ABSTRACT
Human multipotent mesenchymal stromal cells (hMSCs) possess the ability to differentiate into osteoblasts, and they can be utilized as a source for bone regenerative therapy. Osteoinductive pretreatment, which induces the osteoblastic differentiation of hMSCs in vitro, has been widely used for bone tissue engineering prior to cell transplantation. However, the molecular basis of osteoblastic differentiation induced by osteoinductive medium (OIM) is still unknown. Therefore, we used a next‐generation sequencer to investigate the changes in gene expression during the osteoblastic differentiation of hMSCs. The hMSCs used in this study possessed both multipotency and self‐renewal ability. Whole‐transcriptome analysis revealed that the expression of zinc finger and BTB domain containing 16 (ZBTB16) was significantly increased during the osteoblastogenesis of hMSCs. ZBTB16 mRNA and protein expression was enhanced by culturing the hMSCs with OIM. Small interfering RNA (siRNA)‐mediated gene silencing of ZBTB16 decreased the activity of alkaline phosphatase (ALP); the expression of osteogenic genes, such as osteocalcin (OCN) and bone sialoprotein (BSP), and the mineralized nodule formation induced by OIM. siRNA‐mediated gene silencing of Osterix (Osx), which is known as an essential regulator of osteoblastic differentiation, markedly downregulated the expression of ZBTB16. In addition, chromatin immunoprecipitation (ChIP) assays showed that Osx associated with the ZBTB16 promoter region containing the GC‐rich canonical Sp1 sequence, which is the specific Osx binding site. These findings suggest that ZBTB16 acts as a downstream transcriptional regulator of Osx and can be useful as a late marker of osteoblastic differentiation. J. Cell. Biochem. 117: 2423–2434, 2016. © 2016 The Authors. Journal of Cellular Biochemistry published by Wiley Periodicals, Inc.
Keywords: BIOINFORMATICS, CELL DIFFERENTIATION, PERIODONTAL LIGAMENT (PDL), STEM CELL(S), OSTEOGENESIS, MOLECULAR BIOLOGY
A complex network of several transcription factors and signaling proteins regulates osteoblastic differentiation [Zhang, 2010]. Both runt‐related transcription factor two (Runx2) and Osterix (Osx) are important transcription factors for osteoblastic differentiation and bone formation that control the expression of bone‐related genes, such as osteocalcin (OCN), bone sialoprotein (BSP), and osteopontin (OPN) [Xiao et al., 2005; Kim et al., 2006; Matsubara et al., 2008; Niger et al., 2011]. Runx2 null mice are unable to form endochondral and intramembranous bone [Ducy et al., 1997; Komori et al., 1997; Otto et al., 1997]. Osx is specifically expressed in the osteoblasts and osteocytes of all developing bones. Similar to the phenotype of the Runx2 null mice, Osx null mice are also unable to form bone. Although Runx2 is expressed in Osx null mice, Osx is not expressed in Runx2 null mice; therefore, Runx2 is thought to be an upstream regulator of Osx [Nakashima et al., 2002]. Other transcriptional factors, including Dlx5, Twist1, and ATF4, are also involved in osteoblastic differentiation [Acampora et al., 1999; Bialek et al., 2004; Yang et al., 2004]. However, because many studies have indicated that several transcriptional factors regulate osteoblastic differentiation and because in vivo and in vitro data are conflicting in many studies, the mechanisms of osteoblastic differentiation have not been fully elucidated.
Multipotent mesenchymal stromal cells (MSCs) have been isolated from several tissues [Pittenger et al., 1999; Zuk et al., 2002; Lee et al., 2004]. MSCs are capable of differentiating into cells of the mesodermal lineage, such as osteoblasts, chondrocytes, and adipocytes. In addition to multipotency, MSCs are defined as having self‐renewal ability and specific surface marker expression [Dominici et al., 2006]. Furthermore, MSCs stimulate angiogenesis by secreting VEGF and have immunomodulatory properties that are mediated by both cell‐cell contact and secreted bioactive molecules [Caplan and Correa, 2011]. For these reasons, MSCs have been used in several clinical trials and are considered a powerful tool for regenerative therapy and tissue engineering [Trubiani et al., 2008; Iwata et al., 2009]. In fact, several cellular bone matrices, which contain living cells, are commercially available for clinical use [Skovrlj et al., 2014].
Previous studies have indicated that osteoinductive medium (OIM) containing ascorbic acid (AA), β‐glycerophosphate (β‐GP), and dexamethasone (Dex) induces the osteoblastic differentiation of MSCs in vitro [Gay et al., 2007]. However, the mechanisms by which OIM enhances osteoblastic differentiation and regulates osteogenic transcriptional factors are not well understood. In our recent study, high‐throughput mRNA sequencing (RNA‐seq) of hMSCs revealed the involvement of the WNT signaling pathway during OIM‐mediated osteoblastic differentiation [Yamada et al., 2013]. We also observed that several transcriptional factors were drastically increased in response to OIM‐mediated osteoblastic differentiation. After we reviewed RNA‐seq results [Yamada et al., 2013], we focused our attention on Zinc Finger and BTB Domain‐Containing 16 (ZBTB16), also known as promyelotic leukemia zinc finger (PLZF), which was especially upregulated when hMSCs were cultured in OIM.
Here, we report that ZBTB16 functions as a key regulator of osteoblast and mature osteoblast differentiation and matrix mineralization in hMSCs. In addition, the expression of ZBTB16 may be used as a late marker of osteoblastic differentiation. Furthermore, our results indicated that the expression of ZBTB16 during osteoblastic differentiation depends on Osterix but not on RUNX2.
MATERIALS AND METHODS
The experimental protocol was approved by the ethics committee of Tokyo Women's Medical University. All of the subjects signed informed consent forms approving the donation of their teeth that were extracted for impaction reasons.
CELL CULTURE
Human periodontal ligament (PDL)‐derived MSCs were obtained and prepared as previously described [Iwata et al., 2010]; these cells showed the hMSC phenotype defined by Dominici et al. (2006). Briefly, PDL tissues were separated from the mid‐third of the extracted human teeth and dispersed in alpha‐minimum essential medium with GlutaMAX (α‐MEM) (Gibco, Life Technologies, Carlsbad) containing 1% sulbactam/ampicillin (UNASYN‐S KIT; Pfizer, Tokyo, Japan), 1,200 PU/mL dispase (Sanko Junyaku, Tokyo, Japan), and 0.8 PZ‐U/mL collagenase type 1 (Serva Electrophoresis, Heidelberg, Germany). A single‐cell suspension was spread in a T25 Primaria culture flask (Falcon; Passage 0), and the cells were cultured in complete medium (α‐MEM supplemented with 100 U/mL penicillin and 100 mg/mL streptomycin containing 10% fetal bovine serum [FBS; Moregate Biotech, Queensland, Australia]) at 37°C in a humidified atmosphere of 95% air and 5% CO2 . hMSCs between the third and sixth passages were used for experimentation. Human bone marrow‐derived MSCs (hBMMSCs) from three different lots were purchased from Lonza (Lonza, Walkersville, MD). These cells were cultured in complete medium as described above.
FLOW CYTOMETRIC ASSAY
A total of 5 × 105 cells was suspended in 100 µL of Dulbecco's phosphate‐buffered saline (PBS; Gibco) containing 10 µg/mL of each specific antibody. To detect the expression of surface markers, fluorescein isothiocyanate (FITC)‐ or phycoerythrin (PE)‐conjugated antibodies against CD11b, CD14, CD29, CD34, CD45, CD73, CD90, and CD105 (BD Biosciences, San Jose, CA) were used. FITC‐conjugated non‐specific mouse IgG (BD Biosciences) and PE‐conjugated non‐specific mouse IgG (R and D Systems) were employed as isotype controls. After the cells were incubated for 30 min at 4°C, they were washed with PBS and suspended in 300 µL of PBS for further analysis. Cell fluorescence was determined using a flow cytometer (Epics‐XL; Beckman Coulter, Fullerton, CA).
DIFFERENTIATION ASSAY
To study osteogenesis, hMSCs were seeded onto 100 mm dishes (Primaria; Corning, NY) at a density of 50 cells/dish and cultured in complete medium for 14 d, after which the medium was changed to OIM. This medium consisted of complete medium supplemented with 82 µg/mL L‐ascorbic acid phosphate magnesium salt (AA; Wako Pure Chemical, Tokyo, Japan), 10 mmol/L β‐glycerophosphate (βGP; Sigma–Aldrich, St. Louis, MO), and 10 nmol/L dexamethasone (DEX; Fuji Pharma, Tokyo, Japan). After the cells were incubated for an additional 21 d, they were stained with 1% alizarin red S solution (Wako Pure Chemical). To quantify the intensity of alizarin red S staining, the bound staining was eluted with 10% (wt/vol) cetylpyridinium chloride (Sigma–Aldrich), and the alizarin red S dye in the samples was quantified by measuring the absorbance at a wavelength of 570 nm.
To study adipogenesis, hMSCs were seeded onto 100 mm dishes at a density of 50 cells/dish and cultured in complete medium for 14 d, after which the medium was changed to adipogenic medium. This medium consisted of complete medium supplemented with 100 nmol/L DEX, 0.5 mmol/L isobutyl‐1‐methyl xanthine (Sigma–Aldrich), and 50 mmol/L indomethacin (Wako Pure Chemical). After the adipogenic cultures were incubated for an additional 21 d, they were fixed in 4% paraformaldehyde (Wako Pure Chemical) and stained with fresh Oil Red O solution.
To study chondrogenesis, hMSCs were placed in a 15 mL polypropylene tube (BD Biosciences) and centrifuged at 450g for 10 min. The formed pellet was cultured in chondrogenic medium, which consisted of high‐glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 500 ng/mL recombinant human bone morphogenetic protein 2 (BMP‐2) (R and D Systems, Minneapolis, MN), 10 ng/mL transforming growth factor β3 (R and D Systems), 10 nM DEX, 50 mg/ml AA, 40 mg/mL proline, 100 mg/mL pyruvate, and 50 mg/ml ITS1Premix (BD Biosciences), and the medium was changed every 3 d. After the pellets were incubated for 21 d, they were embedded in paraffin and cut into 5 µm tissue sections. These sections were stained with toluidine blue.
PREPARATION OF THE TOTAL RNA LIBRARY AND SEQUENCING
For these studies, hMSCs were isolated from three different subjects and cultured using three different methods: (1) hMSCs were cultured in complete medium for 4 d (OIM [−] 4 d), (2) hMSCs were cultured with OIM for 4 d (OIM [+] 4 d), and (3) hMSCs were cultured with OIM for 14 d (OIM [+] 14 d). Total RNA was purified and concentrated using a RiboMinus Eukaryote Kit (Invitrogen, Life Technologies, Carlsbad, CA) and RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA). Ribosomal RNA‐depleted total RNA was utilized to construct a library using a SOLiD Total RNA‐seq Kit (Life Technologies), and then the library was sequenced using the SOLiD System (Life Technologies). All of the procedures were performed according to the manufacturers’ instructions.
WHOLE‐TRANSCRIPTOME ANALYSIS
The SOLiD system generated 94.4–118.2 million single‐ended 50 bp reads from nine samples (MSCs were isolated from three different subjects, and RNA samples were isolated from three groups, OIM [−] 4 d, OIM [+] 4 d, and OIM [+] 14 d). We used the Tophat2 (v. 2.0.14)‐Cufflinks (v.2.2.1) pipeline [Trapnell et al., 2012] to gather the uniquely mapped reads to the human reference genome (hg19) and to quantify RefSeq gene expression as the unit of fragments per kilobase of exon per million mapped reads (FPKM). After combining nine samples using Cuffmerge, we performed Cuffdiff processing in the Cufflinks package to detect differentially expressed genes (DEGs) between the tissues via two‐group t‐tests coupled to a Benjamini–Hochberg correction (q‐value) as previously described [Park et al., 2016]. We defined DEGs when the fold change of the FPKMs was >2.0 and the q‐value was <0.05. The human reference genome and the RefSeq annotation were downloaded from http://genome.ucsc.edu/. The RNA‐seq datasets generated in this study have been deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive (DRA) under accession number DRA003917.
ALKALINE PHOSPHATASE (ALP) ACTIVITY ASSAY
hMSCs were seeded onto a 96‐well plate at a density of 1 × 104 cells/well and cultured in complete medium for 48 h. The medium was changed to complete medium or OIM. After the hMSCs were incubated for 5 d, the medium in the culture dish was removed, and the hMSCs were washed twice with sterile saline. ALP activity in the hMSCs was evaluated using a LabAssay ALP Kit (Wako Pure Chemical). The absorbance was measured at a wavelength of 405 nm using a plate reader (Model 450; Bio‐Rad, Hercules, CA).
MEASUREMENT OF CELL VIABILITY
Cell viability was measured by MTS assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay; Promega, Madison, WI). hMSCs were seeded on a 96‐well plate at a density of 5 × 103 cells/well and cultured for 24 h in antibiotic‐free α‐MEM containing 10% FBS. After small interfering RNA (siRNA) transfection was performed (the details of these methods are described below; 2.10), the cells were further cultured with or without OIM for 5 d. The absorbance was measured at a wavelength of 490 nm after 1 h according to the manufacturer's instructions.
RNA ISOLATION AND REAL‐TIME RT‐PCR
hMSCs were seeded onto 6‐well plates at a density of 4 × 104 cells/well and cultured for 2 d in complete medium. The medium was changed to complete medium or OIM after 4, 7, and 14 d. Total RNA was isolated using the QIAshredder and an RNeasy Plus Mini Kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized from 500 ng of total RNA using a Superscript VILO cDNA Synthesis Kit (Invitrogen). mRNA expression levels were quantitatively analyzed by real‐time RT‐PCR using the StepOnePlus Real‐Time PCR system (Applied Biosystems, Foster City, CA). The primers used were as follows: ZBTB16 (Hs00957433_m1), Osx (Hs00541729_m1), RUNX2 (Hs00231692_m1), OCN (Hs01587814_g1), BSP (Hs00173720_m1), and β‐actin (43263251E; all of the probes were obtained from Applied Biosystems). The mRNA expression values were normalized to β‐actin, and the fold changes were calculated using the values obtained by the ΔCT method at each time point [Livak and Schmittgen, 2001].
WESTERN BLOT ANALYSIS
After hMSCs were cultured with or without OIM for 4, 7, and 14 d, then the cells were washed twice with cold PBS (Gibco) and lysed for 15 min on ice with Cell Lysis Buffer (Cell Signaling Technology, Danvers, MA), respectively. The lysed cells were collected using a cell scraper, sonicated using a Bioruptor (Cosmo Bio, Tokyo, Japan), and centrifuged at 14,000g for 15 min at 4°C. The protein concentrations were determined using a Qubit Protein Assay Kit and a Qubit fluorometer (Invitrogen). The proteins were prepared for electrophoresis by adding NuPAGE LDS Sample Buffer (Invitrogen), heating the samples to 70°C for 10 min, and then loading 30 µg of each sample onto NuPAGE Novex 4–12% Bis–Tris Mini Gels (Invitrogen), which were run at 200 V for 35 min in NuPAGE MES SDS Running Buffer (Invitrogen). The gels were transferred to nitrocellulose membranes using the iBlot Dry Blotting System (Invitrogen). The membranes were blocked for 1 h in Tris‐buffered saline/Tween (TBS‐T; 20 mmol/L Tris–HCl, 150 mmol/L NaCl [pH 7.5], and 0.05% Tween 20) containing 3% ECL Prime Blocking Reagent (GE Healthcare, Buckinghamshire, UK). The blots were subsequently incubated with anti‐ZBTB16 (1:500, Sigma–Aldrich) or anti‐β‐actin (1:1000, Cell Signaling) in blocking buffer overnight at 4°C, washed three times for 20 min with TBS‐T, and incubated with anti‐rabbit IgG (the secondary antibody; 1:15,000) for 30 min at room temperature. After the membranes were incubated with the secondary antibody, they were washed three times for 20 min with TBS‐T. The immunoreactive bands were detected using ECL Prime Western Blotting Detection Reagent (GE Healthcare) followed by image analysis (LAS‐4000 Mini; Fujifilm, Tokyo, Japan).
SMALL INTERFERING RNA (siRNA) TRANSFECTION
siRNAs targeting ZBTB16 (Silencer Pre‐designed siRNA: s15200), Osx (s42459 and s42458), and RUNX2 (s2455) as well as the non‐targeting control siRNA (Silencer Select Negative Control #1 siRNA: 4390843) were purchased from Applied Biosystems. For the ALP activity assay, MTS assay and differentiation assay, hMSCs were seeded on 96‐well plates at a density of 5 × 103 cells/well and cultured for 24 h in antibiotic‐free α‐MEM containing 10% FBS. When the cells were 30–50% confluent, siRNA transfection was performed. siRNA for ZBTB16 (30 µmol/L) was mixed with 0.2 µL of Lipofectamine RNAiMAX reagent (Invitrogen) in 20 µL of Opti‐MEM Reduced‐Serum Medium (Invitrogen). After the mixture was incubated for 20 min at room temperature to form a complex, it was added to the cell culture. After 6 h, the medium was changed, and the cells were treated with or without OIM for 3, 5, and 21 d.
For real‐time RT‐PCR and Western blot analyses, hMSCs were seeded on six‐well plates at a density of 2 × 104 cells/well and cultured for 24 h in antibiotic‐free α‐MEM containing 10% FBS. The cells were transiently transfected with siZBTB16 (30 µmol/L), siOsx (30 µmol/L), and siRUNX2 (30 µmol/L) as described above. After 6 h, the medium was changed, and the cells were treated with or without OIM for 3 and 7 d. Then, total RNA and protein from hMSCs were isolated as described above.
CHROMATIN IMMUNOPRECIPITATION (ChIP) ASSAY
hMSCs were seeded onto 100 mm culture dishes at a density of 2 × 105 cells/dish and cultured for 2 d in complete medium. Then, the medium was changed to OIM supplemented with recombinant human BMP‐6 (rh‐BMP‐6: 200 ng/mL: R and D Systems) for 7 d.
ChIP was performed using the MAGnify Chromatin Immunoprecipitation System (Life Technologies) according to the manufacturer's protocol and using ChIP grade antibodies against Osx (Abcam: ab22552), ZBTB16 (Santa Cruz, PLZF D‐9: sc‐28319), and normal rabbit IgG (Invitrogen).
The purified DNA samples were analyzed by PCR using primers of the proximal ZBTB16 promoter region containing the Sp1 sequence. The primer pairs were as follows: ZBTB16 promoter P1, 5′‐CGCCAGCACTAAAGATGGA‐3′ (forward) and 5′‐CCTCTCTGCTGCGAGGTTAG‐3′ (reverse); ZBTB16 promoter P2, 5′‐ACTGTTTCCACCCAGATTGC‐3′ (forward) and 5′‐ACACCCGTTTTTAGCTGTCG‐3′ (reverse); negative control P3, 5′‐AAGAGTTGCAACATTCCATTCA‐3′ (forward) and 5′‐CTCAGCTTGGTCACCAGGTA‐3′ (reverse); Osx promoter P1, 5′‐TGTCAGTGCGTTCCAGTCTC‐3′ (forward) and 5′‐GCTCCACTCCTGTTCCACTC‐3′; (reverse); and negative control P2, 5′‐AATGGTTCCTGTGGTTCAGC‐3′ (forward) and 5′‐TAAACCCCCTAGGCATCTGG‐3′ (reverse).
STATISTICAL AND BIOINFORMATICS ANALYSIS
Clustering analysis was conducted using the function hclust of the R programming language (http://www.r-project.org/). Euclidean distances among the FPKMs were adjusted by quantile normalization, and mean centering was clustered by the hierarchical average linkage method. Gene Ontology (GO) biological process terms were analyzed using the PANTHER classification system (http://www.pantherdb.org/) with the annotation dataset GO biological process complete. We addressed over‐ and under‐represented GO terms in each expression cluster with Bonferroni's corrected P value (<0.05). In the statistical analyses, all of the values are reported as the mean ± standard deviation (SD). The differences between the groups were assessed by Student's t‐test or t‐test followed by Bonferroni's multiple comparison correction procedure. Values of P < 0.05 and P < 0.01 were considered significant.
RESULTS
hMSCs DERIVED FROM PDL POSSESS MSC‐LIKE PROPERTIES
First, we verified that the cells used in this study corresponded to the minimal criteria of MSCs defined by Dominici et al. (2006). Flow cytometric analysis confirmed that these hMSCs expressed characteristic antigens of MSC markers, including CD29+, CD44+, CD73+, CD90+, CD105+, CD11b−, CD14−, CD34,− and CD45 (Fig. 1A). These cells were also able to form colonies (Fig. 1B). To confirm the multipotency of these cells, we performed differentiation assays using differentiation medium for osteogenesis, adipogenesis, and chondrogenesis. When the hMSCs were cultured with OIM, calcium deposits were detected by alizarin red S‐positive staining (Fig. 1C). When hMSCs were cultured with adipogenesis differentiation medium, these cells showed Oil Red O‐positive lipid droplets (Fig. 1D). When hMSCs were cultured with chondrogenesis differentiation medium, matrix proteoglycan was detected by toluidine blue staining (Fig. 1E). These results indicated that these hMSCs derived from PDL possessed MSC‐like properties, including specific surface antigen expression, colony‐forming and plastic adherent abilities, and multipotential differentiation capabilities.
Figure 1.
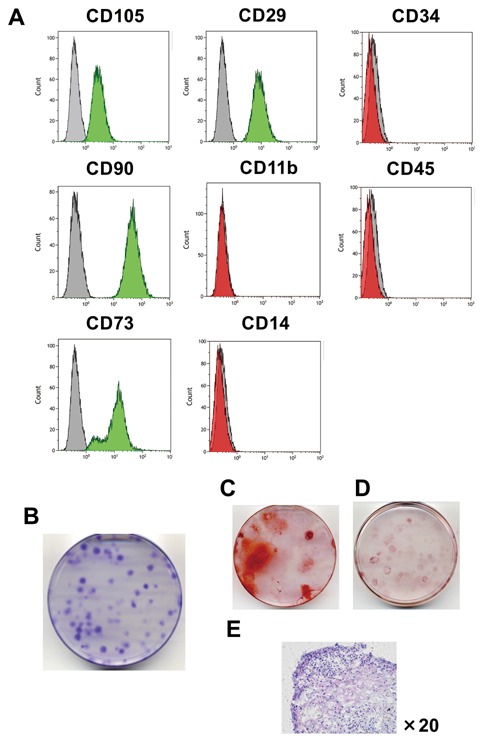
Characterization of hMSCs derived from PDL in vitro. (A) hMSCs were stained with specific antibodies and were analyzed for protein expression by flow cytometry. Green‐ (>95%) or red‐ (<5%) filled areas show the fluorescence intensity of hMSCs with the indicated antigens. Shaded areas show the fluorescence intensity of hMSCs with isotype controls (negative control). (B) Colony‐forming assay and (C)–(E) differentiation assay of hMSCs. (C) hMSCs positively stained with alizarin red S. This figure shows results for cells cultured with OIM for 21 d. (D) hMSCs were positively stained with Oil Red O. This figure shows the results for cells cultured with adipogenic medium for 21 d. (E) hMSCs were positively stained with toluidine blue. This figure shows cells cultured with chondrogenic medium for 21 d. These figures show the representative data from three different samples; a similar tendency was observed in other hMSCs.
ZBTB16 WAS SIGNIFICANTLY UPREGULATED BY OIM
To characterize the transcriptome landscape, we first analyzed the RNA‐seq data of hMSCs from three donors cultured with or without OIM (AA + β‐GP + Dex) for 4 or 14 d and identified 11,695 genes (FPKM > 1.0). Using this expression profile, we detected 15 gene clusters in the hMSCs that exhibited distinct expression patterns upon osteoblastic differentiation (Fig. 2A). Genes in each cluster were significantly enriched by specific GO terms. For instance, the term metabolic processes detected from the hMSCs cultured without OIM for 4 d (Cluster 7 and 11) was comprised of genes different from that were highly expressed in hMSCs cultured with OIM for 4 and 14 d (Cluster 9, 13, 14, and 15). In addition, the genes transiently expressed in hMSCs cultured with OIM for 4 d (Cluster 2) were likely to be involved in activation of the cell cycle process. These results suggested that during hMSC differentiation, the replacement and onset of biological processes are governed by specific gene sets that are wired into a regulatory network that remains unknown.
Figure 2.
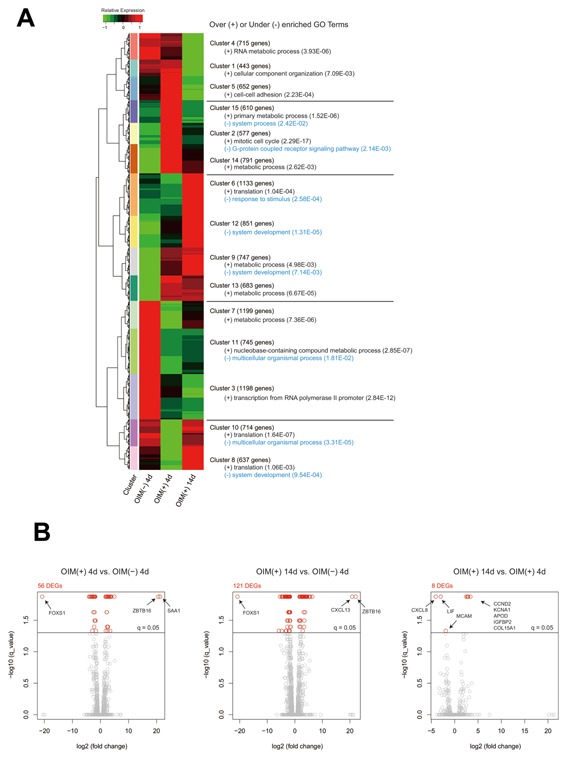
Gene expression patterns, Gene Ontology (GO) term enrichment, and differentially expressed genes (DEGs) among the hMSCs. (A) The whole‐transcriptome analysis (FPKM > 1.0) detected 11,695 genes that were categorized into 15 patterns by a hierarchical clustering approach. The genes in each pattern were over‐ and under‐represented by specific GO biological process terms (P < 0.05). (B) The statistical test detected 132 DEGs in total among the three tissues using a threshold (q‐value < 0.05) and a more than two‐fold change in FPKMs. Left: FPKM of OIM (+) 4 d over FPKM of OIM (−) 4 d. Middle: FPKM of OIM (+) 14 d over FPKM of OIM (−) 4 d. Right: FPKM of OIM (+) 14 d over FPKM of OIM (+) 4 d. The gray and red circles represent non‐DEGs and DEGs, respectively. OIM (−) 4 d, hMSCs cultured without OIM for 4 d; OIM (+) 4 d, hMSCs cultured with OIM for 4 d; OIM (+) 14 d, hMSCs cultured with OIM for 14 d.
To detect potential key regulators during differentiation, we next investigated the DEGs among the cells (fold change of FPKMs > 2.0, q‐value < 0.05) and identified 132 genes (Fig. 2B). In particular, ZBTB16, also known as PLZF, was remarkably upregulated along with signaling molecules (e.g., SAA1 and CXCL13) in the cells cultured with OIM for 4 and 14 d. Interestingly, the forkhead family transcription factor FOXS1 was significantly downregulated in these cells and may reflect the replacement of biological processes that we observed in the GO term analysis, that is, switching the transcriptional activity of FOXS1 to that of ZBTB16, which should be further studied elsewhere.
ZBTB16 WAS ONLY DETECTED WHEN hMSCs WERE CULTURED WITH OIM
We next determined the effect of OIM stimulation on ZBTB16 expression in hMSCs. When hMSCs were cultured with normal medium, ZBTB16 expression was not detected. In contrast, ZBTB16 expression was significantly enhanced when hMSCs were cultured in OIM for 14 d in all of the samples (Fig. 3A). To investigate whether ZBTB16 expression induced by OIM is specific to hMSCs derived from PDL, we performed the same assay using hBMMSCs. ZBTB16 expression in hBMMSCs was also significantly enhanced when the cells were cultured with OIM for 14 d (Fig. 3B). As shown in Figure 3C, the protein level of ZBTB16 was also enhanced and maximized on d 14 when the hMSCs were cultured with OIM.
Figure 3.

ZBTB16 expression depends on OIM during osteoblastic differentiation. (A) Three hMSCs (hMSCs #1, #2, and #3) were cultured with or without OIM (containing AA, β‐GP, and Dex) for 4, 7, and 14 d. The mRNA levels of ZBTB16 were evaluated by TaqMan assays. N.D.: not detectable. The bars represent the mean ± S.D. of triplicate wells. The results are representative of three independent experiments. β‐Actin was used as an internal control. (B) hBMMSCs were cultured with or without OIM for 4, 7, and 14 d. (C) Western blot analysis of ZBTB16 using cell lysates from hMSCs cultured with or without OIM for 4, 7, and 14 d. β‐Actin was used as an internal control. Representative data from three different samples are shown in (B) and (C); a similar tendency was observed in other samples. The experiments shown in Figure 3A and B were performed in triplicate.
ZBTB16 KNOCKDOWN INHIBITED THE OIM‐INDUCED OSTEOBLASTIC DIFFERENTIATION OF hMSCs
OIM‐induced ZBTB16 expression decreased by approximately 90% after siZBTB16 transfection, compared with the siControl (Fig. 4A). The transfection of siZBTB16 suppressed ALP activity compared with the non‐siRNA or siControl when hMSCs were cultured with OIM (Fig. 4B); a similar phenomenon was also observed in hBMMSCs (Fig. 4C). MTS assay showed that siZBTB16 transfection did not affect the viability of the hMSCs (Fig. 4D). As shown in Figure 3E, mineralized nodule formation, which was visualized by alizarin red S staining, increased when hMSCs were cultured with OIM. However, the stained areas were decreased upon siZBTB16 transfection compared with non‐siRNA and siControl transfection when hMSCs were cultured with OIM (Fig. 4E and F). Furthermore, the expression of the osteoblastic markers OCN and BSP was significantly decreased by siZBTB16 (Fig. 4G).
Figure 4.
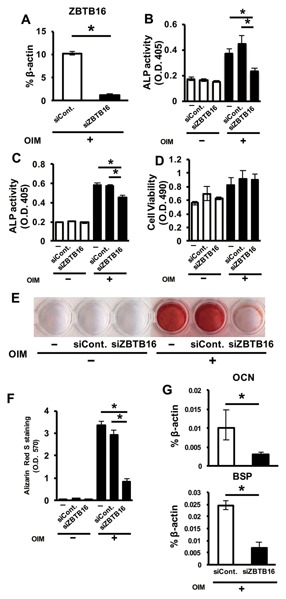
The inhibitory effect of siZBTB16 on ALP activity, mineralized nodule formation, and osteogenic gene expression in hMSCs. (A) The efficacy of the siRNA inhibiting ZBTB16 mRNA expression. (B) The effect of siRNA targeting ZBTB16 in hMSCs on the osteoblastic differentiation of hMSCs. The ALP activity of hMSCs after transfection and culture with or without OIM for 5 d. (C) The effect of siRNA targeting ZBTB16 on the osteoblastic differentiation of hBMMSCs. The ALP activity of hBMMSCs after they were transfected and cultured with or without OIM for 5 d. (D) The cell viability of hMSCs was measured by MTS assay after they were transfected and cultured with or without OIM for 5 d. *P < 0.05, compared with non‐siRNA and siControl. (E) Alizarin red S staining of hMSCs after the cells were transfected with siZBTB16 and cultured with or without OIM for 21 d. (F) Bound stain was eluted with a solution of 10% cetylpyridinium chloride and quantified by measuring the absorbance at a wavelength of 570 nm using a plate reader. (G) Effect of a siRNA targeting ZBTB16 in hMSCs on OCN and BSP expression after hMSCs were transfected and cultured with OIM for 5 d. *P < 0.05, compared with siControl. (−), non‐siRNA; siCont., siControl. These figures show representative data from three different samples; a similar tendency was observed in other hMSCs. The experiments shown in Figure 4A, B–D, F, and G were performed in triplicate.
OSTERIX IS NECESSARY FOR OIM‐INDUCED ZBTB16 EXPRESSION IN hMSCs
To investigate the relation between ZBTB16 and other osteogenic transcription factors, hMSCs were transfected with siRNAs targeting RUNX2 and Osx. After transfection, the knockdown efficiencies of these siRNAs were verified by real‐time RT‐PCR. Osx and RUNX2 expression levels were decreased by approximately 70–80% following transfection with siOsx and siRUNX2 (Fig. 5A and B), respectively, compared with the siControl. Two different siRNAs targeting Osterix (siOsx1 and 2) markedly suppressed ZBTB16 expression compared with the siControl (Fig. 5A). However, siRUNX2 did not suppress ZBTB16 expression (Fig. 5B). Similar to the results shown by gene expression analysis, ZBTB16 protein levels were decreased by both siOsx and siZBTB16 (Fig. 5C).
Figure 5.
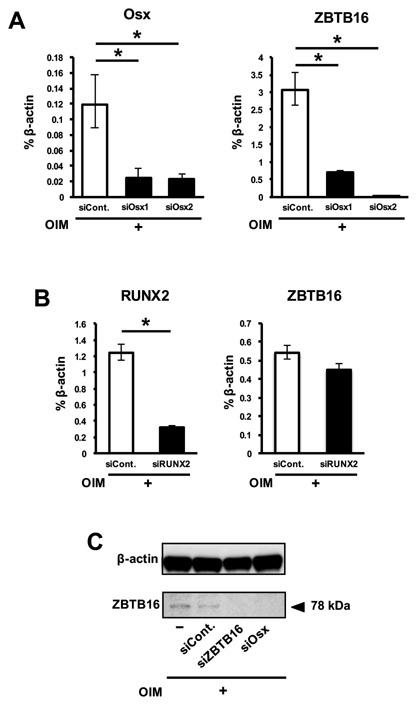
The mRNA and protein levels of ZBTB16 were decreased by siOsx. (A) hMSCs were transfected with non (siCont.)‐ or Osx (siOsx1 and siOsx2)‐specific siRNA and cultured with OIM. RNA was extracted, and real‐time RT‐PCR was performed for Osx and ZBTB16. (B) hMSCs were transfected with non (siCont.)‐ or RUNX2 (siRUNX2)‐specific siRNA and cultured with OIM. RNA was extracted, and real‐time RT‐PCR was performed for RUNX2 and ZBTB16. (C) Western blot analysis of ZBTB16 using cell lysates from hMSCs after they were transfected and cultured with OIM. *P < 0.05, compared with siControl. These figures show representative data from three different samples; a similar tendency was observed in other hMSCs. The experiments shown in Figure 5A and B were performed in triplicate.
OSTERIX BINDS TO THE SP1‐BINDING SITE OF THE ZBTB16 PROMOTER REGION DURING OSTEOBLASTIC DIFFERENTIATION
To confirm that Osx may be an upstream regulator of ZBTB16 during osteoblastic differentiation, we performed a ChIP assay in hMSCs. First, we selectively designed the primer of the proximal ZBTB16 promoter region containing two Sp1 sequences within 1 kbp from the transcription start site (Fig. 6A). As shown in Figure 6B, Osx associated with the ZBTB16 promoter region containing the two Sp1 sites. However, Osx did not associate with the ZBTB16 distal 3–4 kbp promoter region without the SP1 site sequence (negative control region). Furthermore, because previous studies have shown that ZBTB16 binds to the Osx promoter region [Felthaus et al., 2014], we also examined this point by performing a ChIP assay. Interestingly, ZBTB16 was also associated with the binding site in the Osx promoter region (Supplemental Fig. S1A and B). These data suggested that ZBTB16 and Osx might mutually bind to each other's promoters.
Figure 6.
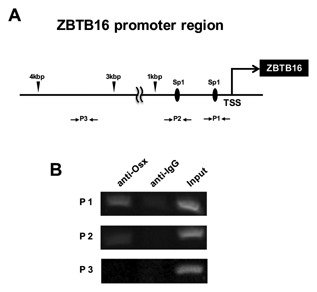
Osx binds to the Sp1 sequence of the ZBTB16 promoter region. (A) The diagram shows the location of the primers used to amplify the Sp1 or the non‐Sp1 site of the ZBTB16 promoter region in the ChIP assay. TSS, transcription start site of ZBTB16; P1, primer 1 set; P2, primer 2 set; P3 (negative control), primer 3 set. (B) hMSCs were cultured with OIM and rhBMP‐6 (200 ng/ml) for 7 d. ChIP assay was performed with antibodies against Osx and IgG (negative control). Ten percent of the input was loaded as a control. This figure shows representative data from three different samples; a similar tendency was observed in other hMSCs.
DISCUSSION
Although several studies have suggested that, culturing MSCs with osteoinductive supplements enhances bone regeneration in vivo [Yoon et al., 2007; Di Bella et al., 2008; Iwata et al., 2009], the mechanisms of OIM‐induced osteoblastic differentiation have not been clarified. Because several transcriptional factors, including Runx2 and Osx, are known to regulate differentiation into the osteoblastic lineage [Komori, 2006], we focused on OIM‐induced transcriptional factors in hMSCs. To investigate possible mechanisms, the change in gene expression was analyzed using a next‐generation sequencer for whole‐transcriptome analysis. In the analysis, ZBTB16 expression was significantly increased in the OIM treatment group (d 4 or 14) compared to those in the non‐treatment group (d 4). Therefore, we investigated the role of ZBTB16 during osteoblastic differentiation in hMSCs.
ZBTB16 is a zinc finger transcriptional factor that contains a BTB/POZ domain for protein‐protein interactions at the N‐terminus and nine Kruppel‐type zinc finger domains for DNA binding at the C‐terminus [Zhang et al., 1999]. Previous reports identified ZBTB16 as a spermatogonia‐specific transcriptional factor in the testis that is required to regulate self‐renewal and maintenance of the stem cell pool [Costoya et al., 2004] or as a transcriptional signature of natural killer T cells that directs their innate‐like effector differentiation during thymus development [Savage et al., 2008]. Some studies have shown that ZBTB16 is involved in skeletal development and osteoblastic differentiation. ZBTB16‐null mice display patterning defects that affect all skeletal structures of the limb [Barna et al., 2000], and ZBTB16 cooperates with Gli3 to promote the limb fate [Barna et al., 2005]. Furthermore, ZBTB16 knockdown results in the downregulation of osteoblastic genes such as ALP, COL1A1, OCN, and RUNX2 in hMSCs [Ikeda et al., 2005]. Therefore, researchers have suggested that ZBTB16 is involved in early osteoblastic differentiation as an upstream regulator of RUNX2 [Ikeda et al., 2005]. In dental follicle cells (DFCs), ZBTB16 was strongly upregulated after treatment with dexamethasone and ZBTB16 overexpression increased ALP, and Osx expression [Felthaus et al., 2014]. Similar results were also observed in our study. ZBTB16 mRNA and protein expression was not detected without OIM but was detected when the cells were cultured with OIM, and the levels increased for 2 weeks.
To analyze the functional roles of ZBTB16 during osteoblastic differentiation, siRNA knockdown experiments were performed. Both the ALP activity and the mRNA expression of OCN, and BSP were decreased by siZBTB16. In addition, mineralized nodule formation was also decreased, suggesting that ZBTB16 positively affects the osteoblastic differentiation of hMSCs.
Next, we investigated the interaction of ZBTB16 and other osteogenic transcriptional factors. Generally, Runx2 and Osx are thought to be master key regulators of osteoblastic differentiation [Komori, 2006; Zhang, 2010]. In our research, because ZBTB16 expression was increased during osteoblastic differentiation (Fig. 2A), we hypothesized that RUNX2 and Osx regulated ZBTB16 expression.
Previous studies have suggested that Osx is a downstream target gene of Runx2 and is required for the final commitment of the osteoblast lineage [Nishio et al., 2006; Sinha and Zhou, 2013]. However, in our study, although Osx knockdown decreased ZBTB16 expression, RUNX2 knockdown did not affect ZBTB16 expression. These results suggested that, OIM‐induced ZBTB16 expression depends on Osx but is independent of RUNX2. Other studies have also reported that BMP‐2‐induced Osx expression is independent of Runx2 [Lee et al., 2003; Ulsamer et al., 2008]. Furthermore, it has been reported that the BMP signal induced Runx2‐deficient cell lines to express markers related to osteoblastic differentiation, such as ALP, OCN, and Osx, via a Runx2‐independent pathway [Liu et al., 2007].
Next, ChIP assay was performed to confirm whether Osx is an upstream regulator of ZBTB16. It has been reported that Osx interacts with the promoters of osteogenic genes such as COL1A1, BSP, or OCN [Koga et al., 2005; Ortuño et al., 2010; Sinha et al., 2010; Ortuño et al., 2013]. In these cases, Osx binds the GC‐rich canonical Sp1 sequences of the osteoblastic gene promoters and regulates the transcription of these genes.
Similarly, our results showed that Osx binds to the Sp1 sequences of the ZBTB16 promoter region. These results suggested that, Osx might be an upstream regulator of ZBTB16 during osteoblastic differentiation. However, a previous study reported that ZBTB16 is involved in early osteoblastic differentiation as an upstream regulator gene of RUNX2 [Ikeda et al., 2005]. A recent study reported that, ZBTB16 binds to the promoter region of Osx but not that of RUNX2 [Felthaus et al., 2014]. The results of the ChIP assay in our study also confirmed that ZBTB16 bound the promoter region of Osx. However, Osx expression was not affected by siZBTB16, although ZBTB16 bound the promoter region of Osx (Supplemental Fig. S2). Some of the discrepancies among these findings may be caused by the different cells and methodologies used. Furthermore, because the transcriptional network is complicated, it is possible that ZBTB16 regulates RUNX2 expression when MSCs differentiate into osteoblast precursor cells (early phase) and that Osx regulates ZBTB16 expression when osteoblasts differentiate into mature osteoblasts (late phase). In addition, ZBTB16 regulates the mature osteoblastic marker genes, such as OCN and BSP, and may interact with Osx expression by binding to this promoter region. Hence, although a positive role of ZBTB16 in osteoblastogenesis has already been reported in different in vitro models, the observation from our study that Osx is a potential upstream regulator of ZBTB16 provides new insight into the regulatory networks during osteoblastic differentiation in hMSCs.
In our study, when hMSCs were cultured with OIM only, Osx expression was low (Supplemental Fig. S3A). It is well known that BMPs are potent regulators of osteoblastic differentiation and induce Osx expression [Cheng et al., 2003; Yagi et al., 2003]. Furthermore, some researchers have suggested that BMP‐6 induces Osx expression during osteoblastic differentiation and enhances calcified nodule formation in human cells [Friedman et al., 2006; Zhu et al., 2012; Hakki et al., 2014]. Therefore, to increase the amount of Osx protein obtained from cell culture, hMSCs were cultured in OIM supplemented with BMP‐6 (200 ng/mL) (Supplemental Fig. S3A).
Given that BMP‐6 enhanced Osx expression and that ZBTB16 was not affected by BMP‐6 in our study (Supplemental Fig. S3B), it is possible that Osx activation by BMP‐6 promotes the transcriptional activity of ZBTB16 through interactions with co‐binding factors. In the future, we will elucidate how Osx interacts with other transcriptional factors in the ZBTB16 promoter region to regulate ZBTB16 transcription and osteoblastic differentiation.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Supporting Figure Legends.
Supporting Figure S1.
Supporting Figure S2.
Supporting Figure S3.
ACKNOWLEDGMENTS
This study was supported by the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program of the Project for Developing Innovation Systems “Cell Sheet Tissue Engineering Center (CSTEC)” funded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, a bilateral joint research project between Japan and Korea funded by the Japan Society for the Promotion of Science (JSPS), JSPS KAKENHI Grant Number 15K11224, and Japan Agency for Medical Research and Development (AMED).
Conflict of interests: The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
REFERENCES
- Acampora D, Merlo GR, Paleari L, Zerega B, Postiglione MP, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. 1999. Craniofacial, vestibular, and bone defects in mice lacking the distal‐less‐related gene Dlx5. Development 126(17):3795–3809. [DOI] [PubMed] [Google Scholar]
- Barna M, Hawe N, Niswander L, Pandolfi PP. 2000. Plzf regulates limb and axial skeletal patterning. Nat Genet 25(2):166–172. [DOI] [PubMed] [Google Scholar]
- Barna M, Pandolfi PP, Niswander L. 2005. Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature 436(7048):277–281. [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. 2004. A twist code determines the onset of osteoblast differentiation. Dev Cell 6(3):423–435. [DOI] [PubMed] [Google Scholar]
- Caplan AI, Correa D. 2011. The MSC: An injury drugstore. Cell Stem Cell 9(1):11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Jiang W, Phillips FM, Haydon RC, Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichakarn P, Szatkowski JP, Park JY, He TC. 2003. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J Bone Joint Surg Am 85:1544–1552. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. 2004. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36:653–659. [DOI] [PubMed] [Google Scholar]
- Di Bella C, Farlie P, Penington AJ. 2008. Bone regeneration in a rabbit critical‐sized skull defect using autologous adipose‐derived cells. Tissue Eng Part A 14(4):483–490. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper‐Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. 2006. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8(4):315–317. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. 1997. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 89(5):747–754. [DOI] [PubMed] [Google Scholar]
- Felthaus O, Gosau M, Klein S, Prantl L, Reichert TE, Schmalz G, Morsczeck C. 2014. Dexamethasone‐related osteogenic differentiation of dental follicle cells depends on ZBTB16 but not Runx2. Cell Tissue Res 357(3):695–705. [DOI] [PubMed] [Google Scholar]
- Friedman MS, Long MW, Hankenson KD. 2006. Osteogenic differentiation of human mesenchymal stem cells is regulated by bone morphogenetic protein‐6. J Cell Biochem 98:538–554. [DOI] [PubMed] [Google Scholar]
- Gay IC, Chen S, MacDougall M. 2007. Isolation and characterization of multipotent human periodontal ligament stem cells. Orthod Craniofacial Res 10:149–160. [DOI] [PubMed] [Google Scholar]
- Hakki SS, Bozkurt B, Hakki EE, Kayis SA, Turac G, Yilmaz I, Karaoz E. 2014. Bone morphogenetic protein‐2, ‐6, and ‐7 differently regulate osteogenic differentiation of human periodontal ligament stem cells. J Biomed Mater Res Part B 102B(1):119–130. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Yoshida K, Tsukahara S, Sakamoto Y, Tanaka H, Furukawa K, Inoue I. 2005. The promyelotic leukemia zinc finger promotes osteoblastic differentiation of human mesenchymal stem cells as an upstream regulator of CBFA1. J Biol Chem 280(9):8523–8530. [DOI] [PubMed] [Google Scholar]
- Iwata T, Yamato M, Tsuchioka H, Takagi R, Mukobata S, Washio K, Okano T, Ishikawa I. 2009. Periodontal regeneration with multi‐layered periodontal ligament‐derived cell sheets in a canine model. Biomaterials 30(14):2716–2723. [DOI] [PubMed] [Google Scholar]
- Iwata T, Yamato M, Zhang Z, Mukobata S, Washio K, Ando T, Feijen J, Okano T, Ishikawa I. 2010. Validation of human periodontal ligament‐derived cells as a reliable source for cytotherapeutic use. J Clin Periodontol 37(12):1088–1099. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Kim HN, Park EK, Lee BH, Ryoo HM, Kim SY, Kim IS, Stein JL, Lian JB, Stein GS, van Wijnen AJ, Choi JY. 2006. The bone‐related Zn finger transcription factor Osterix promotes proliferation of mesenchymal cells. Gene 366:145–151. [DOI] [PubMed] [Google Scholar]
- Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H. 2005. NFAT and Osterix cooperatively regulate bone formation. Nat Med 11(8):880–885. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89(5):755–764. [DOI] [PubMed] [Google Scholar]
- Komori T. 2006. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem 99:1233–1239. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. 2003. BMP‐2‐induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun 309(3):689–694. [DOI] [PubMed] [Google Scholar]
- Lee OK, Kuo TK, Chen WM, Lee KD, Hsieh SL, Chen TH. 2004. Isolation of multipotent mesenchymal stem cells from umbilical cord blood. Blood 103(5):1669–1675. [DOI] [PubMed] [Google Scholar]
- Liu T, Gao Y, Sakamoto K, Minamizato T, Furukawa K, Tsukazaki T, Shibata Y, Bessho K, Komori T, Yamaguchi A. 2007. BMP‐2 promotes differentiation of osteoblasts and chondroblasts in Runx2‐deficient cell lines. J Cell Physiol 211(3):728–735. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐ time quantitative PCR and the 2‐ΔΔCT Method. Methods 25(4):402–408. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, Aburatani H, Nishimura R, Yoneda T. 2008. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem 283:29119–29125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. 2002. The novel Zinc Finger‐containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell 108(1):17–29. [DOI] [PubMed] [Google Scholar]
- Niger C, Lima F, Yoo DJ, Gupta RR, Buo AM, Hebert C, Stains JP. 2011. The transcriptional activity of osterix requires the recruitment of Sp1 to the osteocalcin proximal promoter. Bone 49:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio Y, Dong Y, Paris M, O'Keefe RJ, Schwarz EM, Drissi H. 2006. Runx2‐mediated regulation of the zinc finger Osterix/Sp7 gene. Gene 372:62–70. [DOI] [PubMed] [Google Scholar]
- Ortuño MJ, Ruiz‐Gaspà S, Rodríguez‐Carballo E, Susperregui AR, Bartrons R, Rosa JL, Ventura F. 2010. P38 regulates expression of osteoblast‐specific genes by phosphorylation of osterix. J Biol Chem 285:31985–31994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortuño MJ, Susperregui AR, Artigas N, Rosa JL, Ventura F. 2013. Osterix induces Col1a1 gene expression through binding to Sp1 sites in the bone enhancer and proximal promoter regions. Bone 52(2):548–556. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89(5):765–771. [DOI] [PubMed] [Google Scholar]
- Park SJ, Saito‐Adachi M, Komiyama Y, Nakai K. 2016. Advances, practice, and clinical perspectives in high‐throughput sequencing. Oral Dis 22(5):353–364. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284(5411):143–147. [DOI] [PubMed] [Google Scholar]
- Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29:391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha KM, Zhou X. 2013. Genetic and molecular control of osterix in skeletal formation. J Cell Biochem 114(5):975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. 2010. Regulation of the osteoblast‐specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J 29(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovrlj B, Guzman JZ, Al Maaieh M, Cho SK, Iatridis JC, Qureshi SA. 2014. Cellular bone matrices: Viable stem cell‐containing bone graft substitutes. Spine J 14:2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubiani O, Orsini G, Zini N, Di Iorio D, Piccirilli M, Piattelli A, Caputi S. 2008. Regenerative potential of human periodontal ligament derived stem cells on three‐dimensional biomaterials: A morphological report. J Biomed Mater Res A 87(4):986–993. [DOI] [PubMed] [Google Scholar]
- Ulsamer A, Ortuño MJ, Ruiz S, Susperregui AR, Osses N, Rosa JL, Ventura F. 2008. BMP‐2 Induces Osterix expression through up‐regulation of Dlx5 and its phosphorylation by p38. J Biol Chem 283(7):3816–3826. [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. 2005. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast‐specific Osteocalcin gene expression. J Biol Chem 280:30689–30696. [DOI] [PubMed] [Google Scholar]
- Yagi K, Tsuji K, Nifuji A, Shinomiya K, Nakashima K, DeCrombrugghe B, Noda M. 2003. Bone morphogenetic protein‐2 enhances osterix gene expression in chondrocytes. J Cell Biochem 88:1077–1083. [DOI] [PubMed] [Google Scholar]
- Yamada A, Iwata T, Yamato M, Okano T, Izumi Y. 2013. Diverse functions of secreted frizzled‐related proteins in the osteoblastogenesis of human multipotent mesenchymal stromal cells. Biomaterials 34(13):3270–3278. [DOI] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone‐Corsi P, Townes TM, Hanauer A, Karsenty G. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: Implication for Coffin‐Lowry syndrome. Cell 117(3):387–398. [DOI] [PubMed] [Google Scholar]
- Yoon E, Dhar S, Chun DE, Gharibjanian NA, Evans GR. 2007. In vivo osteogenic potential of human adipose‐derived stem cells/poly lactide‐co‐glycolic acid constructs for bone regeneration in a rat critical‐sized calvarial defect model. Tissue Eng 13(3):619–627. [DOI] [PubMed] [Google Scholar]
- Zhang T, Xiong H, Kan LX, Zhang CK, Jiao XF, Fu G, Zhang QH, Lu L, Tong JH, Gu BW, Yu M, Liu JX, Licht J, Waxman S, Zelent A, Chen E, Chen SJ. 1999. Genomic sequence, structural organization, molecular evolution, and aberrant rearrangement of promyelocytic leukemia zinc finger gene. Proc Natl Acad Sci USA 96(20):11422–11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. 2010. Transcriptional regulation of bone formation by the osteoblast‐specific transcription factor Osx. J Orthop Surg Res 5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Friedman MS, Luo W, Woolf P, Hankenson KD. 2012. The transcription factor osterix (SP7) regulates BMP6‐induced human osteoblast differentiation. J Cell Physiol 227(6):2677–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. 2002. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell 13(12):4279–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Supporting Figure Legends.
Supporting Figure S1.
Supporting Figure S2.
Supporting Figure S3.


