Abstract
The advent of high‐throughput sequencing technologies coupled with new detection methods of RNA modifications has enabled investigation of a new layer of gene regulation − the epitranscriptome. With over 100 known RNA modifications, understanding the repertoire of RNA modifications is a huge undertaking. This review summarizes what is known about RNA modifications with an emphasis on discoveries in plants. RNA ribose modifications, base methylations and pseudouridylation are required for normal development in Arabidopsis, as mutations in the enzymes modifying them have diverse effects on plant development and stress responses. These modifications can regulate RNA structure, turnover and translation. Transfer RNA and ribosomal RNA modifications have been mapped extensively and their functions investigated in many organisms, including plants. Recent work exploring the locations, functions and targeting of N6‐methyladenosine (m6A), 5‐methylcytosine (m5C), pseudouridine (Ψ), and additional modifications in mRNAs and ncRNAs are highlighted, as well as those previously known on tRNAs and rRNAs. Many questions remain as to the exact mechanisms of targeting and functions of specific modified sites and whether these modifications have distinct functions in the different classes of RNAs.
Keywords: RNA modifications, epitranscriptome, RNA 5‐methylcytosine (m5C), N6‐methyladenosine (m6A), Pseudouridine (Ψ), Arabidopsis
Iain Robert Searle
Edited by: Zhizhong Gong, China Agricultural University, China
INTRODUCTION
Chemical modifications of DNA and proteins such as histones have been established as important regulators of gene expression, eukaryotic development and stress responses (Suzuki and Bird 2008; Lawrence et al. 2016). More recently, a new level of gene regulation, the epitranscriptome, or RNA modifications has gained interest and momentum. There are over 100 different RNA modifications found in different RNA species, the most abundant and most intensively studied are transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). Loss of modifications on tRNAs and rRNAs is linked to multiple human diseases (Blanco and Frye 2014; Torres et al. 2014) and detrimental effects on development and stress responses in other organisms, including plants, underscoring their vital roles (Motorin and Helm 2011; El Yacoubi et al. 2012). However, new functions and interactions are also being discovered for RNA modifications in mRNAs and other non‐coding RNAs (ncRNAs) such as long non‐coding RNAs (lncRNAs), micro RNAs (miRNAs) and other small RNAs.
Although the presence of RNA modifications such as the 5’ cap structure and internal N6‐methyladenosine (m6A) in mRNAs has been known for decades (Desrosiers et al. 1974; Perry and Kelley 1974; Dubin and Taylor 1975; Shatkin 1976), the flood gates have only just been opened for a new wave of research describing other modifications and their impact on gene regulation. Several recently developed high‐throughput sequencing methods for detecting RNA modifications have allowed investigation of low abundance mRNA and ncRNAs on an unprecedented scale thereby enabling deciphering of their functions in RNA metabolism, gene regulation, translation, development and stress responses (methods are reviewed in Shafik et al. 2016). Interestingly, many of the RNA modifications and the enzymes responsible for ‘writing’ and ‘reading’ the modifications are conserved across the three domains of life (Jackman and Alfonzo 2013), suggesting important, conserved biological functions of this added layer of complexity and flexibility for RNA regulation.
In this article, we provide an update on the research on tRNAs and rRNAs, highlighting the discoveries in plants before discussing the recent studies investigating the dynamic role of RNA modifications in regulating the function of tRNAs and rRNAs, and the epitranscriptomic landscape of other classes of RNA, including mRNAs and lncRNAs. Continuing on from Fray and Simpson (2015), this review extends and discusses recent developments utilizing transcriptome‐wide sequencing to explore the RNA modification landscapes of N6‐methyladenosine (m6A), 5‐methylcytosine (m5C), pseudouridine (Ψ) and other modifications which perturb Watson‐Crick base pairing. Finally, we will discuss conclusions and future perspectives to shed light on the Arabidopsis epitranscriptome.
PLANT TRANSFER RNA MODIFICATIONS AND THEIR FUNCTIONS
Transfer RNAs are considered the most heavily modified types of RNA, and these modifications are highly conserved in bacteria, yeast, mammals and plants, consistent with their central role in translation. At least 92 unique chemical modifications have been identified in tRNAs with varied chemical properties and effects on the stability and function of tRNAs (Machnicka et al. 2013). Transfer RNA modifications include RNA editing of adenosine to inosine (A‐I), methylation or acetylation of RNA bases, isomerization or reduction of uridine to pseudouridine (Ψ) or dihydrouridine (D) to name a few. These post‐transcriptional modifications can occur on the base, or on the ribose sugar backbone of the RNA molecule.
Transfer RNAs can vary in length from 70–90 nucleotides long with RNA modifications occurring at different positions on the iconic clover‐leaf secondary structure. The functional roles of tRNA modifications are determined by their position on the clover leaf structure and by the chemical properties of the RNA modification. The functions of these modifications affecting tRNA biogenesis can be divided into three major groups, (i) modifications that affect amino‐acylation on the acceptor stem; (ii) modifications on or near the anticodon loop can affect anti‐codon binding, wobble base pairing and frame shifting; and (iii) other positions on D‐stem, TΨC stem and variable loop/junction affecting stability, structure, translation and tRNA cleavage/degradation. The positions and types of modifications and the enzymes responsible for mediating them throughout different domains of life have been reviewed extensively in (Phizicky and Hopper 2010; El Yacoubi et al. 2012; Towns and Begley 2012). Here we focus on tRNA modifications and tRNA modifying enzymes investigated in plants.
The identities of RNA modifications present in tRNAs of several plant species (Arabidopsis, tobacco, maize, hybrid aspen and wheat) have been investigated using a combination of chromatography and mass spectroscopy techniques on purified tRNAs (Shugart 1972; Jones and Scott 1981; Chen et al. 2010; Hienzsch et al. 2013). In two independent studies on Arabidopsis tRNAs, a total of 26 known tRNA RNA modifications were identified and four novel, potentially plant specific RNA modifications (Chen et al. 2010; Hienzsch et al. 2013). In addition, bioinformatics approaches have been used to predict hundreds of RNA base modification sites in Arabidopsis miRNAs and tRNAs, based on the ability of certain RNA modifications to introduce mismatches in sequences after reverse transcription (Iida et al. 2009). Evidence has also shown dynamic regulation of tRNA modifications. When comparing different plant tissues, differences in the abundance and types of tRNA modifications were found when comparing different plant tissues and cell cultures (Jones and Scott 1981; Hienzsch et al. 2013) and new and old leaves (Shugart 1972). Recently, the tRNA modification 2’‐0‐cytosine methylation (Cm) was shown to be increased in response to pathogen infection in Arabidopsis (Ramirez et al. 2015). In other organisms, several tRNA modifications were shown to be induced under stress conditions such as oxidative stress (Chan et al. 2010; Chan et al. 2012), nutrient starvation (Preston et al. 2013) and toxins (Hertz et al. 2014).
Identification of RNA modifications present in tRNAs is only half the story − the tRNA modifying enzymes, or ‘writers’ are just beginning to be characterized in plants. A combination of bioinformatics and reverse genetics approaches have been used to predict and identify tRNA modifying enzymes in Arabidopsis (Golovko et al. 2002; Chen et al. 2006; Miyawaki et al. 2006; Pavlopoulou and Kossida 2009; Zhou et al. 2009; Chen et al. 2010; Hu et al. 2010; Mehlgarten et al. 2010; Leihne et al. 2011; Zhou et al. 2013; Burgess et al. 2015; Ramirez et al. 2015). Transfer RNA modifying enzymes have been characterized for mediating base methylations of guanine and cytosine residues, and modifying 2′‐O‐ribose methylations.
RNA methylation of guanine and cytosine residues commonly occurs in Arabidopsis tRNAs and have roles in mediating RNA structure and stabilization through for example Mg2+ binding (Chen et al. 1993; David et al. 2016). Based on homologous genes in yeast, three guanosine transfer RNA methyltransferase (TRM) enzymes have been identified in Arabidopsis, namely AtTRM10 (At5g47680), AtTRM11 (At3g26410) and AtTRM82 (At1g03110), which mediate m1G, m2G and m7G in tRNAs, respectively (Chen et al. 2010). Of these, a biological role in plant development was only identified for AtTRM11, as the mutant showed an early‐flowering phenotype. 5‐methylcytosine in tRNAs is mediated by Arabidopsis transfer RNA aspartic acid methyltransferase 1 (TRDMT1, At5g25480) at position 38 and by tRNA specific methyltransferase 4B (TRM4B, At2g22400) at the variable loop/TΨC stem junction (Goll et al. 2006; Burgess et al. 2015). Loss of both TRDMT1 and TRM4B results in increased sensitivity to the antibiotic Hygromycin B, suggesting roles for these modifications in translation (Burgess et al. 2015). Similar functions were found for TRM4 in yeast (Wu et al. 1998) and translation efficiency was reduced in mammals (Tuorto et al. 2012). Another tRNA methylation modification is 2′‐O‐ribose methylation. Recently, an Arabidopsis homolog of yeast TRM7 (At5g01230), a 2′‐O‐ribose methyltransferase, was identified to be required for efficient immune response to Pseudomonas syringae (Ramirez et al. 2015).
Several modifications in the anticodon loop fine‐tune translation by reducing frame shift mutations and mediating codon binding stringency at the third ‘wobble’ base pair position. RNA editing of adenosine to inosine (A‐I) at the first position of the anticodon allows a single tRNA to decode multiple codons for the same amino acid, because I can base pair with A, C or U. RNA editing (A‐I) by AtTAD1 (homologous to yeast Tad1p tRNA‐specific adenosine deaminase) (At1g01760) at the position 3’‐adjacent to the anticodon in nuclear tRNAAla(AGC) has been shown to be required for efficient translation under stress conditions, as Arabidopsis attad1 mutants have reduced biomass when exposed to heat and cold stress treatments (Zhou et al. 2013). The molecular function of this specific RNA editing event is unclear. A conserved multi‐protein Elongator complex mediates acetylation of histones and tRNA wobble uridine modifications (Mehlgarten et al. 2010). Four components of the Elongator complex have been characterized in plants, demonstrating roles for the Elongator complex in ABA and oxidative stress response in Arabidopsis (Chen et al. 2006; Zhou et al. 2009). In addition, Elongator mutants such as atelp1 (At5g13680) display pleiotropic growth defects (Chen et al. 2010).
RIBOSOMAL RNA MODIFICATIONS AND FUNCTIONS IN ARABIDOPSIS
Ribosomes are multi‐subunit complexes of non‐coding ribosomal RNAs and proteins. In eukaryotes, three of the four rRNAs present in the small and large rRNA subunits are encoded in a single, polycistronic, pre‐rRNA transcript. Multiple processing steps involving cleavage and RNA modifications are required for maturation and assembly of the rRNAs with ribosomal proteins (Henras et al. 2015). Ribosomal RNA modifications tend to be clustered around conserved structural and functional regions of the ribosomes such as the peptidyl transferase center (PTC) and are required for efficient translation (Decatur and Fournier 2002). Ribosomal RNAs contain three broad types of RNA modifications, ribose methylation, pseudouridylation and several types of base methylations (e.g. m5C, m3U, m6A) reviewed in (Decatur and Fournier 2002; Baxter‐Roshek et al. 2007).
The most abundant rRNA modifications are Ψ and 2′‐O‐ribose methylations. The majority of these rRNA modifications are mediated by small nucleolar ribonucleoprotein complexes (snoRNPs) composed of multiple conserved proteins and a small nucleolar RNA (snoRNA), which directs sequence‐specific targeting. These two modifications are guided by two different classes of snoRNAs, (i) box‐C/D snoRNAs which guide 2′‐O‐ribose methylations mediated by the methyltransferase NOP1 (yeast)/Fibrillarin (human) and (ii) box‐H/ACA snoRNAs which direct conversion of uridine to pseudouridine by Cbf5/NAP57/Dyskerin (human) (Kiss 2001; Brown et al. 2003). Three genes encoding homologues of the essential yeast and human Fibrillarin 2′‐O‐ribose methyltransferase were identified in Arabidopsis, AtFIB1 (At5g52470), AtFIB2 (At4g25630) and AtFIB3 (At5g52490) (Barneche et al. 2000; Pih et al. 2000). Of these three genes, transcripts were only detected from AtFIB1 and AtFIB2, and both proteins are able to partially complement a conditional yeast NOP1/Fibrillarin mutant. This suggests that the Arabidopsis 2′‐O‐ribose methyltransferase snoRNPs might be heterogeneous, and contain either AtFIB1 or AtFIB2, and these different snoRNPs may have specialized functions in plants. Similarly, removal of rRNA pseudouridylation in yeast and Arabidopsis by deletion of CBF5, is also lethal (Lermontova et al. 2007) while defects in the human homolog Dyskerin result in dyskeratosis congenita, a disease characterized by abnormal skin pigmentation and bone marrow failure (Heiss et al. 1998). Moreover, patients with this condition were recently found to have reduced Ψ in rRNA and the ncRNA telomerase component TERC (Telomerase RNA component) (Schwartz et al. 2014a). Another chloroplast specific rRNA Ψ synthase was identified in Arabidopsis in a suppressor screen for mutants complementing a chloroplast variegation mutation, SUPPRESSOR OF VARIAGATION1 (SVR1, At2g39140) (Yu et al. 2008). Arabidopsis svr1 mutants are small and pale green, with defects in chloroplast rRNA processing and translation. SVR1 is predicted to target chloroplast rRNA Ψ independently of a snoRNA guide in a similar manner to other tRNA and mitochondrial rRNA Ψ synthases from yeast and bacteria (Ansmant et al. 2000).
Unlike most of the rRNA Ψ and 2′‐O‐ribose methylations, which are catalyzed by snoRNPs, the rRNA base methylations are all performed by site‐specific base methyltransferases. The nuclear large subunit 25S rRNA in yeast and Arabidopsis contains two m5C sites, which are methylated by RNA methyltransferases RCM1 (rRNA cytosine methyltransferase 1) and NOP2 (nucleolar protein 2) (Sharma et al. 2013; Gigova et al. 2014). The two methylation sites have roles in antibiotic sensitivity and rRNA biogenesis and processing in yeast, respectively (Hong et al. 1997; Sharma et al. 2013). In Arabidopsis the RCM1 homolog, NOP2/Sun domain protein 5 (NSUN5), was found to methylate the orthologous position in 25S rRNA (Burgess et al. 2015). The second m5C site unexpectedly remained unchanged in single mutants for all three Arabidopsis NOP2 homologs, NOP2A (At5g55920), NOP2B (At4g26600) and NOP2C (At1g06560), as nop2a mutants have a leaf phenotype (Fujikura et al. 2009; Burgess et al. 2015). The unchanged methylation level in the single mutants may suggest functional redundancy (Burgess et al. 2015). Two adjacent adenosines are N‐6 dimethylated (m2 6A) in small subunit rRNAs of eukaryotes and prokaryotes by adenosine dimethyl transferase 1 (DIM1) homologs. Similar to the case of NOP2, the Arabidopsis genome encodes three rRNA dimethyl transferase enzymes: DIM1A (At2g47420), the nuclear 18S rRNA dimethyl transferase required for organized root growth and epidermal patterning (Wieckowski and Schiefelbein 2012), DIM1B (At5g66360), the mitochondrial rRNA dimethyl transferase (Richter et al. 2010) and DIM1C/PALEFACE1 (At1g01860), which is located in the chloroplast and is required for chloroplast development in the cold (Tokuhisa et al. 1998).
In organisms such as yeast, with only one copy of DIM1 and NOP2, loss of either of these enzymes results in lethality (Lafontaine et al. 1994; Hong et al. 1997). Surprisingly, the presence of catalytically inactivated modifying enzymes rescues the phenotype in several organisms, suggesting other important roles for DIM1 and NOP2 in ribosome biogenesis (Lafontaine et al. 1995; King and Redman 2002; Zorbas et al. 2015). Arabidopsis dim1a and nop2a mutants both display small, malformed leaves, slow growth and other phenotypes (Fujikura et al. 2009; Wieckowski and Schiefelbein 2012), reminiscent of many other Arabidopsis mutants with roles in rRNA biogenesis and of ribosomal protein mutants (Nishimura et al. 2005; Byrne 2009; Abbasi et al. 2010), pointing to additional functions besides RNA methylation for these proteins in plants. Another predicted rRNA m5C methyltransferase is Arabidopsis RNA methyltransferase (RNMT, At3g13180), which is related to the bacterial Fmu 16S rRNA methyltransferase (Pavlopoulou and Kossida 2009). Arabidopsis rnmt mutants have reduced global cytosine methylation, however, the specific nucleotide position is yet to be identified (Hebrard et al. 2013). In addition to m5C and m2 6A, Arabidopsis rRNA also contains several m6A base methylations (Wan et al. 2015).
MESSENGER RNA AND OTHER NON‐CODING RNA MODIFICATIONS
In the following sections we review and discuss the RNA modifications discovered in Arabidopsis, animal, yeast and bacterial epitranscriptomes to date and their diverse functions. Recently, two modifications have been identified transcriptome‐wide using direct detection methods, m6A (Immunoprecipitation and next generation sequencing) and m5C (RNA Bisulfite sequencing) in plants (Luo et al. 2014; Li et al. 2014b; Wan et al. 2015; David et al. 2016). The first discovered and globally most abundant RNA modification, pseudouridine (Ψ) has been mapped transcriptome‐wide by several recent studies in mammals and yeast (Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014a; Li et al. 2015). Although Ψ sites have not been mapped in plants transcriptome‐wide to date, the enzymatic functions required for transcriptome‐wide Ψ have been investigated in Arabidopsis (Lermontova et al. 2007; Yu et al. 2008; Chen et al. 2010). Interestingly, these high throughput studies have revealed m6A, m5C and Ψ to show distinct distribution patterns along mRNA transcripts and are associated with specific functions as discussed in the following sections (Figure 1). Additional modifications in the Arabidopsis epitranscriptome such as 3‐methyl cytosine (m3C) and 1‐methyl guanosine (m1G) have been computationally predicted transcriptome‐wide based on common nucleotide substitution and reverse transcription errors caused by these modifications during RNA‐seq library preparation (Ryvkin et al. 2013; Vandivier et al. 2015). The modifications m6A, m5C and Ψ are unable to be detected using this method, as they do not alter Watson‐Crick base pairing.
Figure 1.
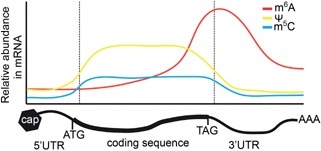
RNA modifications in messenger RNAs have distinct deposition patterns Shown is a pictorial representation of relative abundance of the RNA modifications m6A, m5C and Ψ along mRNA transcripts. These representations are based on transcriptome‐wide RNA bisulfite sequencing data for m5C and antibody data for m6A in animals and plants. The Ψ abundance is based on a combination of Ψ‐seq and antibody enrichment data from animals. m6A is lowly abundant along coding sequences and enriched at long last exons and at the start of 3′UTR's. While the majority of m5C sites are detected in the coding sequence of mRNA transcripts, m5C sites are statistically enriched in 3′UTR's. For Ψ, the modified sites are evenly distributed along the coding sequence, but are statistically underrepresented in 5′ UTRs.
N6‐METHYLADENOSINE (m6A)
m6A ‘writers’
Although the presence of m6A in mRNAs was first discovered in the 1970s (Desrosiers et al. 1974; Perry and Kelley 1974), many questions still remain unanswered about the roles of m6A in protein coding transcripts. While Ψ is the globally most abundant RNA modification, m6A is the most highly abundant RNA modification in mRNAs and is enriched in poly‐adenylated RNA fractions in plants and animals and has recently also been identified in bacterial mRNAs (Zhong et al. 2008; Meyer et al. 2012; Deng et al. 2015). A multi‐protein complex mediates these m6A sites. The catalytic core is composed of a heterodimer of methyltransferase like 3 (METTL3) and methyltransferase like 14 (METTL14) in mammals (Bokar et al. 1994; Liu et al. 2014). Recently, the mammalian splicing factor Wilm's tumor 1 associating protein (WTAP) and KIAA1429 were identified as additional components of the m6A ‘writer’ complex (Ping et al. 2014; Schwartz et al. 2014b). WTAP may have roles in targeting the m6A activity of METTL3 and METTL14, in a site‐specific manner, as m6A sites were mediated by WTAP dependent or independent mechanisms (Schwartz et al. 2014b).
Likewise, the Arabidopsis m6A ‘writer’ complex contains the adenosine methyltransferase MTA (At4g10760), which is predicted to form a heterodimer with MTB (At4g09980) (Bujnicki et al. 2002; Zhong et al. 2008). The Arabidopsis homolog of mammalian WTAP is known as Arabidopsis thaliana FKBP12 interacting protein 37 (AtFIP37, At3g54170), and was identified as a binding partner of MTA several years prior to similar studies in mammals (Faure et al. 1998; Zhong et al. 2008). This is shown in Figure 2A. Further studies are required to identify additional components and interacting proteins. All three known components of the Arabidopsis m6A writer complex are essential, as loss results in embryo lethality (Bujnicki et al. 2002; Vespa et al. 2004; Zhong et al. 2008). The lethality of mta mutants in Arabidopsis can be rescued by expressing MTA during embryo development using the ABI3 promoter (Bodi et al. 2012). Use of this system allowed investigation of the requirement for m6A in vegetative development, floral architecture and cell specification. The importance of m6A methylation for gene regulation is underscored by disorders caused by loss of m6A ‘writer’ complex components in human, yeast, mouse and fly (Clancy et al. 2002; Hongay and Orr‐Weaver 2011; Bodi et al. 2012; Wang et al. 2014b; Chen et al. 2015b).
Figure 2.
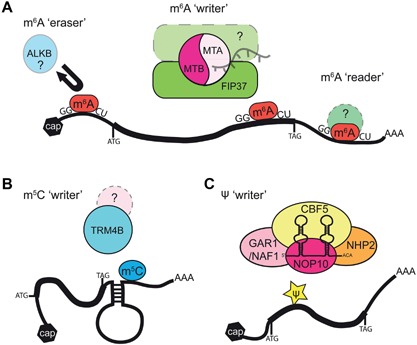
Distinct catalytic and targeting mechanisms of different RNA modifications in Arabidopsis (A) The predicted Arabidopsis m6A ‘writer’ complex is composed of a heterodimer of MTA and MTB, bound to AtFIP37 and potentially other, uncharacterized proteins. In mammals, miRNAs are able to guide the m6A ‘writer’ complex, however, it is not known if this targeting mechanism is conserved in plants. Potential m6A ‘erasers’ and ‘readers’ have been predicted in Arabidopsis and await further characterization. (B) A model for targeting of m5C methylation by TRM4B, based on RNA structure and potentially the presence of other RNA modifications. (C) Proposed H/ACA snoRNP Ψ ‘writer’ complex in Arabidopsis contains a guide H/ACA box snoRNA and the proteins GAR1 (At3g03920/At5g18180) or NAF1 (At1g03530), NHP2 (At5g08180), NOP10 (At2g20490) and the Ψ synthase AtCBF5 (At3g57150).
In order to elucidate why m6A is essential to plant development, three independent studies have mapped m6A epitranscriptomes in Arabidopsis and rice (Luo et al. 2014; Li et al. 2014b; Wan et al. 2015). As reported in the earlier studies in mammalian mRNAs, m6A sites were found to occur all along transcripts, with low signals observed across coding sequences and high enrichment in 3′UTRs and around stop codons (Dominissini et al. 2012; Meyer et al. 2012). Specifically in plants, there was a slight enrichment for m6A peaks at the start codon (Luo et al. 2014; Li et al. 2014b). However, a more recent study with greater sequencing depth and stringency conditions for m6A antibody binding, did not detect enrichment at start codons in Arabidopsis (Wan et al. 2015). Thousands of methylated transcripts were detected in different tissue types in Arabidopsis and rice and even in different Arabidopsis ecotypes. While many sites were specific to a particular tissue or ecotype, a large number of these sites were also conserved, even between animals and plants (Luo et al. 2014; Wan et al. 2015). As the deposition patterns and even specific m6A sites are conserved, the targeting mechanisms of the m6A ‘writer’ complex and functions of m6A are also likely conserved (Figure 2A).
In support of this, transcriptome‐wide mapping studies of m6A have confirmed earlier reports that the m6A ‘writer’ complex methylates sites within a highly conserved consensus sequence ‘RRACH’, (R = A/G and H = A/C/U) and this mostly occurs in GAC or less commonly in the AAC context (Wei and Moss 1977; Csepany et al. 1990). This consensus sequence is conserved in yeast, mammals and plants (Dominissini et al. 2012; Meyer et al. 2012; Schwartz et al. 2013; Luo et al. 2014; Li et al. 2014b; Wan et al. 2015). Interestingly, this is not the case for prokaryotes as unique distribution patterns and potential targeting were discovered in bacteria, as unlike animals and plants, m6A is enriched in coding sequences and at a novel ‘GCCAG’ consensus sequence (Deng et al. 2015). The mechanism of targeting and the functional significance of the distributions of m6A methylation on mRNA remain to be elucidated. While the highly conserved eukaryotic consensus sequence ‘RRACH’ is present many times in the transcriptome, the mechanism for determining which of these sites are methylated, remains unknown.
m6A ‘erasers’ − reversible RNA methylation
One intriguing mechanism for regulating m6A deposition is through the active removal of m6A in mRNAs. Two m6A demethylases have been characterized in mammals, namely Fat mass and obesity associated protein (FTO) and Alkylation repair homologue protein 5 (ALKBH5) (Jia et al. 2011; Zheng et al. 2013). These demethylases are part of the Escherichia coli ALKB dioxygenase homologs (ALKBH) family. The founding member, E. coli ALKB mediates oxidative demethylation of nucleic acid bases in DNA and RNA (Aas et al. 2003). Based on sequence homology, 13 ALKBH family proteins were predicted in Arabidopsis (Mielecki et al. 2012). These proteins showed diverse subcellular localizations, suggesting specialized functions in different cell compartments and hence potential layers of regulation for m6A demethylation in plants (Mielecki et al. 2012). While the identity of the Arabidopsis m6A demethylase(s) are still undetermined, they are expected to cause gross development defects, reminiscent of their animal homologs. In mice, loss of FTO leads to defects in alternative splicing and adipogenesis (Zhao et al. 2014), while loss of ALKBH5 affects mRNA processing in human cells and leads to male infertility in mice (Zheng et al. 2013). While the m6A ‘writer’ complex and demethylase ‘erasers’ act in concert to dynamically regulate m6A, additional RNA binding proteins or ‘readers’ are thought to decide the fate of m6A methylated transcripts.
m6A ‘readers’ − consequences for m6A on RNAs
The presence of m6A can influence RNA metabolism by regulating binding of modified RNA with proteins (m6A ‘readers’) and can also alter local RNA structure, leading to alternate outcomes for methylated and non‐methylated transcripts. Several classes of m6A ‘reader’ proteins have been identified in animals, such as YTH domain proteins, serine/arginine‐rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) (Dominissini et al. 2012; Schwartz et al. 2013; Zhao et al. 2014; Wang et al. 2014a; Schwartz et al. 2014b; Alarcon et al. 2015a). Potential homologs of these m6A ‘readers’ have been identified in plants, suggesting conserved functions (Lorkovic and Barta 2002).
YTH domain containing proteins have been identified as a class of RNA binding proteins that preferentially bind m6A methylated RNA in mammals and yeast (Dominissini et al. 2012; Schwartz et al. 2013; Wang et al. 2014a; Schwartz et al. 2014b). The first m6A ‘reader’ to be characterized was YTH Domain Family 2 (YTHDF2), which was shown to bind thousands of m6A containing mRNAs in the cytoplasm and deliver them to processing bodies for degradation (Wang et al. 2014a). This discovery explains the negative correlation of m6A with mRNA abundance in both plants and animals as m6A is used as a mark for rapid turn‐over of mRNAs (Li et al. 2014b; Schwartz et al. 2014b; Wang et al. 2014a; Wan et al. 2015). The sub‐cellular locations of m6A readers is crucial for the outcome of m6A on RNAs and this is clearly demonstrated by YTHDF2 (Zhou et al. 2015). While cytoplasmic YTHDF2 leads to mRNA decay, heat shock induces YTHDF2 to re‐localize to the nucleus. YTHDF2 is then able to compete with the nuclear ‘eraser’ FTO for binding of m6A sites, leading to increased 5’UTR methylation of newly transcribed, heat stress responsive mRNAs. The increased methylation leads to increased translation initiation independent of the 5’ cap, allowing selective mRNA translation under heat shock stress. In addition, the m6A reader YTHDF1 has also been shown to increase translation initiation of transcripts harboring m6A sites (Wang et al. 2015). This occurs in the cytoplasm, leading to competition with the mRNA degrading cytoplasmic YTHDF2. This competition is thought to allow fast responses and regulation of mRNA abundance and translation through m6A methylation.
Plant proteins containing this conserved YTH domain are expected to mediate similar functions for m6A in RNA. The Arabidopsis genome encodes 13 predicted YTH domain containing proteins, which may be responsible for ‘reading’ the m6A code and regulating RNA metabolism (Li et al. 2014a). One such protein is the Arabidopsis homologue of Cleavage and Polyadenylation Specificity Factor 30 (CPSF30, At1g30460), which is required in plants and mammals for polyadenylation and 3′end formation (Thomas et al. 2012; Chan et al. 2014). Intriguingly, while the presence of the YTH domain of Arabidopsis AtCPSF30 is dependent on alternative splicing, the YTH domain is completely absent in yeast and mammalian CPSF30 homologs (Delaney et al. 2006; Hunt et al. 2012; Chakrabarti and Hunt 2015). Furthermore, AtCPSF30 has also been shown to be involved in oxidative stress responses (Zhang et al. 2008) and is required for programmed cell death and immunity in Arabidopsis (Bruggeman et al. 2014). These functions are independent of the YTH domain and raise questions about the possible roles of YTH domain‐containing AtCPSF30 in regulating m6A containing RNAs.
In addition to YTH domain proteins, two other classes of potential m6A ‘readers’, namely SR proteins and hnRNPs, have been investigated. One SR protein, SR Splicing Factor 2 (SRSF2), was shown to preferentially bind mRNAs containing m6A sites, leading to increased inclusion of target exons during splicing when the ‘eraser’ FTO is depleted (Zhao et al. 2014). SRSF2 RNA binding sites tend to overlap with m6A sites, however, it is unclear if SRSF2 binds m6A directly, or indirectly through interactions with other proteins. Recently, SRSF3 and SRSF10 were found to competitively bind YTHDC1, which directly binds m6A, to regulate mRNA splicing (Xiao et al. 2016). While the interaction between SRSF3 and YTHDC1 promotes SRSF3 binding to RNA target sites, YTHDC1 binding of SRSF10 inhibits SRSF10 binding to RNA target sites. In combination, these events result in exon inclusion, while successful SRSF10 binding to RNA results in exon exclusion. Similarly, over expressing the predicted Arabidopsis SRSF2 ortholog AtSRp30 (At1g09140) demonstrated its function in regulating splicing (Lopato et al. 1999). Further studies are required to determine if m6A deposition directly or indirectly affects the activities of the eighteen SR proteins in Arabidopsis (Lorkovic and Barta 2002). Other m6A ‘readers’ include hnRNPs, which have diverse roles in RNA processing and export (Lorkovic et al. 2000).
Recently, m6A methylation was shown to be required for the biogenesis and function of a subset of miRNAs, and this is mediated in part through the m6A ‘reader’ HNRNPA2B1 and m6A ‘anti‐reader’ human antigen R (HuR) in animals. miRNA processing and abundance is deregulated when either the m6A ‘writer’ METTL3 or the m6A ‘eraser’ FTO were perturbed, demonstrating a role for m6A in miRNA biogenesis (Berulava et al. 2015; Alarcon et al. 2015b). The m6A mark in miRNAs is important, as nuclear HNRNPA2B1 binds a subset of m6A containing pri‐miRNAs and recruits the Microprocessor complex to cleave pri‐miRNAs into pre‐miRNAs (Alarcon et al. 2015a). In addition, m6A methylation is required for efficient regulation of a sub‐set of miRNA target transcripts. m6A methylation can aid miRNA driven degradation of mRNAs, as m6A blocks binding of the m6A ‘anti‐reader’ HuR, allowing miRNAs access to their target sites in mRNAs (Wang et al. 2014b). While m6A methylation regulates the biogenesis of a subset of miRNAs, some miRNAs are also able to affect targeting of m6A methylation. Artificial and endogenous miRNAs were recently shown to target m6A deposition and increase m6A abundance by guiding and modulating METTL3 binding to mRNAs (Chen et al. 2015b). This proposed mechanism of miRNAs guiding m6A sites is supported as animal miRNA target sites are highly enriched at m6A methylated regions, however, no such correlation was identified in Arabidopsis (Luo et al. 2014; Chen et al. 2015b). Is another class of small guide RNAs mediating m6A targeting in plants? These intriguing findings raise many questions about possible roles of m6A in regulating miRNAs in plants, and how the plant m6A ‘writer’ complex is targeted to mRNAs.
Another interesting example of an hnRNP m6A ‘reader’ is HNRNPC. HNRNPC does not bind m6A, however, it requires m6A methylation of mRNA and lncRNA targets such as Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) to alter local RNA structure in order to facilitate RNA binding (Liu et al. 2015). This RNA remodeling, or ‘m6A‐switch’, is achieved through the ability of m6A to disrupt adenosines from forming non‐Watson‐Crick G:A base pairs and also destabilizes A:U base pairs (Roost et al. 2015). It remains to be seen if the nine hnRNPs in Arabidopsis genome also show such diverse interactions with m6A as a dance partner (Lorkovic et al. 2000).
Additional functions for m6A methylation in reprogramming, organ differentiation and cell division functions are conserved, as indicated by the Arabidopsis mta phenotype and the roles of m6A in inducing pluripotent cells in mammals and sporulation in yeast (Clancy et al. 2002; Bodi et al. 2012; Wang et al. 2014b; Chen et al. 2015b). Furthermore, m6A was shown to regulate the mammalian circadian clock, as loss of RNA m6A methylation slows RNA processing resulting in delayed release of mature clock transcripts from the nucleus (Fustin et al. 2013). m6A may also play a role in regulating the plant circadian clock, as several transcripts regulating the Arabidopsis clock were highly methylated (Wan et al. 2015). Functions for m6A in splicing intron retention, polyadenylation, microRNA regulation, reprogramming and stress responses in plants warrants further investigation. Recently developed, single nucleotide resolution approaches to mapping m6A epitranscriptomes will enable further characterization of the functions of this mark in plants (Linder et al. 2015; Chen et al. 2015a).
5‐METHYLCYTOSINE (M5C)
m5C ‘writers’
While the functions of m5C as an epigenetic mark in DNA have been studied intensively, the role of m5C in RNA is less well studied. The importance of m5C has been established for tRNAs and rRNAs (Motorin et al. 2010), but functions are still being investigated for other RNAs such as mRNAs and lncRNAs. m5C was first identified transcriptome‐wide using RNA Bisulfite sequencing (bsRNA‐seq) in human (HeLa) cells, uncovering over 10,000 m5C sites (Squires et al. 2012). This prompted the development of additional techniques that enrich the direct RNA targets of specific RNA methyltransferases using RNA immunoprecipitation (Khoddami and Cairns 2013; Hussain et al. 2013a). Recently, the Arabidopsis m5C landscape was mapped using bsRNA‐seq in several tissue types and RNA methyltransferase mutants, identifying hundreds of m5C sites (David et al. 2016).
Together, these studies identified two m5C ‘writers’ that catalyze methylation in mRNAs and other classes of RNAs; the first RNA methyltransferase is tRNA specific methyltransferase 4 (TRM4) otherwise known as NOP2/Sun domain protein 2 (NSUN2), in yeast and animals respectively. NSUN2 plays broad roles in many organisms for mediating oxidative stress tolerance and balancing stem cell self‐renewal and differentiation. This is demonstrated in nsun2 mutant mice presenting with epidermal differentiation defects, male infertility and small size, which is thought to be due to a reduction in stem cell proliferation (Blanco et al. 2011; Hussain et al. 2013b). Furthermore, NSUN2 depletion in humans leads to mild microcephaly, short stature and neurological disorders (Abbasi‐Moheb et al. 2012; Khan et al. 2012; Martinez et al. 2012; Fahiminiya et al. 2014). Loss of nsun2 leads to increased tRNA cleavage under oxidative stress and these cleavage products are thought to cause these neuro‐developmental disorders (Blanco et al. 2014). These roles are also conserved in plants, as Arabidopsis trm4b mutants display shorter primary roots, which is linked to a reduced capacity for cells to divide in the root meristem (David et al. 2016). Furthermore, trm4b mutants are also more sensitive to oxidative stress and have reduced stability of non‐methylated tRNAs. However, it is difficult to tease apart the contributions of tRNA and mRNA methylations to these biological functions, as NSUN2/TRM4B methylates both these classes of RNAs.
The second m5C ‘writer’ shown to target mRNAs is Transfer RNA aspartic acid methyltransferase 1 (TRDMT1) also known as DNA methyltransferase 2 (DNMT2). TRDMT1 was previously thought to methylate DNA, due to its structural similarity to DNA methyltransferases, however it is now regarded as an RNA methyltransferase (Goll et al. 2006). In plants and animals, depletion of TRDMT1 is not phenotypically evident under controlled conditions (Goll et al. 2006). However, the functions of TRDMT1 become apparent under stress conditions such as oxidative and heat stress in Drosophila (Schaefer et al. 2010). Stress induced cleavage of tRNAs in TRDMT1 mutants also leads to inhibition of Dicer‐2 functions (Durdevic et al. 2013b). Furthermore, TRDMT1 is required for efficient immune response against viruses in Drosophila (Durdevic et al. 2013a). In contrast, depletion of TRDMT1 in zebrafish leads to gross morphological defects (Rai et al. 2007). NSUN2 mediates many more m5C sites in the transcriptome than TRDMT1. Only two TRDMT1 mRNA targets were identified in human cells, type I cytokeratin KRT18 mRNA and KRT18 pseudogene mRNA and this methylation was not conserved in mouse (Khoddami and Cairns 2013). This minor role for TRDMT1 in mediating m5C transcriptome‐wide seems to be conserved in plants, as only tRNA targets were identified (Burgess et al. 2015; David et al. 2016).
LOCATIONS, FUNCTIONS AND TARGETING OF m5C
In order to determine potential functions of m5C in RNA, transcriptome‐wide deposition patterns of this mark were analyzed in human cancer cells. Methylated sites are statistically enriched in ncRNAs compared to mRNAs (Squires et al. 2012). Within mRNAs, m5C sites are observed in higher numbers than expected for untranslated regions and are relatively depleted in coding regions, when normalized for length and sequence coverage (Squires et al. 2012). Moreover, m5C candidate sites in 3′UTRs are associated with binding regions for the Argonaute I–IV proteins, which are involved in miRNA mediated decay and translational inhibition, suggesting possible roles for m5C in mediating miRNA activity (Squires et al. 2012). Although further experiments are required to clearly determine the m5C and Argonaute association. Additional functions for m5C in increasing mRNAs half‐life have been proposed, as synthetic m5C methylated mRNAs exhibit increased stability (Warren et al. 2010). This does not seem to be the case for the majority of mRNAs, as methylation levels do not strongly correlate with gross changes in transcript abundance in mammals or plants (Hussain et al. 2013a; David et al. 2016). Furthermore, no major changes in global mRNA abundance were observed in mouse nsun2 mutants (Tuorto et al. 2012; Hussain et al. 2013b).
In contrast to mRNAs, several functions for m5C have been investigated in ncRNAs. Vault ncRNAs were identified as NSUN2‐specific m5C targets (Hussain et al. 2013a). Loss of m5C in vault ncRNAs leads to processing into small RNAs which can be incorporated into Argonaute complexes to regulate genes, in a manner similar to miRNAs. In addition to roles for m5C in small ncRNAs, functions for this modification have been elucidated for long ncRNAs. The 5′ A‐region of the lncRNA X‐inactive specific transcript (XIST) contains five m5C sites, which were shown to inhibit binding of the Polycomb repressive complex 2 (PRC2) in vitro (Amort et al. 2013). It remains to be determined if the PRC2 complex acts globally as an m5C ‘anti‐reader’ in both plants and animals.
Analysis of m5C transcriptome‐wide has shown that only approximately 0.4% of cytosines are methylated in mRNA, suggesting precise targeting of m5C to select target sites (Squires et al. 2012). In archaea, m5C was located in a consensus motif of AUCGANGU in mRNAs, providing a potential targeting mechanism for archaeal m5C ‘writers’ (Edelheit et al. 2013). In contrast, no such consensus target sequences were identified for m5C sites in animals or plants (Squires et al. 2012; Hussain et al. 2013a; David et al. 2016). As a general consensus sequence has not been identified, it is hypothesized that additional factors such as local RNA structure and RNA binding proteins may regulate the site selection of TRM4 and TRDMT1 (Figure 2B).
Many questions remain unanswered such as the targeting mechanism of m5C ‘writers’ and the functions of m5C in mRNAs and other non‐coding RNAs. The identification of potential m5C ‘readers’ and ‘erasers’ using techniques such as m5C RNA bait to immuno‐precipitate m5C binding proteins, similar to those performed for m6A should lead to future insights into how m5C sculpts the epitranscriptome.
PSEUDOURIDYLATION (Ψ)
Isomerization of uridine to Ψ was the first RNA modification to be discovered, and is also the most abundant (Charette and Gray 2000; Ge and Yu 2013). As discussed earlier, Ψ is common in tRNAs and rRNAs and also in spliceosomal snRNAs, however, it is an open question whether Ψ is present on Arabidopsis mRNAs. Recently, four research groups independently investigated Ψ transcriptome wide at single‐nucleotide resolution in yeast, human and mouse cells using modified approaches to Ψ‐sequencing (Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014a; Li et al. 2015). Using these transcriptome‐wide approaches, they were able to confirm known Ψ sites and cognate Ψ synthases in tRNAs, rRNAs, snRNAs and snoRNAs and extend the known sites to mRNAs and lncRNAs such as XIST and MALAT1. As the components required for Ψ are conserved in Arabidopsis, it seems more than likely that Ψ also occurs in plant mRNAs (Lermontova et al. 2007; Yu et al. 2008; Chen et al. 2010) (Figure 2C).
Ψ synthases are targeted to specific sites in RNAs through two mechanisms (1) snoRNA guided H/ACA snoRNPs containing CBF5/Dyskerin and (2) snoRNA independent Pseudouridine synthases (PUS). Using a combination of deletion and knock down mutants for PUS proteins and CBF5/Dyskerin in yeast and human, mRNA Ψ sites were found to be dependent on Ψ synthases using both snoRNA dependent and independent mechanisms.
While the targeting mechanism for Ψ by H/ACA snoRNPs is based on the snoRNA guide, and synthetic snoRNAs have been successfully designed to target Ψ at novel sites (Karijolich and Yu 2011), the targeting mechanisms of PUS proteins to RNA are less understood. Transcriptome‐wide identification of Ψ sites in several yeast PUS deletion mutants allowed analysis and confirmation of sequence consensus sites preferred by specific PUS enzymes. In particular, yeast PUS4 mediated Ψ occurs at ‘GUΨC/NANNC’ consensus sites, while yeast PUS7 Ψ sites generally occur at the consensus ‘UGΨA/R’. Not all sites with these consensus sequences are modified, suggesting other, additional factors mediating targeting. For example, the structure, as opposed to the sequence, of the tRNASer anticodon and TΨC stem loops were required for human PUS1 targeting (Sibert and Patton 2012).
Ψ sites mediated by these enzymes were located all along mRNA transcripts, with no positional bias found in coding sequences in any of the four transcriptome‐wide studies (Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014a; Li et al. 2015). However, while Ψ sites were under represented in 3’UTRs of yeast and human cervical cancer (HeLa) cells (Carlile et al. 2014), a chemical pulldown method, which enriched for Ψ sites prior to sequencing found that Ψ sites were under represented in 5′UTRs of mouse and human (H36KT) cells (Li et al. 2015). Prior enrichment of Ψ sites enabled the identification of thousands of sites transcriptome‐wide, compared to other studies finding only hundreds of sites.
Dynamic regulation of Ψ sites was conserved across species, as tissue specific and stress responsive Ψ sites were identified in animals and yeast. Strong, stimuli‐specific patterns of Ψ were induced for heat shock, addition of a viral mimic and oxidative stress (Schwartz et al. 2014a; Li et al. 2015). Ψ sites were also regulated by different cellular growth rates and nutrient availability (Carlile et al. 2014). Interestingly, Schwartz et al. (2014a) show that PUS7 Ψ mediates heat sensitivity in yeast, as yeast mutants have increased heat sensitivity and >200 PUS7 dependent Ψ sites are induced by heat shock. Ψ transcripts were expressed at higher levels in wild type than in PUS7 mutants during heat shock, suggesting a role for Ψ in stabilizing specific mRNAs in stress conditions.
The function of Ψ in mRNAs is unclear, however, Ψ is thought to help stabilize RNAs by promoting base stacking, pairing, and conformational stability. Ψ may also affect the translation of modified mRNAs (Davis 1995). For example, Ψ has been shown to convert nonsense codons into sense codons, thus ‘rewiring’ the genetic code (Karijolich and Yu 2011). However, the precise role of Ψ in translation is controversial as Ψ has been shown to both aid and inhibit translation in eukaryotic and bacterial systems, respectively, (Kariko et al. 2012; Hoernes et al. 2015). The locations and roles of this modification in Arabidopsis mRNAs is yet to be discovered.
OTHER MODIFICATIONS IN THE ARABIDOPSIS EPITRANSCRIPTOME
Of the over 100 RNA modifications discovered in RNA, high‐throughput methods of detection have been limited to detecting only a small subset of these modifications. Recently, a new hybrid method has been introduced, referred to as HAMR (High‐throughput Annotation of Modified Ribonucleotides), which is able to detect and predict modifications that affect Watson‐Crick base pairing transcriptome‐wide (Ryvkin et al. 2013). HAMR was trained using data from well characterized yeast tRNA modifications to predict the identity of several RNA modifications. Subsequently, HAMR was put to use on the Arabidopsis epitranscriptome, and several types of RNA modifications that perturb reverse transcription were predicted in all types of RNA classes (Vandivier et al. 2015).
Three types of RNA‐seq datasets were tested and compared in this study, (i) polyadenylated, (ii) small RNAs and (iii) degrading RNA. RNA modifications were enriched in Arabidopsis exons and 3’UTRs of uncapped, degrading mRNA and lncRNA transcripts and the same enrichment pattern was detected in two human cell lines, suggesting broad conservation and possible regulatory functions of these RNA modifications. It remains to be determined if RNA modifications are targeted to degrading transcripts, or if the RNA modifications serve as signals to mark transcripts for degradation. In addition, RNA modifications predicted by HAMR in stable mRNAs from the polyadenylated RNA‐seq data sets were enriched within introns that were annotated to be alternatively spliced in both plants and humans.
Distributions of specific RNA modifications were specific to different types of RNAs and depended on whether the transcripts were undergoing degradation. For example, degrading mRNA transcripts had much higher predicted levels of dihydrouridylation (D), N6‐isopentenyladenosylation (i6A) and threonylcarbamoyladenosylation (t6A) than stable mRNAs. Uncapped, degrading transcripts involved in various stress responses were enriched for HAMR‐predicted modifications, suggesting possible roles in gene regulation and stress responses for these mRNA modifications in Arabidopsis.
FUTURE DIRECTIONS AND CONCLUSIONS
The four base constituents of RNA are modified by over 100 different RNA modifications. This additional complexity of RNA is essential for basic functions, such as gene regulation and translation. The Arabidopsis epitranscriptome has now been mapped for several RNA modifications, which occur in different locations across transcripts, are inducible in response to abiotic and biotic stresses and have diverse roles in plant development, ranging from subtle (m5C) to dramatic (m6A) effects on plant growth. While the RNA modifying ‘writers’ have been investigated in plants, studies on potential ‘erasers’ and ‘readers’ are lacking. The Arabidopsis genome encodes over 200 RNA binding proteins which serve as potential readers and effectors of outcomes for RNA modifications (Lorkovic and Barta 2002). Furthermore, potential Arabidopsis ‘erasers’ from the ALKBH family are yet to be explored for roles in plant development and mediating dynamic regulation of RNA modifications (Mielecki et al. 2012). The ALKBH family of dioxygenases has diverse substrate specificities and are not limited to demethylation of adenosine (Aas et al. 2003; Jia et al. 2011). Specific Arabidopsis ALKBH family proteins may also remove additional RNA modifications. Further research is needed to elucidate the mechanisms and functional roles of mRNA modifications such as alternative splicing, and stress responses. Using small RNA guides, it is possible to artificially induce and block m6A and Ψ in mRNAs (Karijolich and Yu 2011; Chen et al. 2015b). This should enable the study of the specific functions of individual RNA modifications. There are many different ways that RNA modifications can affect RNA structure and interactions between RNA, RNA and proteins and even potentially RNA‐DNA interactions (Figure 3). The next steps for deciphering the Arabidopsis epitranscriptome include Ψ‐seq, mapping 2′‐O‐ribose methylations (Karijolich and Yu 2011; Birkedal et al. 2015), mapping N1‐methyladenosine (m1A) (Dominissini et al. 2016), single‐nucleotide resolution mapping of m6A (Ke et al. 2015), and determining potential reversibility, and the elusive targeting mechanism(s) for RNA modifications.
Figure 3.
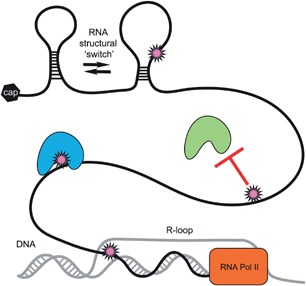
Potential functions of RNA modifications in mediating interactions between nucleic acids and nucleic acids and proteins RNA modifications have diverse chemical properties and can have different effects on RNA interactions. A spikey, pink ball is used to represent a generic RNA modification. RNA modifications can regulate protein binding to RNA through remodeling of local RNA structure (e.g. ‘m6A switches’), increasing or inhibiting protein binding. In addition, RNA modifications could potentially affect other types of interactions, such as R‐loops, which are RNA‐DNA hybrids.
ACKNOWLEDGEMENTS
This research was supported by ARC grants DP110103805 and FT13100525 awarded to I.S. and an APA and a GRDC PhD top‐up scholarship awarded to A.B.
Burgess A, David R, Searle IR (2016) Deciphering the epitranscriptome: A green perspective. J Integr Plant Biol 58: 822–835
Available online on May 12, 2016 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE ( 2003) Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863 [DOI] [PubMed] [Google Scholar]
- Abbasi‐Moheb L, Mertel S, Gonsior M, Nouri‐Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli‐Nieh S, Cremer K, Weissmann R, Tzschach A, Garshasbi M, Abedini SS, Najmabadi H, Ropers HH, Sigrist SJ, Kuss AW ( 2012) Mutations in NSUN2 cause autosomal‐recessive intellectual disability. Am J Hum Genet 90: 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi N, Kim HB, Park NI, Kim HS, Kim YK, Park YI, Choi SB ( 2010) APUM23, a nucleolar Puf domain protein, is involved in pre‐ribosomal RNA processing and normal growth patterning in Arabidopsis . Plant J 64: 960–976 [DOI] [PubMed] [Google Scholar]
- Alarcon CR, Goodarzi H, Lee H, Liu XH, Tavazoie S, Tavazoie SF ( 2015a) HNRNPA2B1 is a mediator of m(6)A‐dependent nuclear RNA processing events. Cell 162: 1299–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF ( 2015b) N6‐methyladenosine marks primary microRNAs for processing. Nature 519: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T, Souliere MF, Wille A, Jia XY, Fiegl H, Worle H, Micura R, Lusser A ( 2013) Long non‐coding RNAs as targets for cytosine methylation. RNA Biol 10: 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansmant I, Massenet S, Grosjean H, Motorin Y, Branlant C ( 2000) Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of psi(2819) in 21S mitochondrial ribosomal RNA. Nucleic Acids Res 28: 1941–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche F, Steinmetz F, Echeverria M ( 2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana . J Biol Chem 275: 27212–27220 [DOI] [PubMed] [Google Scholar]
- Baxter‐Roshek JL, Petrov AN, Dinman JD ( 2007) Optimization of ribosome structure and function by rRNA base modification. PLoS ONE 2: e174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berulava T, Rahmann S, Rademacher K, Klein‐Hitpass L, Horsthemke B ( 2015) N6‐adenosine methylation in MiRNAs. PLoS ONE 10: e0118438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal U, Christensen‐Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H ( 2015) Profiling of ribose methylations in RNA by high‐throughput sequencing. Angew Chem Int Ed Engl 54: 451–455 [DOI] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, Kellner S, Holter SM, Garrett L, Wurst W, Becker L, Klopstock T, Fuchs H, Gailus‐Durner V, de Angelis MH, Karadottir RT, Helm M, Ule J, Gleeson JG, Odom DT, Frye M ( 2014) Aberrant methylation of tRNAs links cellular stress to neuro‐developmental disorders. EMBO J 33: 2020–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Frye M ( 2014) Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol 31: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Kurowski A, Nichols J, Watt FM, Benitah SA, Frye M ( 2011) The RNA‐methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet 7: e1002403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Zhong SL, Mehra S, Song J, Graham N, Li HY, May S, Fray RG ( 2012) Adenosine methylation in Arabidopsis mRNA is associated with the 3 ' end and reduced levels cause developmental defects. Front Plant Sci 3: 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Rath‐Shambaugh ME, Ludwiczak R, Narayan P, Rottman F ( 1994) Characterization and partial purification of mRNA N6‐adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem 269: 17697–17704 [PubMed] [Google Scholar]
- Brown JW, Echeverria M, Qu LH ( 2003) Plant snoRNAs: Functional evolution and new modes of gene expression. Trends Plant Sci 8: 42–49 [DOI] [PubMed] [Google Scholar]
- Bruggeman Q, Garmier M, de Bont L, Soubigou‐Taconnat L, Mazubert C, Benhamed M, Raynaud C, Bergounioux C, Delarue M ( 2014) The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: A key factor of programmed cell death and a regulator of immunity in Arabidopsis . Plant Physiol 165: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Blumenthal RM ( 2002) Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT‐A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol 55: 431–444 [DOI] [PubMed] [Google Scholar]
- Burgess AL, David R, Searle IR ( 2015) Conservation of tRNA and rRNA 5‐methylcytosine in the kingdom Plantae. BMC Plant Biol 15: 199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME ( 2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Carlile TM, Rojas‐Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV ( 2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M, Hunt AG ( 2015) CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules 5: 1151–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ ( 2010) A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CTY, Pang YLJ, Deng WJ, Babu IR, Dyavaiah M, Begley TJ, Dedon PC ( 2012) Reprogramming of tRNA modifications controls the oxidative stress response by codon‐biased translation of proteins. Nat Commun 3: 937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR, 3rd , Ule J, Manley JL, Shi Y ( 2014) CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3' processing. Genes Dev 28: 2370–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, Gray MW ( 2000) Pseudouridine in RNA: What, where, how, and why. IUBMB Life 49: 341–351 [DOI] [PubMed] [Google Scholar]
- Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, Han D, Dominissini D, Dai Q, Pan T, He C ( 2015a) High‐resolution N(6) ‐methyladenosine (m(6) A) map using photo‐crosslinking‐assisted m(6) A sequencing. Angew Chem Int Ed Engl 54: 1587–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Jager G, Zheng B ( 2010) Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana . BMC Plant Biol 10: 201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, Li A, Yang Y, Jin KX, Zhao X, Li Y, Ping XL, Lai WY, Wu LG, Jiang G, Wang HL, Sang L, Wang XJ, Yang YG, Zhou Q ( 2015b) m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16: 289–301 [DOI] [PubMed] [Google Scholar]
- Chen Y, Sierzputowskagracz H, Guenther R, Everett K, Agris PF ( 1993) 5‐methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem‐loop of yeast phenylalanine transfer‐RNA. Biochemistry 32: 10249–10253 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu JK, Gong Z ( 2006) Mutations in ABO1/ELO2, a subunit of holo‐Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana . Mol Cell Biol 26: 6902–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA ( 2002) Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6‐methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res 30: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csepany T, Lin A, Baldick CJ, Jr. , Beemon K ( 1990) Sequence specificity of mRNA N6‐adenosine methyltransferase. J Biol Chem 265: 20117–20122 [PubMed] [Google Scholar]
- David R, Burgess AL, Parker BJ, Pulsford KE, Sibbritt T, Preiss T, Searle IR ( 2016) Transcriptome‐wide mapping of RNA 5‐methylcytosine in Arabidopsis mRNAs and ncRNAs. Genome Res dx.doi.org/10.6084/m9.figshare.3408193.v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR ( 1995) Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ ( 2002) rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 [DOI] [PubMed] [Google Scholar]
- Delaney KJ, Xu R, Zhang J, Li QQ, Yun KY, Falcone DL, Hunt AG ( 2006) Calmodulin interacts with and regulates the RNA‐binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol 140: 1507–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C ( 2015) Widespread occurrence of N6‐methyladenosine in bacterial mRNA. Nucleic Acids Res 43: 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F ( 1974) Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 71: 3971–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, Salmon‐Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob‐Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G ( 2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature 485: 201–206 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch‐Moshkovitz S, Peer E, Kol N, Ben‐Haim MS, Dai Q, Di Segni A, Salmon‐Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Dore LC, Amariglio N, Rechavi G, He C ( 2016) The dynamic N1‐methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin DT, Taylor RH ( 1975) The methylation state of poly A‐containing messenger RNA from cultured hamster cells. Nucleic Acids Res 2: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, Schaefer M ( 2013a) Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep 14: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M ( 2013b) The RNA methyltransferase Dnmt2 is required for efficient dicer‐2‐dependent siRNA pathway activity in Drosophila. Cell Rep 4: 931–937 [DOI] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R ( 2013) Transcriptome‐wide mapping of 5‐methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9: e1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crecy‐Lagard V ( 2012) Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 46: 69–95 [DOI] [PubMed] [Google Scholar]
- Fahiminiya S, Almuriekhi M, Nawaz Z, Staffa A, Lepage P, Ali R, Hashim L, Schwartzentruber J, Abu Khadija K, Zaineddin S, Gamal H, Majewski J, Ben‐Omran T ( 2014) Whole exome sequencing unravels disease‐causing genes in consanguineous families in Qatar. Clin Genet 86: 134–141 [DOI] [PubMed] [Google Scholar]
- Faure JD, Gingerich D, Howell SH ( 1998) An Arabidopsis immunophilin, AtFKBP12, binds to AtFIP37 (FKBP interacting protein) in an interaction that is disrupted by FK506. Plant J 15: 783–789 [DOI] [PubMed] [Google Scholar]
- Fray RG, Simpson GG ( 2015) The Arabidopsis epitranscriptome. Curr Opin Plant Biol 27: 17–21 [DOI] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H ( 2009) Coordination of cell proliferation and cell expansion mediated by ribosome‐related processes in the leaves of Arabidopsis thaliana . Plant J 59: 499–508 [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, Okamura H ( 2013) RNA‐methylation‐dependent RNA processing controls the speed of the circadian clock. Cell 155: 793–806 [DOI] [PubMed] [Google Scholar]
- Ge JH, Yu YT ( 2013) RNA pseudouridylation: New insights into an old modification. Trends Biochem Sci 38: 210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigova A, Duggimpudi S, Pollex T, Schaefer M, Kos M ( 2014) A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 20: 1632–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang XY, Golic KG, Jacobsen SE, Bestor TH ( 2006) Methylation of tRNA(AsP) by the DNA methyltransferase homolog Dnmt2. Science 311: 395–398 [DOI] [PubMed] [Google Scholar]
- Golovko A, Sitbon F, Tillberg E, Nicander B ( 2002) Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana . Plant Mol Biol 49: 161–169 [DOI] [PubMed] [Google Scholar]
- Hebrard C, Trap‐Gentil MV, Lafon‐Placette C, Delaunay A, Joseph C, Lefebvre M, Barnes S, Maury S ( 2013) Identification of differentially methylated regions during vernalization revealed a role for RNA methyltransferases in bolting. J Exp Bot 64: 651–663 [DOI] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I ( 1998) X‐linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Henras AK, Plisson‐Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE ( 2015) An overview of pre‐ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA 6: 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz R, Tovy A, Kirschenbaum M, Geffen M, Nozaki T, Adir N, Ankri S ( 2014) The entamoeba histolytica Dnmt2 homolog (ehmeth) confers resistance to nitrosative stress. Eukaryotic Cell 13: 494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hienzsch A, Deiml C, Reiter V, Carell T ( 2013) Total synthesis of the hypermodified RNA bases wybutosine and hydroxywybutosine and their quantification together with other modified RNA bases in plant materials. Chemistry 19: 4244–4248 [DOI] [PubMed] [Google Scholar]
- Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, Huttenhofer A, Erlacher MD ( 2015) Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res 44: 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Brockenbrough JS, Wu P, Aris JP ( 1997) Nop2p is required for pre‐rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol 17: 378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Orr‐Weaver TL ( 2011) Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci USA 108: 14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Qin Z, Wang M, Xu C, Feng G, Liu J, Meng Z, Hu Y ( 2010) The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J 61: 600–610 [DOI] [PubMed] [Google Scholar]
- Hunt AG, Xing D, Li QQ ( 2012) Plant polyadenylation factors: Conservation and variety in the polyadenylation complex in plants. BMC Genomics 13: 641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, Frye M ( 2013a) NSun2‐mediated cytosine‐5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 4: 255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M ( 2013b) The mouse cytosine‐5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 33: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Jin HL, Zhu JK ( 2009) Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana . BMC Genomics 10: 155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Alfonzo JD ( 2013) Transfer RNA modifications: Nature's combinatorial chemistry playground. Wires RNA 4: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C ( 2011) N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol 7: 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LH, Scott TK ( 1981) Transfer ribonucleic acid modification and its relationship to tumorous and nontumorous plant growth. Plant Physiol 67: 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Yu YT ( 2011) Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474: 395–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K, Muramatsu H, Keller JM, Weissman D ( 2012) Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine‐containing mRNA encoding erythropoietin. Mol Ther 20: 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker‐Scharff I, Moore MJ, Park CY, Vagbo CB, Kussnierczyk A, Klungland A, Darnell JE, Jr. , Darnell RB ( 2015) A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev 29: 2037–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, Ishak GE, Doherty D, Weksberg R, Ayub M, Windpassinger C, Ibrahim S, Frye M, Ansar M, Vincent JB ( 2012) Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal‐recessive intellectual disability. Am J Hum Genet 90: 856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoddami V, Cairns BR ( 2013) Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol 31: 458–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MY, Redman KL ( 2002) RNA methyltransferases utilize two cysteine residues in the formation of 5‐methylcytosine. Biochemistry 41: 11218–11225 [DOI] [PubMed] [Google Scholar]
- Kiss T ( 2001) Small nucleolar RNA‐guided post‐transcriptional modification of cellular RNAs. EMBO J 20: 3617–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D, Delcour J, Glasser AL, Desgres J, Vandenhaute J ( 1994) The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3'‐terminal loop of 18 S rRNA is essential in yeast. J Mol Biol 241: 492–497 [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Vandenhaute J, Tollervey D ( 1995) The 18S rRNA dimethylase Dim1p is required for pre‐ribosomal RNA processing in yeast. Genes Dev 9: 2470–2481 [DOI] [PubMed] [Google Scholar]
- Lawrence M, Daujat S, Schneider R ( 2016) Lateral thinking: How histone modifications regulate gene expression. Trends Genet 32: 42–56 [DOI] [PubMed] [Google Scholar]
- Leihne V, Kirpekar F, Vagbo CB, van den Born E, Krokan HE, Grini PE, Meza TJ, Falnes PO ( 2011) Roles of Trm9‐and ALKBH8‐like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res 39: 7688–7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Schubert V, Bornke F, Macas J, Schubert I ( 2007) Arabidopsis CBF5 interacts with the H/ACA snoRNP assembly factor NAF1. Plant Mol Biol 65: 615–626 [DOI] [PubMed] [Google Scholar]
- Li D, Zhang H, Hong Y, Huang L, Li X, Zhang Y, Ouyang Z, Song F ( 2014a) Genome‐wide identification, biochemical characterization, and expression analyses of the YTH domain‐containing RNA‐binding protein family in Arabidopsis and rice. Plant Mol Biol Rep 32: 1169–1186 [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C ( 2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: 592–597 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Li C, Hu S, Yu J, Song S ( 2014b) Transcriptome‐wide N(6)‐methyladenosine profiling of rice callus and leaf reveals the presence of tissue‐specific competitors involved in selective mRNA modification. RNA Biol 11: 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin‐George AO, Meydan C, Mason CE, Jaffrey SR ( 2015) Single‐nucleotide‐resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C ( 2014) A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol 10: 93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T ( 2015) N(6)‐methyladenosine‐dependent RNA structural switches regulate RNA‐protein interactions. Nature 518: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato S, Kalyna M, Dorner S, Kobayashi R, Krainer AR, Barta A ( 1999) atSRp30, one of two SF2/ASF‐like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev 13: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Barta A ( 2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA‐binding proteins from the flowering plant Arabidopsis thaliana . Nucleic Acids Res 30: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W ( 2000) Pre‐mRNA splicing in higher plants. Trends Plant Sci 5: 160–167 [DOI] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO ( 2014) Transcriptome‐wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae . PLoS ONE 9: e110799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, He C ( 2014) Unique features of the m6A methylome in Arabidopsis thaliana . Nat Commun 5: 5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin‐Horkawicz S, Rother KM, Helm M, Bujnicki JM, Grosjean H ( 2013) MODOMICS: A database of RNA modification pathways–2013 update. Nucleic Acids Res 41: D262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al‐Gazali L, Gleeson JG ( 2012) Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz‐like syndrome. J Med Genet 49: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, Gong Z, Bystrom AS, Schaffrath R, Breunig KD ( 2010) Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 76: 1082–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR ( 2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149: 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielecki D, Zugaj DL, Muszewska A, Piwowarski J, Chojnacka A, Mielecki M, Nieminuszczy J, Grynberg M, Grzesiuk E ( 2012) Novel AlkB dioxygenases–alternative models for in silico and in vivo studies. PLoS ONE 7: e30588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto‐Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T ( 2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA 103: 16598–16603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Helm M ( 2011) RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2: 611–631 [DOI] [PubMed] [Google Scholar]
- Motorin Y, Lyko F, Helm M ( 2010) 5‐methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res 38: 1415–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K ( 2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin‐mediated gynoecium patterning. Plant Cell 17: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulou A, Kossida S ( 2009) Phylogenetic analysis of the eukaryotic RNA (cytosine‐5)‐methyltransferases. Genomics 93: 350–357 [DOI] [PubMed] [Google Scholar]
- Perry RP, Kelley DE ( 1974) Existence of methylated messenger RNA in mouse L cells. Cell 1: 37–42 [Google Scholar]
- Phizicky EM, Hopper AK ( 2010) tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pih KT, Yi MJ, Liang YS, Shin BJ, Cho MJ, Hwang I, Son D ( 2000) Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis . Plant Physiol 123: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, Zhao X, Li A, Yang Y, Dahal U, Lou XM, Liu X, Huang J, Yuan WP, Zhu XF, Cheng T, Zhao YL, Wang XQ, Danielsen JMR, Liu F, Yang YG ( 2014) Mammalian WTAP is a regulatory subunit of the RNA N6‐methyladenosine methyltransferase. Cell Res 24: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA, D'Silva S, Kon Y, Phizicky EM ( 2013) tRNAHis 5‐methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae. RNA 19: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Chidester S, Zavala CV, Manos EJ, James SR, Karpf AR, Jones DA, Cairns BR ( 2007) Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev 21: 261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez V, Gonzalez B, Lopez A, Castello MJ, Gil MJ, Zheng B, Chen P, Vera P ( 2015) Loss of a conserved tRNA anticodon modification perturbs plant immunity. PLoS Genet 11: e1005586 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Richter U, Kuhn K, Okada S, Brennicke A, Weihe A, Borner T ( 2010) A mitochondrial rRNA dimethyladenosine methyltransferase in Arabidopsis . Plant J 61: 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET ( 2015) Structure and thermodynamics of N6‐methyladenosine in RNA: A spring‐loaded base modification. J Am Chem Soc 137: 2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, Wang LS ( 2013) HAMR: High‐throughput annotation of modified ribonucleotides. RNA 19: 1684–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F ( 2010) RNA methylation by Dnmt2 protects transfer RNAs against stress‐induced cleavage. Genes Dev 24: 1590–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, Carr SA, Lander ES, Fink GR, Regev A ( 2013) High‐resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon‐Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, Fink G, Regev A ( 2014a) Transcriptome‐wide mapping reveals widespread dynamic‐regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter‐Ovanesyan D, Habib N, Cacchiarelli D, Sanjana NE, Freinkman E, Pacold ME, Satija R, Mikkelsen TS, Hacohen N, Zhang F, Carr SA, Lander ES, Regev A ( 2014b) Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep 8: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafik A, Schumann U, Evers M, Sibbritt T, Preiss T ( 2016) The emerging epitranscriptomics of long noncoding RNAs. Biochim Biophys Acta 1859: 59–70 [DOI] [PubMed] [Google Scholar]
- Sharma S, Yang J, Watzinger P, Kotter P, Entian KD ( 2013) Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res 41: 9062–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ ( 1976) Capping of eucaryotic mRNAs. Cell 9: 645–653 [DOI] [PubMed] [Google Scholar]
- Shugart L ( 1972) A possible age‐related modification of phenylalanine transfer RNA from wheat tissue. Exp Gerontol 7: 251–262 [DOI] [PubMed] [Google Scholar]
- Sibert BS, Patton JR ( 2012) Pseudouridine synthase 1: A site‐specific synthase without strict sequence recognition requirements. Nucleic Acids Res 40: 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T ( 2012) Widespread occurrence of 5‐methylcytosine in human coding and non‐coding RNA. Nucleic Acids Res 40: 5023–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A ( 2008) DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Thomas PE, Wu X, Liu M, Gaffney B, Ji G, Li QQ, Hunt AG ( 2012) Genome‐wide control of polyadenylation site choice by CPSF30 in Arabidopsis . Plant Cell 24: 4376–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhisa JG, Vijayan P, Feldmann KA, Browse JA ( 1998) Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA dimethylase. Plant Cell 10: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Batlle E, Ribas de Pouplana L ( 2014) Role of tRNA modifications in human diseases. Trends Mol Med 20: 306–314 [DOI] [PubMed] [Google Scholar]
- Towns WL, Begley TJ ( 2012) Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: Activities, predications, and potential roles in human health. DNA Cell Biol 31: 434–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F ( 2012) RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 19: 900–905 [DOI] [PubMed] [Google Scholar]
- Vandivier LE, Campos R, Kuksa PP, Silverman IM, Wang LS, Gregory BD ( 2015) Chemical modifications mark alternatively spliced and uncapped messenger RNAs in Arabidopsis . Plant Cell 27: 3024–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M ( 2004) The immunophilin‐interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol 134: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Tang K, Zhang D, Xie S, Zhu X, Wang Z, Lang Z ( 2015) Transcriptome‐wide high‐throughput deep m(6)A‐seq reveals unique differential m(6)A methylation patterns between three organs in Arabidopsis thaliana . Genome Biol 16: 272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C ( 2014a) N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature 505: 117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C ( 2015) N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC ( 2014b) N6‐methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ ( 2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7: 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Moss B ( 1977) Nucleotide sequences at the N6‐methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16: 1672–1676 [DOI] [PubMed] [Google Scholar]
- Wieckowski Y, Schiefelbein J ( 2012) Nuclear ribosome biogenesis mediated by the DIM1A rRNA dimethylase is required for organized root growth and epidermal patterning in Arabidopsis . Plant Cell 24: 2839–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Brockenbrough JS, Paddy MR, Aris JP ( 1998) NCL1, a novel gene for a non‐essential nuclear protein in Saccharomyces cerevisiae . Gene 220: 109–117 [DOI] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL, Yang YG ( 2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 61: 507–519 [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S ( 2008) Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2‐mediated leaf variegation in Arabidopsis . Plant Cell 20: 1786–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL ( 2008) A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana . PLoS ONE 3: e2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, Jin KX, Wang X, Huang CM, Fu Y, Ge XM, Song SH, Jeong HS, Yanagisawa H, Niu Y, Jia GF, Wu W, Tong WM, Okamoto A, He C, Rendtlew Danielsen JM, Wang XJ, Yang YG ( 2014) FTO‐dependent demethylation of N6‐methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24: 1403–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu ZK, Bosmans RPG, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia GF, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C ( 2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong SL, Li HY, Bodi Z, Button J, Vespa L, Herzog M, Fray RG ( 2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex‐specific splicing factor. Plant Cell 20: 1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB ( 2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou WB, Karcher D, Bock R ( 2013) Importance of adenosine‐to‐inosine editing adjacent to the anticodon in an Arabidopsis alanine tRNA under environmental stress. Nucleic Acids Res 41: 3362–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hua D, Chen Z, Zhou Z, Gong Z ( 2009) Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis . Plant J 60: 79–90 [DOI] [PubMed] [Google Scholar]
- Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgue‐Hamard V, Lafontaine DL ( 2015) The human 18S rRNA base methyltransferases DIMT1L and WBSCR22‐TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 26: 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]


