Abstract
Background
This study investigated the effects and mechanism of imatinib in inhibiting colon cancer cell proliferation.
Material/Methods
The SW480 cells were divided into 4 imatinib-treated groups: 0 μM, 1.25 μM, 2.5 μM, and 5μM. We analyzed the apoptosis and cell cycle of the 4 groups. The gene and protein expressions of p21, p27, HGF, and GAPDH were measured by RT-PCR and Western blot.
Results
Compared with the 0-μM imatinib-treated group, the apoptosis of 1.25-μM, 2.5-μM, and 5.0-μM treated groups was significantly induced (P<0.05, all). The G1 phase was significantly up-regulated in the 1.25-μM, 2.5-μM, and 5.0-μM treated groups compared with the 0-μM imatinib-treated group (P<0.05, respectively), but the S and G2 phase of 3 imatinib-treated groups were significantly down-regulated (P<0.05, all). The gene and protein expressions of p27 and HGF were significantly different among the 4 groups (P<0.05, all).
Conclusions
Imatinib inhibits proliferation of colon cancer cells by reducing HGF and increasing p27 in a dose-dependent manner.
MeSH Keywords: Colonic Neoplasms, Cyclin-Dependent Kinase Inhibitor p27, Proto-Oncogene Proteins c-met
Background
Colon cancer is a common malignant tumor in the digestive system, and its incidence has been gradually increasing [1]. At present, chemotherapy for colon cancer still uses 5-Fluorouracil as the basis for combination chemotherapy drugs, but adverse effects are worse with this drug and patient tolerance is poor. Although the use of targeted drugs can improve the prognosis of patients with colon cancer, the mortality rate of colon cancer patients is still high, and it is now even more important to find new colon cancer drugs. There have been reports that imatinib had a good therapeutic effect on gastric stromal tumors, leukemia, and gynecological tumors [2–7], but there are no reports about the mechanism of imatinib in colorectal cancer.
At present, HGF is the most widely used growth factor in biological studies, with the C-Met mediated by mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (P13K), and Stat3 pathway [8], promoting the inhibition of cell proliferation, division, and apoptosis. Recent research found HGF is overexpressed in most tumors and is related with tumor cell proliferation. The correlation between imatinib and HGF in colon cancer is unclear.
The aim of this study was to investigate the effect of imatinib on the SW620 colon cancer cell line at different concentrations and times, as well as to explore the mechanism of action.
Material and Methods
Materials
The chemical structure of imatinib (Sigma, with a purity ≥98%) is shown in Figure 1. The colon cancer cell line SW480 was provided by the Shanghai Cell Bank of the Chinese Academy of Sciences. Fetal bovine serum (FBS) was purchased from Hyclone, Leibowitz’s L-15 medium and puromycin were bought from Gibco company, and rabbit anti-GAPDH, anti-HGF, anti-p21, and anti-p27 were purchased from Sigma-Aldrich.
Figure 1.
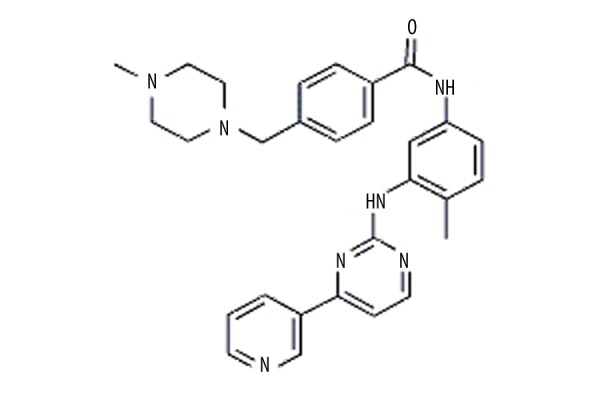
The chemical structure of imatinib.
Cell culture
The SW480 cells were suspended by Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum and cultured in an incubator for a differential plating period of 90 min to allow attachment of the non-myocardial cells. The cardiomyocytes were obtained from the deposit after centrifugation at 1000 rpm for 8 min and plated in 10-cm culture dishes (Falcon) at a density of 1×105 cells/ml. Following incubation for 24 h, the cultures were washed 2 times with phosphate-buffered saline (PBS) to remove the dead and non-adherent cells. The medium was changed every 3 days during incubation, and the treatment factors were administrated on the fifth day. SW480 cells were randomly divided into 4 groups by imatinib dose: 0 μM, 1.25 μM, 2.5 μM, and 5μM for each of 4 different stains.
Apoptosis assay
The apoptotic cells were stained by Annexin V-FITC and the quantification was determined by FACSVerse using FACSuite analysis software (BD Bioscience). In brief, 0-μM, 0.625-μM, 1.25-μM, 2.5-μM, and 5-μM treated cells were harvested and washed with PBS, then added to an annexin V-FITC solution and PI followed by fluorescence-activated cell sorting assay.
Cell cycle analysis
For cell cycle analysis, cells were stained with propidium iodide (PI) using the Cycletest Plus DNA Reagent Kit (BD Biosciences) following the manufacturer’s protocol, and the cell cycle distribution was analyzed by FACSVerse flow cytometer (BD Biosciences). The percentages of cells in G1, S, and G2 phases were counted and compared. The experiments were carried out in triplicate.
Real-time PCR
The cells of the 5 groups were lysed for the expression levels of P 21, P 27, and HGF by reverse transcription and real-time PCR. Total RNA of tissue samples was extracted and reverse transcribed into cDNA following the manufacturer’s protocol. To determine the expression of P 21, P 27, and HGF, quantitative real-time PCR reaction was conducted. All reactions were performed in triplicate, and relative gene expression determinations of P 21, P 27, and HGF were obtained.
Western blot
SDS-PAGE separated proteins were transferred to nitrocellulose membranes and then incubated with 5% blocking reagent for 2 h at room temperature. Subsequently, the membranes were probed with antibodies specific to P27 (1:200, Santa Cruz, USA), P21 (1:500, Santa Cruz, UA) and GAPDH (1:1000, Santa Cruz, USA) overnight at 4°C. The membranes were washed 3 times and incubated with goat anti-mouse P27 (1:1000; Santa Cruz, USA) or goat anti-rabbit IgG HRP (1:1000; Santa Cruz, USA) for 90 min at room temperature followed by enhanced chemiluminescence and exposure to chemiluminescent film.
Statistical analysis
Data are presented as mean ± standard deviation (SD) values. Statistical analyses between the 2 groups were evaluated using one-way ANOVA with SPSS 19.0 (SPSS Inc., Chicago, USA). P<0.05 was considered statistically significant.
Results
Apoptosis assay
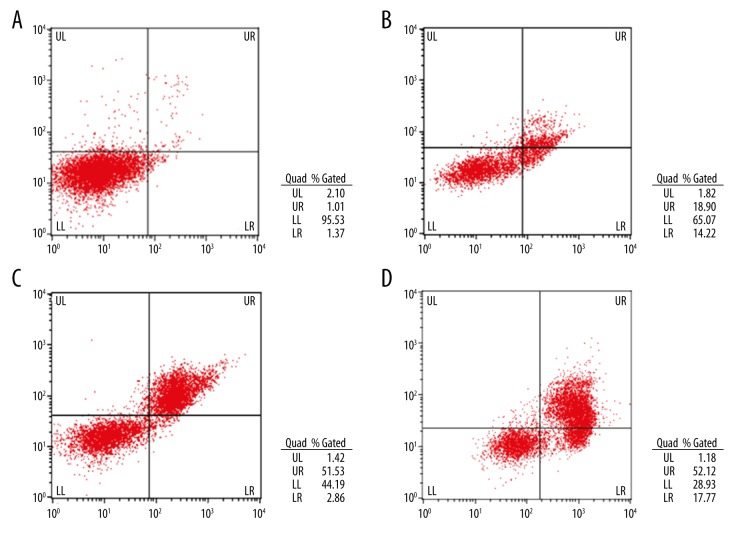
Compared with the 0-μM imatinib-treated group, the apoptosis of 1.25-μM, 2.5-μM, and 5.0-μM treated groups were significantly induced (P<0.05) in a dose-dependent manner (P<0.05) (Figure 2, Table 1).
Figure 2.
(A) Apoptosis assay of the 0-μM imatinib-treated group. (B) Apoptosis assay of the 1.25-μM imatinib-treated group. (C) Apoptosis assay of the 2.5-μM imatinib-treated group. (D) Apoptosis assay of the 5.0-μM imatinib-treated group.
Table 1.
Apotosis assay in four groups(mean ±SD, %).
| Group | UL | UR+LR | LL |
|---|---|---|---|
| 0 μM | 2.51±0.36 | 2.59±0.35 | 94.90±0.55 |
| 1.25 μM | 2.29±0.10 | 33.22±0.52* | 64.49±0.53* |
| 2.5 μM | 2.40±0.59 | 54.30±0.32*,** | 43.30±0.66*,** |
| 5.0 μM | 2.40±0.29 | 68.07±1.10*,**,*** | 29.52±1.01*,**,*** |
P<0.05, there were significantly difference compared with 0 μM;
P<0.05, there were significantly difference compared with 1.25 μM;
P<0.05, there were significantly difference compared with 2.5 μM
Cell cycle
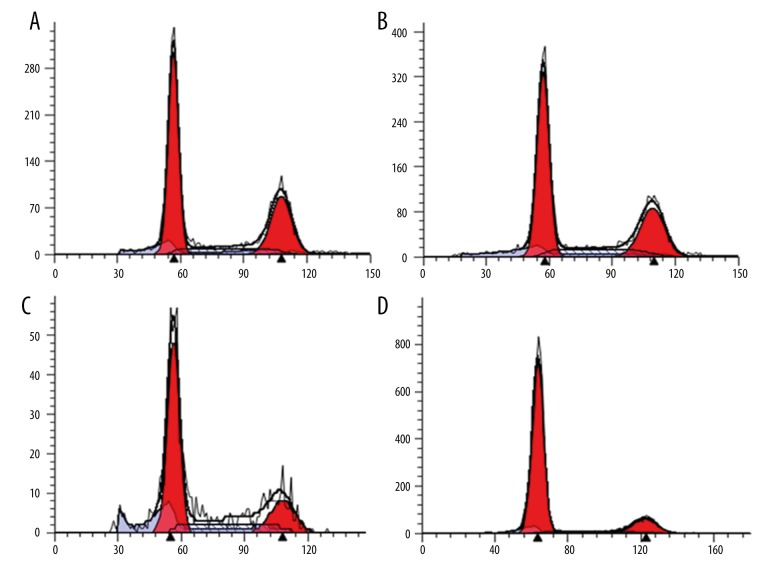
The G1 phase was significantly up-regulated in the 1.25-μM, 2.5-μM, and 5.0-μM treated groups compared with the 0-μM imatinib-treated group (P<0.05), but the S and G2 phases of the 3 imatinib-treated groups were significantly down-regulated (P<0.05), and there were significant differences among the 1.25-μM, 2.5-μM, and 5.0-μM treated groups in G1, S, and G2 phases (P<0.05) (Figure 3, Table 2).
Figure 3.
(A) Cell cycle of the 0-μM imatinib-treated group. (B) Cell cycle of the 1.25-μM imatinib-treated group. (C) Cell cycle of the 2.5-μM imatinib-treated group. (D) Cell cycle of the 5.0-μM imatinib-treated group.
Table 2.
Cell cycle in four groups (mean ±SD, %).
| Group | G1 | S | G2 |
|---|---|---|---|
| 0 μM | 55.67±0.68 | 12.60±2.3 | 31.73±1.72 |
| 1.25 μM | 64.44±0.79* | 8.89±0.79* | 26.67±0.78* |
| 2.5 μM | 75.12±0.79*,** | 6.22±1.66*,** | 18.66±1.09*,** |
| 5.0 μM | 82.51±0.76*,**,*** | 4.37±0.64*,**,*** | 13.12±0.90*,**,*** |
P<0.05, there were significantly difference compared with 0 μM;
P<0.05, there were significantly difference compared with 1.25 μM;
P<0.05, there were significantly difference compared with 2.5 μM.
Relative gene and protein expression
P21 gene and protein expression were not significantly different among the 5 groups. The gene and protein expressions of P27 were significantly up-regulated with increasing imatinib doses (P<0.05); however, the HGF gene and protein expression were significantly down-regulated with increasing imatinib doses (P<0.05) (Figures 4–6).
Figure 4.
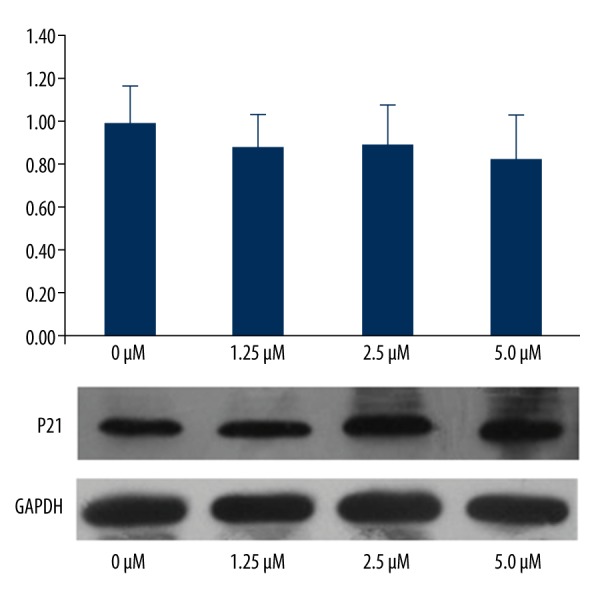
P21 gene and protein expression in the 4 groups.
Figure 5.
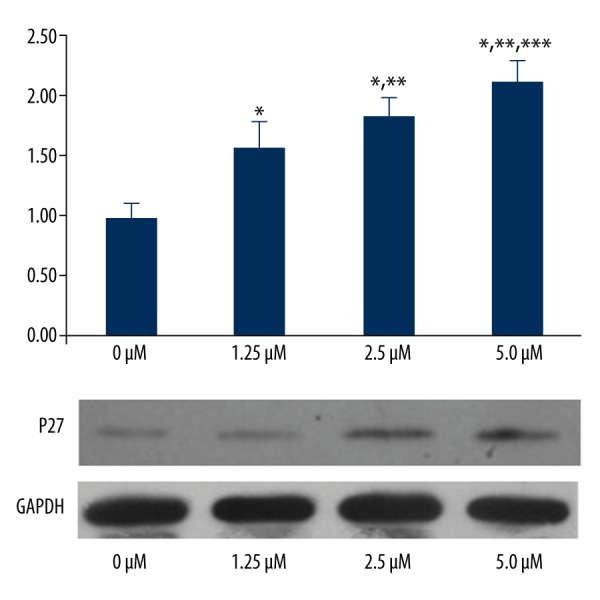
P27 gene and protein expression in the 4 groups. * P<0.05, significantly different compared with 0 μM. ** P<0.05, significantly different compared with 1.25 μM. *** P<0.05, significantly different compared with 2.5 μM.
Figure 6.
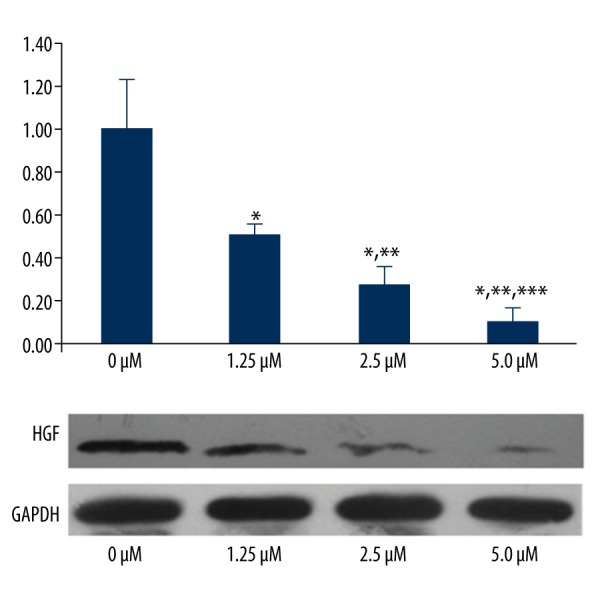
HGF gene and protein expression in the 4 groups. * P<0.05, significantly different compared with 0 μM. ** P<0.05, significantly different compared with 1.25 μM. *** P<0.05, significantly different compared with 2.5 μM.
Discussion
The incident of colorectal cancer ranks third after lung cancer and gastric cancer in China, and the incidence rate is still growing every year [13,14]. Most patients with colon cancer have no symptoms at the early stage, and more than 75% of the patients are diagnosed with advanced cancer [15]. Clinical surgery is the main treatment, but the prognosis is not good.
Some previous studies reported that imatinib inhibits proliferation of cancer cells [16–19]. However, there have been no reports about a relationship between imatinib and colon cancer. In the present study, we found that imatinib had an anti-proliferation effect on colon cancer cells. Recent studies found that tumors are caused by a kind of cell cycle disease. Cell proliferation is achieved through the operation of the cell cycle. The cell cycle is shown in G1, S, G2, and M stains. Some studies found that regulation of the cell cycle can inhibit cell proliferation [20–22]. In this study, we found that imatinib increased S stain and decreased G2 stain in a dose-dependent manner, showing that imatinib might have anti-proliferation effects through inhibiting the cell cycle. Furthermore, we wanted to study the mechanism of imatinib in molecular biology.
P21 and P27 are 2 critical regulators of cell survival and cell cycle by inhibiting both the DNA synthesis regulator proliferating cell nuclear antigen and activation of cyclin D1-CDK4/6 complexes [23–25]. We found that P21 was not significantly different among the 4 groups (with or without imatinib treatment), showing that imatinib had no effects on gene or protein expression of P21. However, our results also showed that imatinib decreased the percentage of cells undergoing apoptosis and promoted G1 phase arrest by stimulating gene and protein expression of P27. With increasing concentrations of imatinib, P27 gene and protein expression also increased. P21 and P 27 were 2 key points in the cyclin-dependent kinase inhibitors (CKIs) signaling pathway, and are associated with the cell cycle. We hypothesize that imatinib may have another pathway in addition to the CKIs signaling pathway.
One of the remaining questions is the underlying mechanisms through which the HGF/c-MET pathway is activated in SW480 cells. Some previous studies have reported activating mutations of the HGF/c-MET pathway in non-small cell lung cancer, hereditary and spontaneous renal carcinomas, hepatocellular carcinomas, gliomas, gastric cancer, squamous cell carcinoma of the head and neck, and breast cancer [26–31]. Results of the present study show that imatinib enhanced HGF gene and protein expression in a dose-dependent manner. This mechanism of imatinib to improve cell apoptosis rate was similar to that reported in previous studies.
Conclusions
Imatinib inhibits cell proliferation of colon cancer by stimulating P27 and HGF expression to enhance the cell apoptosis rate.
Footnotes
Source of support: Departmental sources
References
- 1.Siegel B, Naishadham D, Jemal A. Cancer statistics. Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Tap WD, Schwartz GK. That’s the “GIST” of it: Use of adjuvant imatinib after resection of a primary GI stromal tumor. J Clin Oncol. 2014;32:1543–46. doi: 10.1200/JCO.2013.53.5971. [DOI] [PubMed] [Google Scholar]
- 3.Wang G, Zhao R, Zhao X, et al. MicroRNA-181a enhances the chemotherapeutic sensitivity of chronic myeloid leukemia to imatinib. Oncol Lett. 2015;10:2835–41. doi: 10.3892/ol.2015.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgehama A, Chen W, Pang J, et al. Blockade of the interaction between Bcr-Abl and PTB1B by small molecule SBF-1 to overcomeimatinib-resistance of chronic myeloid leukemia cells. Cancer Lett. 2016;372(1):82–88. doi: 10.1016/j.canlet.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Wafa A, Almedani S, Liehr T, et al. Masked inv dup(22)(q11.23), tetrasomy 8 and trisomy 19 in a blast crisis-chronic myeloid leukemia after interrupted Imatinib-treatment. Mol Cytogenet. 2015;8:98. doi: 10.1186/s13039-015-0204-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu S, Alexiadis M, Fuller PJ. Expression, mutational analysis and in vitro response of imatinib mesylate and nilotinib target genes in ovarian granulosa cell tumors. Gynecol Oncol. 2008;108:182–90. doi: 10.1016/j.ygyno.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Mundhenke C, Weigel MT, Sturner KH, et al. Novel treatment of ovarian cancer cell lines with Imatinib mesylate combined with Paclitaxel and Carboplatin leads to receptor-mediated antiproliferative effects. Cancer Res Clin Oncol. 2008;134:1397–405. doi: 10.1007/s00432-008-0408-0. [DOI] [PubMed] [Google Scholar]
- 8.Furge KA, Zhang YW, Vande Wounde GF. Met receptor tyrosine kinase, enhanced signaling through adapter proteins. Oncogene. 2000;19(49):5582–89. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- 9.Chu JS, Ge FJ, Zhang B, et al. Expression and prognostic value of VEGFR-2,PDGFR-β, and c-Met in advanced hepatocellular carcinoma. J Exp Clin Cancer Res. 2013;32:16. doi: 10.1186/1756-9966-32-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XL, Chen XM, Fang JP, et al. Lentivirus-mediated RNA silencing of c-Met markedly suppresses peritoneal dissemination of gastric cancer in vitro and in vivo. Acta Phamacol Sin. 2012;33:513–22. doi: 10.1038/aps.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Li Q, Zhu L. Expression of the hepatocyte growth factor and c-Met in colon cancer: Correlation with clinicopathological features and overall survival. Tumori. 2012;98:105–12. doi: 10.1177/030089161209800115. [DOI] [PubMed] [Google Scholar]
- 12.Tretiakova M, Salama AK, Karrison T, et al. MET and phosphorylated MET as potential biomarker in lung cancer. J Environ Pathol Toxicol Oncol. 2011;30:341–54. doi: 10.1615/jenvironpatholtoxicoloncol.v30.i4.70. [DOI] [PubMed] [Google Scholar]
- 13.Isik A, Peker K, Firat D, et al. Importance of metastatic lymph node ratio in non-metastatic, lymph node-invaded colon cancer: A clinical trial. Med Sci Monit. 2014;20:1369–75. doi: 10.12659/MSM.890804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isik A, Okan I, Firat D, et al. A new prognostic strategy for gastric carcinoma: Albumin level and metastatic lymph node ratio. Minerva Chir. 2014;69:147–53. [PubMed] [Google Scholar]
- 15.Chalya PL, McHembe MD, Mabula JB, et al. Clinico-pathological patterns and challenges of management of colorectal cancer in a resource-limited setting: A Tanzanian experience. World J Surg Oncol. 2013;88:11–88. doi: 10.1186/1477-7819-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eton O, Billings L, Kim K, et al. Phase II trial of imatinib mesylate(STI-571) in metastatic melanoma (MM) J Clin Oncol. 2004;22:7528–29. [Google Scholar]
- 17.Chou CC, Yang JS, Lu HF, et al. Quercetin-mediated cell cycle arrest and apoptosis involving activation of a caspase cascade through the mitochondrial pathway in human breast cancer MCF-7 cells. Arch Pharm Res. 2010;33:1181–91. doi: 10.1007/s12272-010-0808-y. [DOI] [PubMed] [Google Scholar]
- 18.Jung YH, Heo J, Lee YJ, et al. Quercetin enhances TRAIL-induced apoptosis in prostate cancer cells via increased protein stability of death receptor 5. Life Sci. 2010;86:351–57. doi: 10.1016/j.lfs.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh DK, Lee EJ, Kim HC, et al. Induction of G (1)/S phase arrest and apoptosis by quercetin in human osteosarcoma cells. Arch Pharm Res. 2010;33:781–85. doi: 10.1007/s12272-010-0519-4. [DOI] [PubMed] [Google Scholar]
- 20.Deng QF, Su BO, Zhao YM, et al. Integrin β1-mediated acquired gefitinib resistance in non-small cell lung cancer cells occurs via the phosphoinositide 3-kinase-dependent pathway. Oncol Lett. 2016;11:535–42. doi: 10.3892/ol.2015.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Yin X, Wang W, et al. The effects of buthionine sulfoximine on the proliferation and apoptosis of biliary tract cancer cells induced by cisplatin and gemcitabine. Oncol Lett. 2016;11:474–80. doi: 10.3892/ol.2015.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang XY, Wang YG, Wang YF. Ginsenoside Rb1, Rg1 and three extracts of traditional Chinese medicine attenuate ultraviolet B-induced G1 growth arrest in HaCaT cells and dermal fibroblasts involve down-regulating the expression of p16, p21 and p53. Photodermatol Photolmmunol Photomed. 2011;27:203–12. doi: 10.1111/j.1600-0781.2011.00601.x. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y, Hannon GJ, Zhang H, et al. p21 is auniversal inhibitor of cyclin kinases. Nature. 1993;366:701–4. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 24.Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 25.Han JH, Lee SG, Jung SH, et al. Sesamin inhibits PDGF-mediated proliferation of vascular smooth muscle cells by upregulating p21 and p27. J Agric Food Chem. 2015;63:7317–25. doi: 10.1021/acs.jafc.5b03374. [DOI] [PubMed] [Google Scholar]
- 26.Stella GM, Benvenuti S, Gramaglia D, et al. MET mutations in cancers of unknown primary origin(CUPs) Hum Mutat. 2011;32:44–50. doi: 10.1002/humu.21374. [DOI] [PubMed] [Google Scholar]
- 27.Seiwert TY, Jagadeeswaran R, Faoro L, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009;69:3021–31. doi: 10.1158/0008-5472.CAN-08-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma PC, Tretiakova MS, MacKinnon AC, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008;47:1025–37. doi: 10.1002/gcc.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordano S, Maffe A, Williams TA, et al. Different point mutations in the met oncogene elicit distinct biological properties. FASEB J. 2000;14:399–406. doi: 10.1096/fasebj.14.2.399. [DOI] [PubMed] [Google Scholar]
- 30.Lee JH, Han SU, Cho H, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–53. doi: 10.1038/sj.onc.1203874. [DOI] [PubMed] [Google Scholar]
- 31.Park WS, Dong SM, Kim SY, et al. Somatic mutations in the kinase domain of the Met/hepatocyte growth factor receptor gene in childhood hepatocellular carcinomas. Cancer Res. 1999;59:307–10. [PubMed] [Google Scholar]