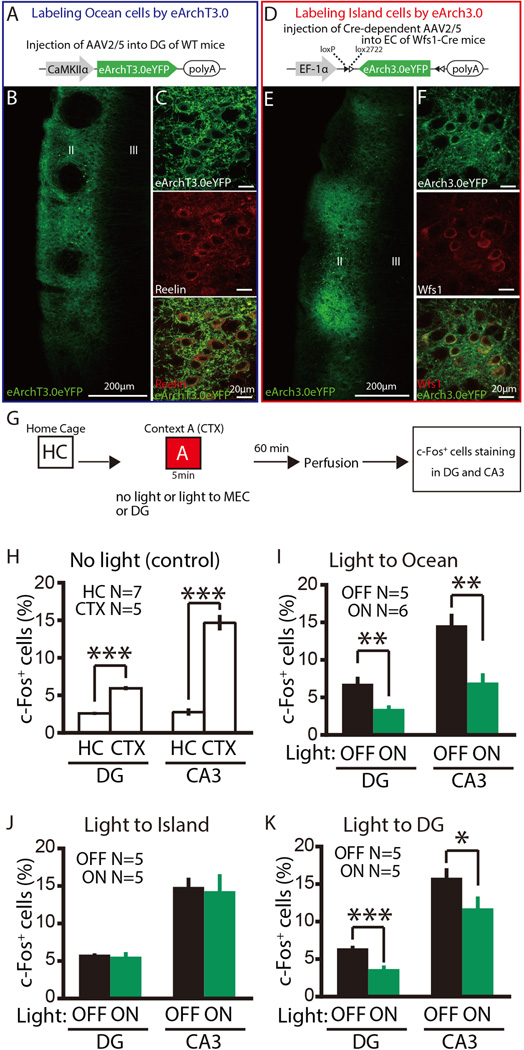
Fig. 5. Ocean cells, but not Island cells, drive context exposure-dependent activation of DG cells and CA3 cells.
(A) Bilateral injection of AAV2/5-CaMKIIα-eArchT3.0eYFP into DG with bilateral implantation of optic fibers into MEC. (B) Parasagittal sections of MEC visualized with eArchT3.0-eYFP-labeled cells (green). (C) A magnified image of a part of (B) after immunostaining with anti-Reelin (red). (D) Bilateral injection of AAV2/9-EF1α-DIO-eArch3.0eYFP into MEC of Wfs1-Cre mice with bilateral implantation of optic fibers. (E) Parasagittal sections of MEC visualized with eArch3.0-eYFP-labeled Island cells (green). (F) A magnified image of a part of (E) after immunostaining with anti-Wfs1 (red). (G) Experimental schedule. (H) Percentages of c-Fos+ cells in DG and CA3 in the novel context exposure group (CTX) and home cage group (HC) of WT mice. (I–K) Percentages of c-Fos+ cells in DG and CA3 of Light-OFF and -ON groups of Ocean CTX mice (I), Island CTX mice (J), or DG CTX mice (K), ***P< 0.001, **P< 0.01, *P< 0.05. Data are represented as mean±SEM. N indicates number of animals.
When AAV2/5-CaMKIIα-eArchT3.0-eYFP (Mattis et al., 2012) was bilaterally injected into DG of C57BL/6 mice (Ocean-eArchT mice) (Figure 5A and 5B), eArchT3.0-eYFP expression was restricted to Reelin+ Ocean cells (Figure 5C). We exposed these mice to Context A while delivering green light bilaterally to MEC during the entire exploration period (Figure 5G). The number of c-Fos+ cells in DG and CA3 areas of the light-ON group was significantly lower than that of the light-OFF group (DG; t9 = 3.38, P < 0.01, CA3; t9 = 4.15, P < 0.01) (Figure 5I). Next, we bilaterally injected AAV2/5-EF1α-DIO-eArch3.0-eYFP into dorsal MEC of Wfs1-Cre mice (Island-eArch mice) (Figure 5D and 5E). eArch3.0-eYFP expression in MECII was restricted to Wfs1+ Island cells (Figure 5F). We exposed these mice to Context A while delivering green light bilaterally to MEC during the entire exploration period (Figure 5G) and found that there was no difference in the number of c-Fos+ cells in either DG or CA3 between the light-ON and light-OFF groups (DG; t8 = 0.41, P > 0.6, CA3; t8 = 0.21, P > 0.8)(Figure 5J). These results indicate that MEC Ocean cells, but not MEC Island cells, drive activation of downstream DG and CA3 cells upon exposure to a novel context (i.e. Context A).
We also expressed eArch specifically in DG cells to block their activity (DG-eArch mice, see Methods, Figure S4) and exposed these mice to a novel context while delivering green light bilaterally to DG during the entire exploration period (Figure 5K). Accordingly, we found that the number of c-Fos+ DG cells in the light-ON group was significantly lower compared to the light-OFF group (DG; t8 = 4.95, P< 0.001). Interestingly, only a small, though significant, reduction in the number of c-Fos+ cells was seen in the CA3 area in the light-ON group compared with the light-OFF group (t8 = 2.59, P< 0.05) (Figure 5K). This contrasts with the highly significant reduction in c-Fos+ seen with Ocean cell inhibition (Figure 5I; 52 % reduction in Ocean-eArchT mice, Figure 5K; 28 % reduction in DG-eArch mice, t9 = 2.26, P< 0.05) and suggests that Ocean cell input contributes to CA3 activity in a novel context. Inputs to CA3 from DG include both developmentally derived old DG-GCs and adult-generated young DG-GCs, which have been shown to play a role in contextual fear memory (Kheirbek et al., 2013). Our eArch-mediated inhibition of DG activity occurs mostly in developmentally derived old DG-GCs, rather than adult-generated young DG-GCs (Figure S4), suggesting that CA3 activity (Figure 5I) in a distinct novel environment may be driven substantially by Ocean cell input via young DG-GCs or via the direct inputs from MEC to CA3 (Steward, 1976; Yeckel and Berger, 1990).
Ocean cells, but not Island cells, contribute to a discriminatory activation of CA3 cells upon serial exposure to distinct contexts
We next investigated potential roles of Ocean and Island cell inputs in a discriminatory activation of CA3 cells upon serial exposures to a pair of contexts. For this purpose, we injected AAV2/9-TRE-mCherry into CA3 of c-fos tTA mice (Reijmers et al., 2007) and exposed them to Context A while off Dox so as to label activated CA3 cells with mCherry (Ramirez et al., 2013). These mice were then immediately placed back on Dox to prevent further labeling of activated cells. The next day, the mice were exposed to Context B, and then euthanized for immunohistochemical detection of endogenous c-Fos in CA3 (Figure 6D). The CA3 cells activated by exposure to Context A are thus identified by mCherry expression and those activated by exposure to Context B are identified by the expression of endogenous c-Fos (Figure 6C) (Denny et al., 2014; Ramirez et al., 2013). We compared the proportion of c-Fos+ CA3 cells (i.e. CA3 cells that were activated by exposure to Context B) in mCherry+ and mCherry− cell populations (i.e. CA3 cells that were activated and unactivated, respectively, by exposure to Context A). Notably, c-Fos expression induced by context exposure in CA3 cells of c-fos-tTA transgenic mice (Figure 6) was higher than that of WT mice (Figure 5), which is consistent with a previous study (Reijmers et al., 2007).
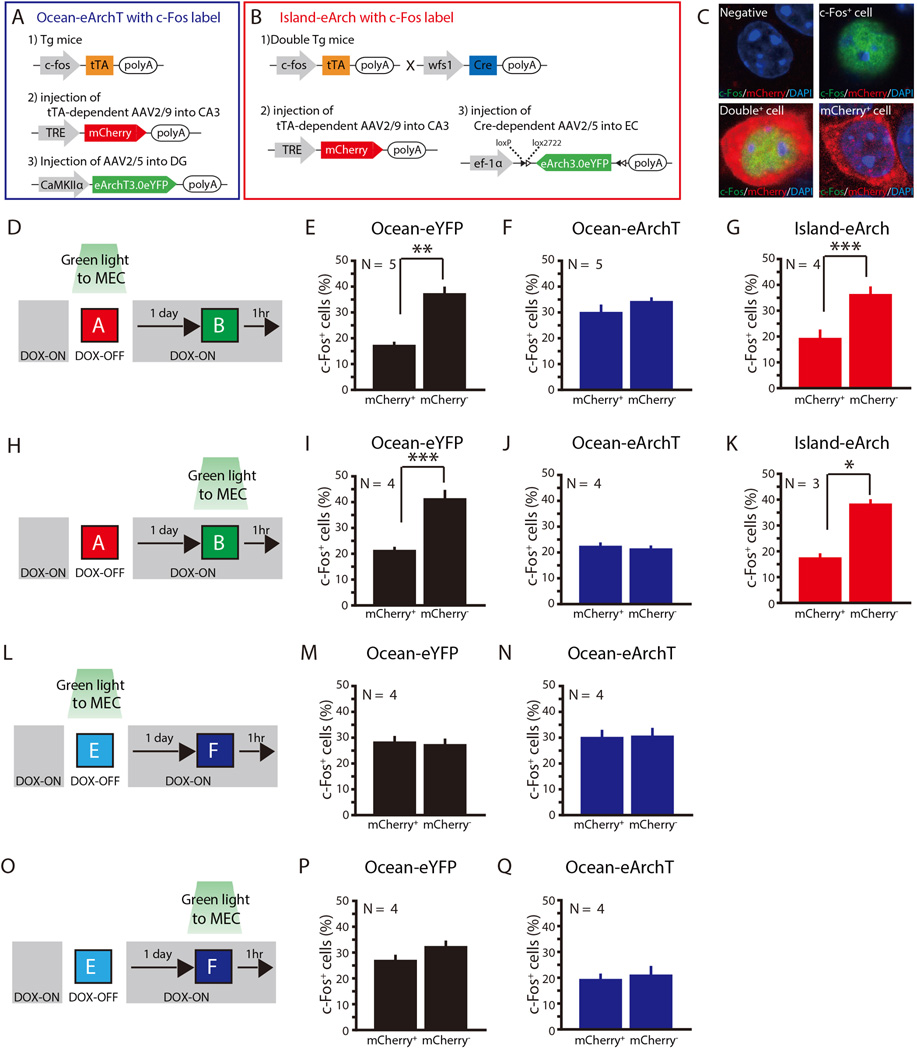
Fig. 6. Ocean cell input, but not Island cell input, is crucial for discriminatory encoding in CA3 for distinct context pairs but not for similar context pairs.
(A) Labeling of c-Fos expressing cells in CA3 combined with optogenetic inhibition of Ocean cells.Bilateral injection of AAV2/5-CaMKIIα-eArchT3.0-eYFP or AAV2/5-CaMKIIα-eYFP (control group; not shown) in DG and AAV2/9-TRE-mCherry in CA3 with bilateral implantation of optic fibers into MEC of c-fos-tTA mice. (B) Labeling of c-Fos-expressed cells in CA3 combined with optogenetic inhibition of Island cells. Bilateral injections of AAV2/5-EF1α-DIOeArch3.0-eYFP in MEC and AAV2/9-TRE-mCherry in CA3, with bilateral implantation of optic fibers into MEC of double transgenic mice (c-fos-tTA mice crossed with Wfs1-Cre mice). (C) Example of c-Fos+ cells, mCherry+ cells, double+ cells, non-labeled cells. (D and H) To label the c-Fos expressed cells in Ocean-eYFP, Ocean-eArchT, and Island-eArch mice (A–B), animals were taken off Dox and exposed to Context A in order to label the cells activated by Context A with mCherry (red). Animals were then put back on Dox and exposed to distinct Context B 24 hours later as activated cells expressed c-Fos (green). Green light was illuminated in MEC during the animal’s exposure to Context A (D) or Context B (H). (E-G) Percentages of c-Fos+ cells in mCherry+ or mCherry− CA3 cells in the three groups of mice related to (D). Total number of mCherry+ cells studied were 254 (E), 122 (F), and 177 (G), and absolute number of mCherrycells studied were 1304 (E), 1432 (F), and 1164 (G) cells respectively. (I–K) Percentages of c-Fos+ cells in mCherry+ or mCherry− CA3 cells in the three groups of mice related to (H). Total number of mCherry+ cells studied were 194 (I), 181 (J), and 141 (K), and absolute number of mCherry- cells studied were 1000 (I), 1002 (J), and 710 (K) cells respectively. (L and O) To label the c-Fos expressed cells in Ocean-eArchT and Ocean-eYFP mice, animals (A) were taken off Dox and exposed to Context E in order to label the cells activated by Context E with mCherry (red). Animals were then put back on Dox and exposed to similar Context F 24 hours later so as to let activated cells express c-Fos (green). Green light was illuminated in MEC during the animal’s exposure to Context E (L) or Context F (O). (M and N) Percentages of c-Fos+ cells in mCherry+ or mCherry− CA3 cells in the two groups of mice related to (L). Total number of mCherry+ cells studied were 273 (M), and 155 (N), and absolute number of mCherry- cells studied were 1349 (M), and 1453 (N) cells respectively. (P and Q) Percentages of c-Fos+ cells in mCherry+ or mCherry− CA3 cells in the two groups of mice related to (O). Total number of mCherry+ cells studied were 319 (P), and 217 (Q), and absolute number of mCherry- cells studied were 1261 (P), and 1204 (Q) cells respectively. ***P< 0.001, **P< 0.01, *P< 0.05. Data are represented as mean±SEM. N indicates number of animals.
Also consistent with previous reports (Niibori et al., 2012; Wintzer et al., 2014), the proportion of c-Fos+ cells in the mCherry+ CA3 cell population in control mice expressing only eYFP (no eArchT) was significantly lower than that of c-Fos+ cells in CA3 mCherry− cell population (paired t test, t4 = −7.37, P< 0.01) (Figure 6E), indicating that Contexts A and B activated largely independent populations of CA3 cells. Inhibiting Ocean cells with eArchT during exposure to Context A resulted in an increase of the proportion of c-Fos+ cells in the mCherry+ cell population that was similar to that in the mCherry− cell population (Figure 6A and 6F), indicating that Ocean cells play a significant role in the discriminatory encoding of the two contexts in CA3. In contrast, inhibition of Island cells with eArchT (Figure 6B) had no effect on the distribution of c-Fos+ cells (paired t test, t3= −12.7, P< 0.001; Figure 6G). Next, we moved the timing of Ocean inhibition to during Context B exposure (Figure 6H–6K). In this case, both eYFP and ArchT-expressing groups started with comparable mCherry+ expression levels upon exposure to Context A. Inhibiting Ocean cells during Context B exposure led to a reduction of the proportion of c-Fos+ cells in the mCherry− cell population to a level similar to that in the mCherry+ cell population, reinforcing the conclusion that MEC Ocean cell input contributes to discriminatory encoding of distinct contexts in CA3. Again, inhibition of Island cells did not show any effect on the distribution of c-Fos+ cells (paired t test, t2= −7.88, P< 0.05; Figure 6K).
Additionally, we examined the role of MEC Ocean cells in the activation of the CA3 cells upon successive exposure to a pair of similar contexts, Context E and Context F (Figure 6L–6Q, Figure S2). In control Ocean-eYFP only mice, the proportion of c-Fos+ cells in the CA3 mCherry+ cell population was not different from that of c-Fos+ cells in CA3 mCherry− cell population (paired t test, t3 = 0.88, P > 0.4) (Figure 6M), indicating that CA3 cells do not discriminate well between the similar context pairs after a one time exposure to each. We also did not observe any changes in the distribution of c-Fos+ cells when Ocean cells were inactivated (paired t test, t3 = 0.35, P>0.5) (Figure 6M–6N). Similar results were obtained when the timing of green light illumination was shifted from the period of Context A to the period of Context B (Figure 6O–6Q). Overall, these data indicate that the Ocean cell input is crucial for the discriminatory response of CA3 cells only when the context pair is sufficiently distinct.
Ocean cells, but not Island cells, facilitate context-specific fear conditioning
We then investigated whether the context discriminatory function of Ocean cell input to CA3-PCs activation could be extended to a behavioral level using context-specific fear conditioning (CFC). For this purpose we subjected Ocean-eArchT mice to CFC while delivering green light bilaterally to MEC during the entire training period (3 min) and monitored the levels of freezing in the conditioned Context A, followed by exposure to a distinct unconditioned Context B (Figure 7A). Control mice expressing only eYFP (no eArchT) exhibited high freezing behavior in the conditioned Context A, and much lower freezing levels in the unconditioned Context B. The freezing behavior of light-inhibited Ocean-eArchT mice was greatly reduced in the conditioned context compared to the control mice (t22 = 2.39, P < 0.05), while there was no significant alteration of freezing level in the distinct unconditioned context (t22 = 0.82, P > 0.4) (Figure 7B). Light had no effect on freezing levels in Island-eArch mice in either the conditioned (t20 = 0.24, P > 0.8) or unconditioned context (t20 = −0.53, P > 0.6) (Figure 7C). We next subjected DG-eArch mice (Figure S4) to CFC while delivering green light bilaterally to DG during the entire training period (Figure 7D). Light-inhibited DG-eArch mice displayed no freezing deficits in the conditioned context (t22 = 0.33, P > 0.7) but interestingly, showed significantly more freezing in the unconditioned context compared to the eYFP control mice (t22 = −2.1, P < 0.05) (Figure 7D) (See Discussion).
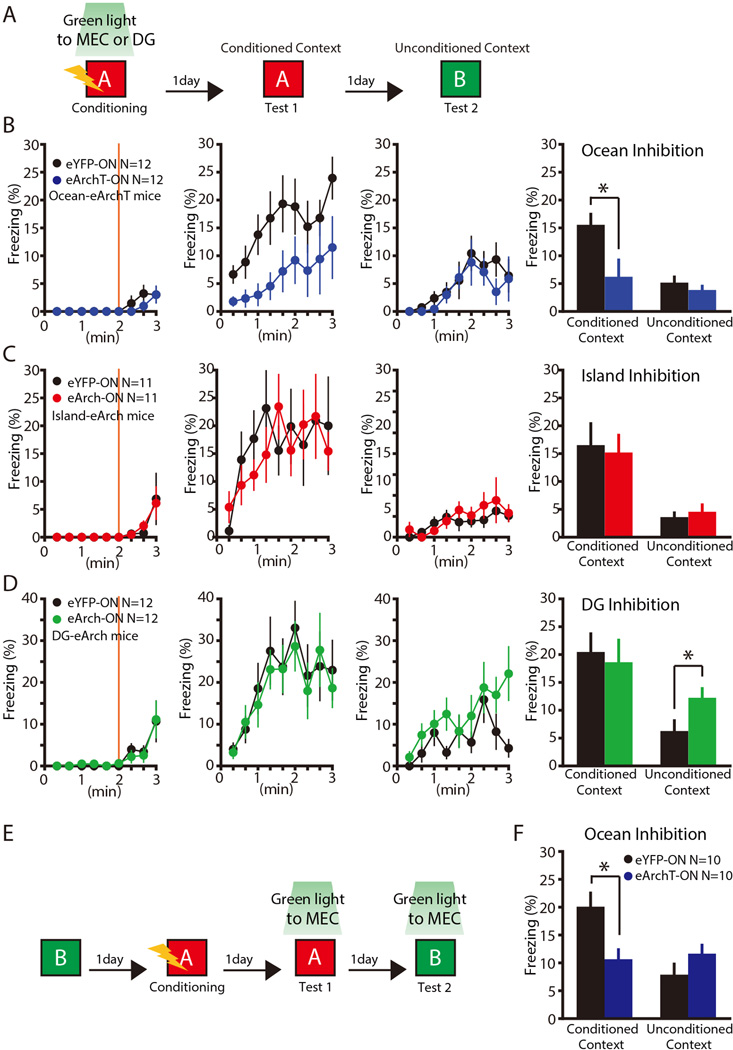
Fig. 7. Ocean cell input, but not Island cell input, facilitates acquisition and retrieval of contextual fear conditioning memory for a pair of distinct contexts.
(A) Experimental schedule for contextual fear conditioning (CFC). (B) Ocean-eArchT mice in CFC. Time course of freezing observed during Conditioning, Test 1, and Test 2. Orange vertical bars represent shock. The far right panel of (B) shows averaged freezing levels during testing on Day 2 and Day 3. (C) Island-eArch mice in CFC. Time course of freezing observed during Conditioning, Test 1, and Test 2 are shown. The far right panel of (C) shows averaged freezing levels during testing on Day 2 and Day 3. (D) DG-eArch mice in CFC. Time course of freezing observed during Conditioning, Test 1, and Test 2 are shown. The far right panel of (D) shows averaged freezing levels during testing on Day 2 and Day 3. (E) Experimental schedule. (F) Averaged freezing levels during test 1(exposed to Context A) and test 2 (exposed to Context B). *P< 0.05. Data are represented as mean±SEM. N indicates number of animals.
In the experiment with Ocean-eArchT mice (Figure 7B), there might have been a floor effect in the freezing levels in Context B, because the freezing levels in Context B without manipulation were already relatively low. To address this potential caveat, we performed an additional experiment by investigating manipulation effects during the recall phase rather than learning phase (Fig. 7E and 7F). For this purpose, we subjected Ocean-eArchT mice to CFC in Context A (Figure 7E). One day after training, we exposed Ocean-eArchT mice to Context A while delivering green light bilaterally to MEC during the entire testing period (Figure 7F). Light-inhibited Ocean-eArchT mice displayed a freezing deficit in the conditioned context compared to the eYFP control mice (t18 = 2.84, P < 0.02) (Figure 7F). In contrast, light had no effect on freezing levels in Ocean-eArchT mice in the unconditioned context (t18 = −1.33, P > 0.19) (Figure 7F). Our results show that MEC Ocean cell input contributes substantially for the recall of the memory of the context (Context A) in which shock was delivered, but has no detectable role in the recall of generalized fear memory (Context B).
Trace fear conditioning (TFC) involves temporal association memory because it requires an association of two events—tone and footshock—separated by 20 seconds. Our earlier studies showed that the direct input from MEC layer III to the CA1 area of the hippocampus drives the formation of TFC memory (Suh et al., 2011). MEC layer II Island cells whose primary targets are the inhibitory neurons in the stratum lacunosum of the CA1 region inhibit layer III drive through feedforward inhibition (Kitamura et al., 2014). Consequently, inhibition of Island cell input by ArchT results in a boost of TFC memory (Kitamura et al., 2014). To investigate whether Ocean cells are crucial for TFC, we subjected Ocean-eArchT, Island-eArch, and their respective eYFP-only control mice to TFC while delivering green light bilaterally to MEC during the entire training period (See Methods) (Figure S5A). eArchT-mediated inhibition of Ocean cells had no detectable effect on TFC (Figure S5B–D) whereas, as reported previously (Kitamura et al., 2014), light-inhibited Island-eArch mice showed enhanced freezing amplitudes and post-tone freezing duration compared with the control eYFP mice (Figure S5E–G).
Discussion
In this study, we showed that Ocean cells, excitatory stellate neurons in MEC layer II projecting to DG and CA3, rapidly form a distinct representation of a novel context and drive context-specific activation of downstream CA3 cells as well as context-specific fear conditioning. In contrast, Island cells, excitatory pyramidal neurons in MEC layer II projecting into CA1, do not show context-specific encoding and are dispensable for context-specific memory. Thus Ocean cells provide contextual information to DG and CA3, but Island cells do not.
It has been thought that the DG-CA3 circuit generates context-specific information and contributes to the discrimination of contexts, spaces, and events (Bakker et al., 2008; Leutgeb et al., 2007; Marr, 1971; O'Reilly and McClelland, 1994; Treves and Rolls, 1994). Some lesion and spatially-restricted NMDA receptor knockout mouse studies have supported this concept (Gilbert et al., 2001; Kesner et al., 2000; McHugh et al., 2007). Here we found that integrated sensory information about a specific context is already present in some MECII Ocean cells, such that these cells can rapidly respond to distinct novel contexts through changes in activity rate, and that the cells holding such information can respond stably over multiple exposures (Figure 2 and Figure 3).
While further studies are necessary to fully characterize the context-specific Ocean cells, several of their features have already emerged. First, these cells respond to a specific context with varying latencies ranging between 10–150 s with a mean of 40–60 s. These values are not significantly different from the latencies of CA1 place cells of about 50 s (Frank et al., 2004) and entorhinal grid cells of 60 s (Hafting et al., 2005), although these comparisons should be taken cautiously because of the different methods of activity monitoring (Ca2+ imaging and tetrode recordings). Another interesting question is what inputs may contribute to the context-specific activity of Ocean cells. Previous studies have identified a diverse set of inputs into MEC, including those from postrhinal cortex, visual cortex, medial septum, para-subiculum, and pre-subiculum (For a review see Canto et al., 2008). Any of these inputs could contribute to the context-specific activity of Ocean cells. Furthermore, the HPC can contribute to this activity via loops of activity going from EC to HPC and back to EC, but our preliminary experiment conducted with pharmacological inhibition of CA1 activity suggests that this contribution is minimal if any (Figure S3). Another interesting question is whether MEC Ocean cells are heterogeneous with respect to their capability of context-specific responsiveness. Our results showed that 50–60% of Ocean cells responded to a given context (Figure 3) and the proportion of non-responding cell population is greater when a pair of more similar contexts was compared (38–40%) than when a pair of more distinct contexts was compared (18–32%) (Figure 3). Thus, we hypothesize that the non-responding cell population is a source of additional context-specific cells and will be recruited as such if the animal is exposed to additional contexts.
Combining the present findings with those previously reported (Denny et al., 2014; Kheirbek et al., 2013; Nakashiba et al., 2012), the following mutually non-exclusive dual mechanisms may underlie the discrimination of contexts. When animals face a relatively novel context, this context rapidly is represented by a distinct population of MEC Ocean cells. The activity of these cells drives context-specific activity in downstream areas like CA3 (Figures 5 and 6) to facilitate the formation of a context-specific memory (Figure 7A and 7B). This drive seems to be delivered to CA3 through adult-generated young DG-GCs and/or the direct input from Ocean cells to CA3 (Steward, 1976; Yeckel and Berger, 1990), because inhibition of developmentally-born old DG-GCs (Figure S3) does not reduce greatly the level of context-exposure dependent activation of CA3 cells (Figure 5K) nor the level of freezing in the conditioned context (Context A) (Figure 7D) whereas inhibition of both developmentally-born old DG-GCs and adult-generated young DG-GCs does reduce the level of freezing in the distinct conditioned context (Kheirbek et al., 2013). Interestingly, we saw increased freezing in the unconditioned context (Context B) in response to inhibition of old DG-GCs (Figure 7D), suggesting that the function of old DG-GCs may be to suppress generalized freezing (i.e. freezing in an unconditioned context). Indeed, one study has reported that memory precision requires feedforward inhibition of old DG-GC input to CA3 (Ruediger et al., 2011).
In contrast, neither Ocean cells (Figure 3L–3Q) nor old DG-GCs (Kheirbek et al., 2012; Nakashiba et al., 2012) appear to contribute significantly to the formation of distinct contextual memories from among similar contexts; this process relies, rather, on the unique properties (Ge et al., 2007; Schmidt-Hieber et al., 2004) and dynamics of adult-born young granule cells which compose just a few percent of the total DG-GCs (Clelland et al., 2009; Creer et al., 2010; Deng et al., 2010; Nakashiba et al., 2012; Sahay et al., 2011; Scobie et al., 2009). The mechanism for discriminating between similar contexts does not seem to occur in CA3 as rapidly as that for discriminating between more distinct contexts (Figure 6). Indeed, repeated exposures and/or association with high-valence stimulation (such as footshocks) is necessary for the gradual formation of memory that discriminates between a pair of more similar contexts (Kheirbek et al., 2012; McHugh et al., 2007; Nakashiba et al., 2012). In natural environments, the similarity or dissimilarity of a given context to other contexts varies broadly and the two mechanisms discussed above likely work together, with differing degrees of contribution to the animal’s overall context discrimination capability.
Our study also reveals the dichotomy of Ocean cells and Island cells of MEC in contextual versus temporal aspects of episodic memory. Ocean cells respond to exposure to a novel context and play a crucial role in encoding contextual information derived from an episode through their projections into DG and CA3. In contrast, Island cells do not respond to contextual exposure in a context-specific manner (Figure 2 and Figure 3) nor are they required for discriminatory encoding of a pair of distinct contexts (Figure 6), nor for context-specific fear conditioning of these distinctive contexts (Figure 7). On the other hand, Island cells drive feed-forward inhibition of CA1 pyramidal cells via their predominating projections to GABAergic interneurons in stratum lacunosum (SL) to inhibit MECIII input into CA1. Whereas MECIII cells drive the formation of trace fear memory (Suh et al., 2011), Island cells suppress the strength and duration of temporal aspects in trace fear memory (Figure S5) (Kitamura et al., 2014). Ocean cells are indifferent in this temporal aspect of episodic memory (Figure S5). Thus, Ocean and Island cells in MECII differentially provide the contextual information and time-related information, respectively to the HPC in order to help form episodic memory.
Experimental Procedures
Animals
All procedures relating to mouse care and treatment conformed to institutional and NIH guidelines. Animals were individually housed in a 12 hour (7am–7pm) light/dark cycle, with food and water ad libitum. For studies of Ocean-GCaMP6f mice, we used 30–40 weeks-old wildtype male C57BL/6 mice (WT). For studies of Island-GCaMP6f mice and Island-eArch mice, we used previously developed Wfs1-Cre transgenic male mice (Kitamura et al., 2014) maintained on a C57BL/6 background (Jackson Lab), adults aged about 30–50 weeks. For studies of Ocean-eArchT mice, we used 15–25 weeks-old wild-type male C57BL/6 mice. For studies of Island-eArch mice, we used 15–25 weeks-old Wfs1-Cre transgenic male mice. For studies of DG granule-eArch3.0-eYFP mice, we used previously developed DOCK10-Cre transgenic 15–25 weeks-old male mice (Kohara et al., 2014). For c-Fos experiments in Figure 3, we used 15–25 weeks-old c-fos-tTA transgenic male mice (Reijmers et al., 2007). In Figure 6, we crossed c-fos-tTA transgenic mice with Wfs1-Cre mice.
Histology and Immunohistochemistry
Mice were transcardially perfused with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS). Brains were post-fixed with the same solution for 24 hours, and then sectioned using a vibratome. For immunohistochemistry (IHC) sliced tissue sections were incubated in 0.3% Triton-X PBS with 5% normal goat serum (NGS) for 1 hour. Primary antibodies were then added to a 5% NGS 0.3% triton-X in PBS solution and incubated overnight at 4°C. Primary antibodies: WFS-1 (Proteintech Group, Inc, 11558-1-AP, 1:500), CalbindinD-28K (Abcam, ab11426, 1:1000) (SWANT, 300, 1:1000), Reelin (MBL International, D223-3, 1:250), NeuN (Millipore, MAB377, 1:1000) (Millipore, ABN78, 1:1000), c-Fos (SANTA CRUZ BIOTECHNOLOGY, SC-52, 1/1000), Prox1 (Invitrogen, P21936, 1/1000), and DCX (Millipore, AB2253, 1/1000). After rinsing with PBS 3 times for 15 min each, sliced tissue sections were subsequently incubated with AlexaFluor405, AlexaFluor488, AlexaFluor546, or AlexaFluor633 conjugated secondary antibodies (Invitrogen, 1:500). Sliced tissue sections were then washed in PBS 3 times for 15 min and mounted in VECTASHIELD medium on glass slides. Some sections were stained by DAPI or Nissl (1:500). Fluorescence images were taken by confocal microscopy using 10×, 20×, 40×, 63× objectives and by fluorescent microscopy using 10×and 20× objectives. Z-projected confocal images were generated by Zenblack.
Preparation of Adeno-Associated Viruses
The AAV2/5-CaMK2α-eYFP was acquired from the University of North Carolina at Chapel Hill (UNC) Vector Core, with a titer of 5.2*10^12 genome copy/ml. The AAV2/5-Syn-DIO-GCaMP6f was generated by and acquired from the University of Pennsylvania Vector Core, with a titer of 1.3*10^13 genome copy/ml. The AAV2/5-Syn-GCaMP6f was generated by and acquired from the University of Pennsylvania Vector Core, with a titer of 1.5*10^13 genome copy/ml. The AAV2/5-CaMK2α-eArchT3.0-eYFP was acquired from the UNC Vector Core, with a titer of 2.5*10^12 genome copy/ml. The AAV2/9-EF1α-DIO-ChR2-eYFP was generated by and acquired from the University of Pennsylvania Vector Core, with a titer of 2.5*10^13 genome copy/ml. The AAV2/5-EF1α-DIO-eYFP (or mCherry) was generated by and acquired from the UNC Vector Core, with a titer of 1.2*10^13 genome copy/ml. The AAV2/5-EF1α-DIOeArch3.0-eYFP was generated by and acquired from the UNC Vector Core, with a titer of 1.6*10^13 genome copy/ml. The AAV2/9-EF1α-DIO-eYFP and AAV2/9-EF1α-DIO-eArch3.0-eYFP were generated by and acquired from the UNC Vector Core. We previously generated the AAV2/9-TRE-ChR2mCherry with a titer of 8.0*10^12 genome copy/ml (Ramirez et al., 2013).
Stereotaxic Surgeries
Stereotactic viral injections, microendoscope implantations, and optic fiber implantations were all performed in accordance with MIT’s CAC guidelines. Mice were anaesthetized using 500 mg/kg avertin. Viruses were injected using a glass micropipette attached to a 10 µl Hamilton microsyringe through a microelectrode holder filled with mineral oil. A microsyringe pump and its controller were used to control the speed of the injection. The needle was slowly lowered to the target site and remained for 10 minutes after the injection.
For Ocean cell-specific GCaMP6f monitoring, unilateral viral delivery into the right dorsal dentate gyrus of the WT male mice was aimed at these coordinates relative to Bregma: AP: −2.0 mm, ML, +1.3 mm, DV, −2.05 mm. WT mice were injected with 150 nl of AAV2/5-Syn-GCaMP6f. One month after AAV injection, we implanted a microendoscope into the dorsal part of MEC (AP: −5.1 mm, ML, ±3.36 mm, DV, −2.20 mm) of the WT mice. For Island cell-specific GCaMP6f monitoring, unilateral viral delivery into the right MEC of Wfs1-Cre male mice was aimed at these coordinates relative to Bregma: AP: −4.85 mm, ML, +3.45 mm, DV, −3.30 mm. Wfs1-Cre mice were injected with 200 nl of AAV2/5-Syn-DIO-GCaMP6f. One month after AAV injection, a microendoscope was implanted into the dorsal part of MEC (AP: −5.1 mm, ML, +3.36 mm, DV, −2.20 mm) of Wfs1-Cre male mice. One month after implantation of the microendoscope, the baseplate for a miniaturized microscope camera (Ziv et al., 2013) was attached above the implanted microendoscope in the mice. After the baseplate surgery, animals were habituated to the attachment of the microscope camera for two weeks.
For behavior experiments in Ocean-eArchT mice, bilateral viral delivery into dorsal DG of the WT male mice was aimed at these coordinates relative to Bregma: AP: −2.0 mm, ML, ±1.3 mm, DV, −2.05 mm. WT male mice were bilaterally injected with 150 nl of AAV2/5-CaMK2αeArchT3.0-eYFP or AAV2/5-CaMK2α-eYFP as control group, and then bilaterally implanted with a Doric patchcord optical fiber (200 µm core diameter) into the dorsal part of MEC (AP: −5.1 mm, ML, ±3.36 mm, DV, −2.20 mm).
For behavior experiments in Island-eArch mice, bilateral viral delivery into MEC of Wfs1-Cre male mice was aimed at these coordinates relative to Bregma: AP: −4.85 mm, ML, ±3.45 mm, DV, −3.30 mm. Wfs1-Cre male mice were bilaterally injected with 200 nl of AAV2/5-EF-1α-DIO-eArch3.0-eYFP or AAV2/5-EF-1α-DIO-eYFP as control group, and were bilaterally implanted with optical fibers into the dorsal part of MEC (AP: −5.1 mm, ML, ±3.36 mm, DV, −2.20 mm). The top part of an Eppendorf tube was inserted to protect the implant and the incision was closed with sutures as per our previous study (Liu et al., 2012).
For behavior experiments in DG-eArch mice, bilateral viral delivery into dorsal DG of Dock10-Cre male mice was aimed at these coordinates relative to Bregma: AP: −2.0 mm, ML, ±1.3 mm, DV, −2.05 mm. The Dock10-Cre male mice were bilaterally injected with 150 nl of AAV2/9-EF1α-DIO-eArch3.0-eYFP or AAV2/9--EF1α-DIO-eYFP as a control group, and were bilaterally implanted with a Doric patchcord optical fiber (200 µm core diameter) into the dorsal DG (AP: −2.0 mm, ML, ±1.3 mm, DV, −1.50 mm). The Dock10-Cre mice were allowed to recover for 5 days before all subsequent experiments.
For CA3 c-Fos labeling experiments in Ocean-eArchT mice (Figure 6), bilateral viral delivery into the dorsal DG of c-fos-tTA transgenic male mice was aimed at these coordinates relative to Bregma: AP: −2.0 mm, ML, ±1.3 mm, DV, −2.05 mm. The c-fos-tTA transgenic male mice were bilaterally injected with 150 nl of AAV2/5-CaMK2α-eArchT3.0eYFP or AAV2/5-CaMK2α-eYFP into the dorsal DG, and then bilaterally injected with 200 nl of AAV2/9-TRE-mCherry into dorsal CA3 (AP: −2.10 mm, ML, ±2.30 mm, DV, −2.30 mm), followed by bilateral implantation of a Doric patchcord optical fiber (200 µm core diameter) into the dorsal part of MEC (AP: −5.1 mm, ML, ±3.36 mm, DV, −2.20 mm). For CA3 c-Fos labeling experiments in Island-eArch mice (Figure 6), bilateral viral delivery into MEC of the c-fos-tTA and Wfs1-Cre double transgenic male mice was aimed at these coordinates relative to Bregma: AP: −4.85 mm, ML, ±3.45 mm, DV, −3.30 mm. The c-fos-tTA transgenic male mice were bilaterally injected with 200 nl of AAV2/5-EF1α-DIO-eArch3.0-eYFP or AAV2/5-EF1α-DIO-eYFP into MEC, and then bilaterally injected with 200 nl of AAV2/9-TRE-mCherry into dorsal CA3 (AP: −2.10 mm, ML, ±2.30 mm, DV, −2.30 mm), followed by bilateral implantation of a Doric patchcord optical fiber (200 µm core diameter) into the dorsal part of MEC (AP: −5.1 mm, ML, ±3.36 mm, DV, −2.20 mm). Mice were given 1.5 mg/kg metacam as analgesic and remained on a heating pad until fully recovered from anesthesia. Mice were allowed to recover for 1 month before all subsequent experiments.
Context Exposure
For Ca2+ imaging experiments during context exposure, we used three different pairs of contexts: distinct pair Context A and B, distinct pair Context C and D, and similar pair Context E and F. Context A was a chamber (29 cm (W) × 23 cm (D) × 20 cm (H)), with distinct visual cues and a grid floor which consisted of 36 stainless steel rods (Context A) (Figure S2). Context B was a chamber (29 cm (W)×23 cm (D) × 20 cm (H)), with different visual cues compared to Context A and a white plastic floor (Context B) (Figure S2). Context C was a cylindrical chamber (diameter 28 cm (D) × 20 cm (H)), with no visual cues on the walls and a white plastic floor (Context C) (Figure S2). Context D was a square chamber (26 cm (W) × 26 cm (D) × 17 cm (H)), with black walls and floors (Context D) (Figure S2). Context E was a chamber with rounded walls (29 cm (W) × 23 cm (D) × 20 cm (H)), with no visual cues on the walls and a grid floor which consisted of 36 stainless steel rods (Context E) (Figure S2). Context F was a rectangular chamber (29 cm (W) × 23 cm (D) × 20 cm (H)), with no-visual cues on the walls and a grid floor which consisted of 36 stainless steel rods (Context F).
Calcium Imaging during Context Exposures
Calcium imaging was performed on male mice in the animal facility during the light cycle. Island and Ocean mice were habituated to human experimenters as well as the experimental room three weeks following AAV injection, microendoscope implant, and baseplate surgeries. Experiments were run in several different pairs of contexts (Figure S2). Ca2+ signals were imaged twice in each context for 5 minutes each time. The chambers were cleaned with 70% ethanol between sessions. Immediately before and after imaging sessions, the mouse rested on a pedestal next to the open field. Calcium events were captured at 20 Hz on an Inscopix miniature microscope. The calcium movie was then motion corrected using Inscopix Mosaic software (correction type: translation and rotation; reference region with spatial mean (r = 20 pixels) subtracted, inverted, and spatial mean applied (r = 5 pixels)). Finally, it was processed by ImageJ (dividing each image, pixel by pixel, by a low-passed (r = 20 pixels) filtered version), and the ΔF/F signal was calculated. Cell locations were carefully selected manually, as small regions of interest (ROIs) (~ 1/3 of cell body size) at the center of the cell bodies. This was done in ImageJ for each processed movie. Approximately 40 cells were selected per behavioral session per animal, and their ΔF/F signals were isolated. Ca2+ events were detected by thresholding (> 3% from the ΔF/F signal at the local maxima of the ΔF/F signal. Cells were said to have responded in a specific context if they had >10 Ca2+ events during exposure to that context. A rate difference index (defined below) was calculated for all cells that had > 10 Ca2+ events during exposure to a context.
Rate Difference Index of Ca2+ Activity and Definition of Context-Specific Cells
We used the following score to determine whether single cells were “context specific”:
Where nevents,X is the number of Ca2+ events in context X. Similar definition for nevents,Y.
The rate difference score was first calculated for every cell in each animal for Context A (first time) versus Context A (second time), and for Context B (first time) versus context B (second time) to determine the natural variability of Ca2+ activity within the same environment. The 99thpercentile of these scores was set as the threshold (0.6). Cells were “context specific” if their difference score for Context A versus Context B exceeded the threshold. In Figure 3, we displayed the cumulative probability of the rate difference indices for same context (e.g. A vs A) and for different contexts (e.g. A vs B). If the distribution shifts to right, this indicates a larger change in the number of Ca2+ events and higher median rate difference index.
Context Exposure and c-Fos+ Cells Counting
Context exposure experiments were performed on male mice aged between 12 and 20 weeks, in the animal facility during the light cycle. In Figure 5, all mice were handled for three days prior to the behavioral experiments. The bilateral optical fiber implants were connected to a 561 nm laser controlled by a function generator. The mice were placed in Context A and allowed to explore for 5 min. Mice received green light stimulation (15 mW, each hemispheres) during the entire period (5 min, Figure 5). Sixty minutes after being exposed to Context A, animals were perfused and their brains were fixed with 4% PFA to detect the c-Fos immunoreactivities in DG and CA3.
For CA3 c-Fos labeling experiments in Ocean-eArchT mice and Island-eArch mice (Figure 6), all mice were handled for three days prior to the behavioral experiments. They were then taken off Dox for 24 hours to open a window of activity-dependent labeling for Context A. Animals were exposed to Context A for 5 min and DOX diets were resumed immediately after exposure to Context A. Twenty-for hours later, animals were exposed to Context B for 5 min. Sixty minutes after exposure to Context B, animals were perfused and their brains were fixed with 4% PFA. Mice received bilateral green light stimulation (15 mW, each hemispheres) during the entire period of exposure to Context A or B (5 min).
In Figure 5, a sampling of c-Fos+ cells was conducted throughout dorsal DG in sagittal sections for the quantification analysis of the number of c-Fos+ cells in DG and CA3 in each group of mice. Ten sagittal hippocampal sections were used to count the number of c-Fos+ cells and DAPI+ cells in DG and CA3 regions. In Figure 6, for quantification analysis of the number of mCherry+ cells and c-Fos+ cells in DG and CA3 region in each group of mice, we randomly chose 10 broad regions of interest (ROI) from which hundreds of cells were to be examined, and took images by confocal microscopy. The total number of examined mice is shown in Figure 6. All counting was performed blind as to the group and condition.
The numbers of mCherry+ cells, c-Fos+ cells and DAPI+ cells in the ROI were counted, and the percentages ratios:
and
were calculated.
The nice property about these two measures is that this quantity #(mCherry+) (or #(mCherry−)) is present in both the denominator and the numerator, and thus, these two ratios are invarianttothe number of mCherry+ (or mCherry−) cells, and any differences in measurements is due to different experimental groups having c-Fos+ populations differently distributed between mCherry+ and mCherry− populations. Detailed information is shown in Table 2 of Supplemental Information.
Calcium Imaging with Pharmacological Manipulation in Hippocampus
Pharmacological manipulation was conducted as previously reported (Kitamura et al., 2009). Fluorescently labelled γ-aminobutyric acid subtype A receptor agonist (FCM, a fluorophore conjugated muscimol, 0.8 mM, Molecular Probe) was bilaterally injected into the dorsal hippocampus (AP: −2.0 mm, ML, ±1.5 mm, DV, −1.50 mm) 30 min before Ca2+ imaging from MEC. Anesthetized recordings in vivo were conducted as previously described (Kitamura et al., 2014).
Fear Conditioning
Fear conditioning was performed on male mice aged between 15 and 25 weeks, in the animal facility during the light cycle with minor modifications to the method described previously (Kitamura et al., 2014). Before fear conditioning, all animals were habituated to human experimenters for 3 days. For contextual fear conditioning, we used two different contexts (Context A and Context B). Context A was a chamber (29 cm (W) × 23 cm (D) × 20 cm (H)) with distinct visual cues and a grid floor which consisted of 36 stainless steel rods (Context A) (Figure S2). Context B was a chamber (29 cm (W) × 23 cm (D) × 20 cm (H)) with different visual cues compared to Context A and a white plastic floor (Context B) (Figure S2). The chambers were cleaned with 70% alcohol prior to the introduction of each individual mouse. In Figure 7, we conducted contextual fear conditioning on Ocean-eArchT mice, Island-eArch mice, and DG-eArch mice. On Day 1, the optical fiber implant was connected to a 561 nm laser controlled by a function generator. The mice were placed in novel Context A and allowed to explore for 120 s, at which point a 2 s, 0.75 mA foot-shock was delivered. Mice remained in the conditioning chamber for a total of 180 s. Mice received green light stimulation (15 mW, each hemisphere) during the entire training period (180 s, Figure 4). On Day 2 (24 hours later), mice were placed into Context A and allowed to explore for 180 s to test contextual memory. On Day 3, mice were placed in distinct Context B and allowed to explore for 180 s to test context specificity.
In Figure S5, we subjected Ocean-eArchT mice, Island-eArch mice, and their respective eYFP control mice to trace fear conditioning. On Day 1, the optical fiber implant was connected to a 561nm laser controlled by a function generator. The mice were placed in novel Context A and allowed to explore for 240 s, at which point a 20 s tone (75 dB, 2000 Hz) was played, followed by a 20 s trace, and then a 2 s, 0.75 mA foot-shock. This was repeated two more times, starting at 402 s and 564 s. Mice received bilateral green light stimulation (15 mW, each hemispheres) during the entire training period (tone, trace and shock, total 42 s, Figure S5). Mice remained in the conditioning chamber for a total of 706 s. On Day 2 (24 hours later), mice were placed in distinct Context B and allowed to explore for 240 s, at which point the same tone as from Day 1 was played for 60 s, followed by 180 s of no-tone (post-tone period). This was repeated two more times and the mice were removed after 960 s in the chamber.
Statistical Analysis
All data are presented as mean ± SEM. N indicates number of animals. n indicates number of cells. Comparisons between two-group data were analyzed by paired t-test or unpaired Student's t test. Comparisons between two-group distribution data were analyzed by Kolmogorov– Smirnov (KS) test. Multiple group comparisons were assessed using a one-way, two-way, or repeated-measures analysis of variance (ANOVA), followed by the post-hoc Scheffe's test when significant main effects or interactions were detected. The null hypothesis was rejected at the P<0.05 level.
Supplementary Material
Acknowledgments
We thank A. Hamalian, C. Ragion, D. Roy, L. Smith, K. Kohara, M. Ragion, S. Perry, C.Puryear, and W. Yu for experimental help, X. Liu for providing AAV2/9-TRE-mCherry virus, J. Young and T. Okuyama for helpful discussions and comments, L. Brenner for paper preparation, and the members of Tonegawa lab for their support. This work was supported by the RIKEN Brain Science Institute (to ST), Howard Hughes Medical Institute (to ST), the Picower Institute Innovation Fund (to ST) and The JPB Foundation (to TK). LJK was supported by an NSF graduate fellowship.
Footnotes
Author Contributions
TK, CS and ST developed the project idea. TK developed the cell-type specific GCAMP-labeling method. TK and JM performed surgery and histology; TK performed the behavior experiments. TK and CS analyzed the data. LJK and MJS helped with analysis. TK, CS and ST wrote the manuscript with input from all authors. ST supervised the entire project.
References
- Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319–335. doi: 10.1002/cne.901240303. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Witter MP. The three-dimensional organization of the hippocampal formation: a review of anatomical data. Neuroscience. 1989;31:571–591. doi: 10.1016/0306-4522(89)90424-7. [DOI] [PubMed] [Google Scholar]
- Bakker A, Kirwan CB, Miller M, Stark CE. Pattern separation in the human hippocampal CA3 and dentate gyrus. Science. 2008;319:1640–1642. doi: 10.1126/science.1152882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto CB, Wouterlood FG, Witter MP. What does the anatomical organization of the entorhinal cortex tell us? Neural Plast. 2008;2008:381243. doi: 10.1155/2008/381243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland CD, Choi M, Romberg C, Clemenson GD, Jr, Fragniere A, Tyers P, Jessberger S, Saksida LM, Barker RA, Gage FH, Bussey TJ. A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science. 2009;325:210–213. doi: 10.1126/science.1173215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creer DJ, Romberg C, Saksida LM, van Praag H, Bussey TJ. Running enhances spatial pattern separation in mice. Proc Natl Acad Sci U S A. 2010;107:2367–2372. doi: 10.1073/pnas.0911725107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci. 2010;11:339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, Hen R. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201. doi: 10.1016/j.neuron.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H. A cortical-hippocampal system for declarative memory. Nat Rev Neurosci. 2000;1:41–50. doi: 10.1038/35036213. [DOI] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- Frank LM, Stanley GB, Brown EN. Hippocampal plasticity across multiple days of exposure to novel environments. J Neurosci. 2004;24:7681–7689. doi: 10.1523/JNEUROSCI.1958-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Yang CH, Hsu KS, Ming GL, Song H. A critical period for enhanced synaptic plasticity in newly generated neurons of the adult brain. Neuron. 2007;54:559–566. doi: 10.1016/j.neuron.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PE, Kesner RP, Lee I. Dissociating hippocampal subregions: double dissociation between dentate gyrus and CA1. Hippocampus. 2001;11:626–636. doi: 10.1002/hipo.1077. [DOI] [PubMed] [Google Scholar]
- Hafting T, Fyhn M, Molden S, Moser MB, Moser EI. Microstructure of a spatial map in the entorhinal cortex. Nature. 2005;436:801–806. doi: 10.1038/nature03721. [DOI] [PubMed] [Google Scholar]
- Kesner RP, Gilbert PE, Wallenstein GV. Testing neural network models of memory with behavioral experiments. Curr Opin Neurobiol. 2000;10:260–265. doi: 10.1016/s0959-4388(00)00067-2. [DOI] [PubMed] [Google Scholar]
- Kheirbek MA, Drew LJ, Burghardt NS, Costantini DO, Tannenholz L, Ahmari SE, Zeng H, Fenton AA, Hen R. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron. 2013;77:955–968. doi: 10.1016/j.neuron.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheirbek MA, Tannenholz L, Hen R. NR2B-dependent plasticity of adult-born granule cells is necessary for context discrimination. J Neurosci. 2012;32:8696–8702. doi: 10.1523/JNEUROSCI.1692-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Pignatelli M, Suh J, Kohara K, Yoshiki A, Abe K, Tonegawa S. Island cells control temporal association memory. Science. 2014;343:896–901. doi: 10.1126/science.1244634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Saitoh Y, Takashima N, Murayama A, Niibori Y, Ageta H, Sekiguchi M, Sugiyama H, Inokuchi K. Adult neurogenesis modulates the hippocampus-dependent period of associative fear memory. Cell. 2009;139:814–827. doi: 10.1016/j.cell.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Kohara K, Pignatelli M, Rivest AJ, Jung HY, Kitamura T, Suh J, Frank D, Kajikawa K, Mise N, Obata Y, et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat Neurosci. 2014;17:269–279. doi: 10.1038/nn.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser MB, Moser EI. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science. 2007;315:961–966. doi: 10.1126/science.1135801. [DOI] [PubMed] [Google Scholar]
- Leutgeb S, Leutgeb JK, Treves A, Moser MB, Moser EI. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science. 2004;305:1295–1298. doi: 10.1126/science.1100265. [DOI] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr D. Simple memory: a theory for archicortex. Philos Trans R Soc Lond B Biol Sci. 1971;262:23–81. doi: 10.1098/rstb.1971.0078. [DOI] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O'Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S. Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science. 2007;317:94–99. doi: 10.1126/science.1140263. [DOI] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ, et al. Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell. 2012;149:188–201. doi: 10.1016/j.cell.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niibori Y, Yu TS, Epp JR, Akers KG, Josselyn SA, Frankland PW. Suppression of adult neurogenesis impairs population coding of similar contexts in hippocampal CA3 region. Nat Commun. 2012;3:1253. doi: 10.1038/ncomms2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly RC, McClelland JL. Hippocampal conjunctive encoding, storage, and recall: avoiding a trade-off. Hippocampus. 1994;4:661–682. doi: 10.1002/hipo.450040605. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Liu X, Lin PA, Suh J, Pignatelli M, Redondo RL, Ryan TJ, Tonegawa S. Creating a false memory in the hippocampus. Science. 2013;341:387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- Ray S, Naumann R, Burgalossi A, Tang Q, Schmidt H, Brecht M. Grid-layout and theta-modulation of layer 2 pyramidal neurons in medial entorhinal cortex. Science. 2014;343:891–896. doi: 10.1126/science.1243028. [DOI] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- Ruediger S, Vittori C, Bednarek E, Genoud C, Strata P, Sacchetti B, Caroni P. Learning-related feedforward inhibitory connectivity growth required for memory precision. Nature. 2011;473:514–518. doi: 10.1038/nature09946. [DOI] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O'Carroll CM, Kheirbek MA, Burghardt NS, Fenton AA, Dranovsky A, Hen R. Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature. 2011;472:466–470. doi: 10.1038/nature09817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarinana J, Kitamura T, Kunzler P, Sultzman L, Tonegawa S. Differential roles of the dopamine 1-class receptors, D1R and D5R, in hippocampal dependent memory. Proc Natl Acad Sci U S A. 2014;111:8245–8250. doi: 10.1073/pnas.1407395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger AR, Cowan WM, Gottlieb DI. An autoradiographic study of the time of origin and the pattern of granule cell migration in the dentate gyrus of the rat. J Comp Neurol. 1975;159:149–175. doi: 10.1002/cne.901590202. [DOI] [PubMed] [Google Scholar]
- Schmidt-Hieber C, Jonas P, Bischofberger J. Enhanced synaptic plasticity in newly generated granule cells of the adult hippocampus. Nature. 2004;429:184–187. doi: 10.1038/nature02553. [DOI] [PubMed] [Google Scholar]
- Scobie KN, Hall BJ, Wilke SA, Klemenhagen KC, Fujii-Kuriyama Y, Ghosh A, Hen R, Sahay A. Kruppel-like factor 9 is necessary for late-phase neuronal maturation in the developing dentate gyrus and during adult hippocampal neurogenesis. J Neurosci. 2009;29:9875–9887. doi: 10.1523/JNEUROSCI.2260-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville WB, Milner B. Loss of recent memory after bilateral hippocampal lesions. J Neurol Neurosurg Psychiatry. 1957;20:11–21. doi: 10.1136/jnnp.20.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Arai Y. Highly polysialylated neural cell adhesion molecule (NCAM-H) is expressed by newly generated granule cells in the dentate gyrus of the adult rat. J Neurosci. 1993;13:2351–2358. doi: 10.1523/JNEUROSCI.13-06-02351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O. Topographic organization of the projections from the entorhinal area to the hippocampal formation of the rat. J Comp Neurol. 1976;167:285–314. doi: 10.1002/cne.901670303. [DOI] [PubMed] [Google Scholar]
- Suh J, Rivest AJ, Nakashiba T, Tominaga T, Tonegawa S. Entorhinal cortex layer III input to the hippocampus is crucial for temporal association memory. Science. 2011;334:1415–1420. doi: 10.1126/science.1210125. [DOI] [PubMed] [Google Scholar]
- Sun C, Kitamura T, Yamamoto J, Martin J, Pignatelli M, Kitch LJ, Schnitzer MJ, Tonegawa S. Distinct speed dependence of entorhinal island and ocean cells, including respective grid cells. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1511668112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treves A, Rolls ET. Computational analysis of the role of the hippocampus in memory. Hippocampus. 1994;4:374–391. doi: 10.1002/hipo.450040319. [DOI] [PubMed] [Google Scholar]
- Tulving E. Episodic memory: from mind to brain. Annu Rev Psychol. 2002;53:1–25. doi: 10.1146/annurev.psych.53.100901.135114. [DOI] [PubMed] [Google Scholar]
- Varga C, Lee SY, Soltesz I. Target-selective GABAergic control of entorhinal cortex output. Nat Neurosci. 2010;13:822–824. doi: 10.1038/nn.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintzer ME, Boehringer R, Polygalov D, McHugh TJ. The hippocampal CA2 ensemble is sensitive to contextual change. J Neurosci. 2014;34:3056–3066. doi: 10.1523/JNEUROSCI.2563-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter MP, Wouterlood FG, Naber PA, Van Haeften T. Anatomical organization of the parahippocampal-hippocampal network. Ann N Y Acad Sci. 2000;911:1–24. doi: 10.1111/j.1749-6632.2000.tb06716.x. [DOI] [PubMed] [Google Scholar]
- Yeckel MF, Berger TW. Feedforward excitation of the hippocampus by afferents from the entorhinal cortex: redefinition of the role of the trisynaptic pathway. Proc Natl Acad Sci U S A. 1990;87:5832–5836. doi: 10.1073/pnas.87.15.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.