Abstract
The cellular response evoked by a hexanuclear platinum complex, Pt6L4 (1), is reported. Compound 1, a 3-nm octahedral cage formed by self-assembly of six Pt(II) centers and four 2,4,6-tris(4-pyridyl)-1,3,5-triazine ligands (L), exhibits promising in vitro potency against a panel of human cancer cell lines. Unlike classical platinum-based anticancer agents, 1 interacts with DNA in a non-covalent, intercalative manner and promotes DNA condensation. In cancer cells, 1 induces DNA damage, upregulates p53, its phosphorylated form phospho-p53 and its downstream effector, p21, as well as both apoptosis and senescence.
Keywords: Self-assembly, Cage, Anticancer, DNA damage
Graphical Abstract
INTRODUCTION
Platinum-based drugs, cisplatin, carboplatin, and oxaliplatin, are routinely used in clinics throughout the world to treat cancer patients.1–3 Approximately 50% of cancer patients undergoing chemotherapy receive one of these platinum agents at some point during the course of their treatment. Despite such widespread use of platinum drugs, drawbacks associated with these agents, such as lack of selectivity, dose-limiting toxic side effects, low bioavailability, and short half-life in blood,4–5 have motivated research into alternative metal-based anticancer agents.6 The search for new chemotherapeutics has included platinum containing compounds, many of them similar in structure to cisplatin. Very few clinically relevant breakthroughs, however, have been made by using this approach. Polynuclear platinum complexes offer an approach to widen the spectrum of activity of current FDA-approved platinum drugs. Here, we combine supramolecular self-assembly and bioinorganic chemistry to develop a hexanuclear platinum compound that potently inhibits proliferation of cancer cells.
Coordination-driven self-assembly is one of the most effective methodologies in inorganic chemistry for preparing supramolecular metal complexes of high structural complexity and diversity.7–10 A large numbers of inorganic supramolecules developed over the past two decades is a testimonial to the rapid growth of this field.7 Inorganic supramolecular structures are also receiving increased attention in drug discovery, owing to their well-defined, unique chemical structures and diverse host-guest chemistry.11 The earliest studies of this kind highlight the unique DNA binding profile and biological activity of a series of metallocylinders.12–16 More recent is work unveiling the in vitro and in vivo anticancer activity of Ru- and Pt-based supramolecules.17–20 We and others have utilized metallocages to encapsulate and deliver biologically active small molecules to cancer cells.21–26 Because the application of inorganic supramolecules in medicine is still in its infancy, fundamental studies aimed at improving our understanding of the biological properties of these complexes are of particular interest.
In this article we present an in-depth mechanistic investigation of the origin of the antiproliferative activity of a self-assembled platinum cage, 1. The cytotoxicity of 1 toward several human cancer lines was determined and the mode of cell death characterized. Detailed biophysical assays were conducted to probe the DNA binding properties of 1.
RESULTS AND DISCUSSION
Synthesis and characterization of the Pt6L4 cage (1)
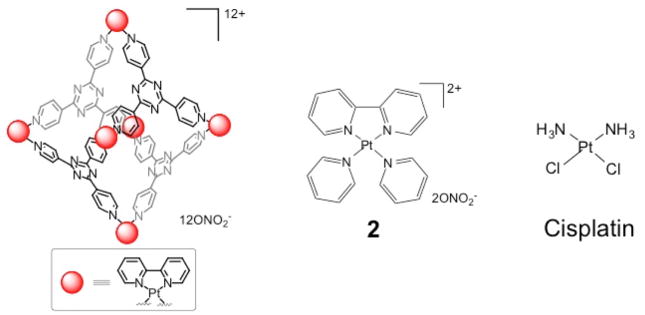
The Pt6L4 cage was prepared using a previously reported procedure.27 The octahedral hexanuclear platinum cage, comprising six Pt(II) centers and four 2,4,6-tris(4-pyridyl)-1,3,5-triazine ligands (L), was self-assembled using a 1-adamantane carboxylic acid template, which was later removed via chloroform extraction. The proton NMR spectrum of 1 (Figure S1) matched that of the previous report and confirmed a 6:4 ratio of Pt(II) centers to pyridyl ligands. Diffusion-Ordered Spectroscopy (DOSY) NMR was used to measure the size of the structure.25,28 The Ddiff value for 1 is 7.8 × 10−11 m2/s in DMSO at R.T. (Figure S2), leading to a hydrodynamic radius (r) of 1.4 nm, calculated using the Stokes-Einstein equation (Ddiff = kBT/6πηr, kB: Boltzmann constant, T: temperature, η: dynamic viscosity). This value agrees with that expected from the crystal structure of the platinum cage.27
Cytoxicity profiles
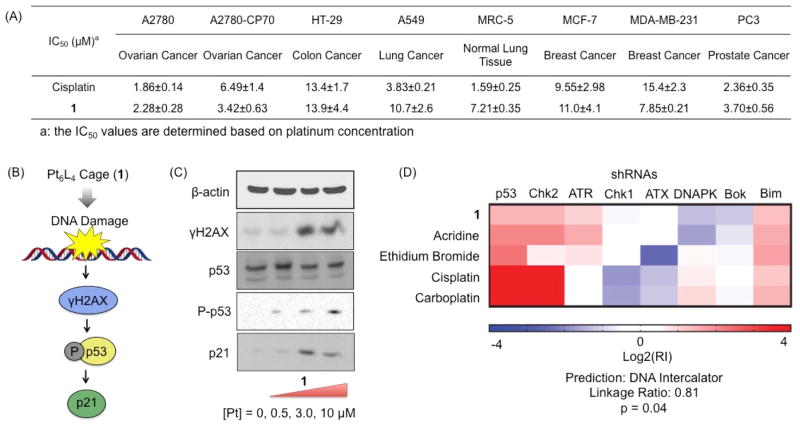
The cytotoxicity of the Pt6L4 cage (1) was assessed in a panel of human cancer and normal cell lines using the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. The cells were incubated with the Pt compounds for 72 h, and the IC50 values (concentrations required to induce 50% viability) were derived from dose–response curves (Figure 1A). All of the IC50 values refer to platinum concentration. In A2780, HT-29, MCF-7, and PC3 cells, 1 exhibits IC50 values comparable to those of cisplatin. Compound 1 displays higher cytotoxicity against A2780CP70 and MDA-MB-231 cells (IC50 = 3.42±0.63 μM for A2780CP70 and IC50 = 7.85±0.21 μM for MD-MBA-231) than cisplatin (IC50 = 6.49±1.40 μM for A2780CP70 and IC50 = 15.4±2.3 μM for MD-MBA-231). Because A2780CP70 and MDA-MB-231 cells are intrinsically resistant to cisplatin, these data show that 1 has the potential to target cispla-tin-resistant cells. To provide insight into the therapeutic potential of 1, we conducted cytotoxicity studies with normal lung fibro-blast MRC5 cells. The platinum cage 1 was reasonably potent toward MRC5 cells (IC50 = 7.21±0.35 μM), but not to the same extent as cisplatin (IC50 = 1.59±0.25 μM).
Figure 1.
Cytotxicity and DNA damage: (A) cytotoxicity profiles of 1 and cisplatin against a panel of human cancer and normal cell lines; (B) DNA damage pathway provoked by 1; (C) Immunoblotting analysis of the biomarkers corresponding to DNA damage pathway; (D) the RNAi signature assay of 1.
Intracellular target
In order to elucidate the mechanism of action, the subcellular target of the platinum cage, 1 was probed.
DNA damage
To confirm genomic DNA as the main cellular target responsible for the cell-killing function of 1, we probed for DNA damage using a number of molecular biological methods. Immunoblotting analysis was conducted to monitor changes in expression of biomarkers related to DNA damage pathways (Figure 1B). A2780 cells incubated with 1 ([Pt] = 10 μM) for 72 h showed a marked increase in phosphorylated H2AX (γH2AX), p53, phosphorylated p53 (P-p53), and p21 (Figure 1C), indicative of DNA damage. An increase in p21 mRNA expression was also observed upon 1 treatment by RT-qPCR (Figure S4). A newly developed RNAi-based signature assay was used to further clarify the intracellular behavior of the cage.29–33 This methodology uses a fluorescence competition assay whereby lymphoma cells that are partially infected with one of eight different short hairpin RNAs (shRNAs). Depending on the drug survival advantage or disadvantage conferred by a given shRNA, shRNA-bearing cells will either enrich or deplete relative to the uninfected population. The collective responses of these cells comprise the signatures, which have been obtained for all classes of clinically used cytotoxic agents. The signature of a new compound is compared to Ovarian cancer cells, A2780 were treated with 1 (10 μM platinum concentration) and, after 5 h incubation, the nucleus, cytosol, mitochondria, membrane were isolated and the content of 1 in each fraction was determined by graphite furnace atomic absorption spectroscopy (GFAAS) (Figure S1). The Pt cage (1) was taken up higher by A2780 cells than cisplatin (Figure S3A). Importantly, 1 was found to penetrate the nucleus, giving it access to genomic DNA (23% of the internalized complex was in the nucleus). Genomic DNA extracted from the nuclei of A2780 cells treated with 1 (10 μM for 5 h followed by incubation in fresh media for an additional 19 h) displayed comparable platinum levels (42.9±17.2 Pt/106 bases) to those extracted from cisplatin treated cells (40.9±1.6 Pt/106 bases). Taken together, the results show that 1 is able to accumulate in the nucleus and target genomic DNA. those of a reference set of drugs using a probabilistic K-nearest-neighbors algorithm. The algorithm then determines whether it belongs to a class in the reference set or a new category that is not represented in the reference set. As shown in Figure 1D, 1 does not share a mechanism of action with cisplatin or carboplatin. Cisplatin, and other DNA cross-linkers in the data set, have high positive shp53:shChk2 log2(RI) values (e.g. 4.2:4.3 for cisplatin), whereas the shp53:shChk2 ratio is 1.5:1.4 for the cage. Against the entire set of drugs, including those with unknown mechanisms, the closest identification was with acridine, a DNA intercalator. The algorithm classified the Pt cage as belonging to the category of DNA intercalators with a p-value of 0.04. Collectively, these results suggest that the Pt cage (1) is a DNA damaging agent and, unlike the classic Pt-based agents, DNA damage is caused by non-covalent interactions.
DNA binding
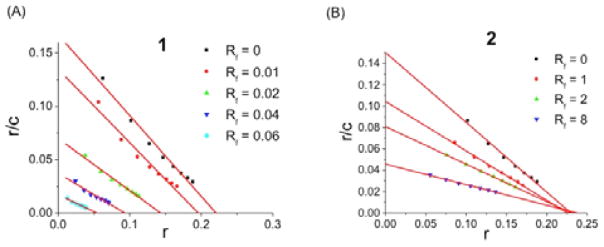
The nature of the non-covalent interaction between DNA and 1 was investigated by analyzing a competitive Scatchard plot.34 This method can determine the DNA binding mode by profiling the inhibition of ethidium bromide binding in the presence of increasing amounts of added competitor. There are four different categories: Type A (competitive), Type D (non-competitive), Type B (both), and Type C (no inhibition). Metallointercalators generally belong to Type A. As shown in Figure 2A, 1 displays Type B behavior, which is a combination of competitive (change of slope) and noncompetitive (change of intercept on the abscissa) inhibition. Competitive inhibition, characteristic of intercalation, is likely to involve the platinum-bipyridine corners. To evaluate this hypothesis, a mononuclear analog [Pt(2,2′-bipyridyl)(pyridine)2](NO3)2 (2), reminiscent of the platinum-bipyridine corners found in 1, was prepared and subjected to a Scatchard analysis.35 As anticipated, 2 exhibited Type A behavior (Figure 2B). To interpret the noncompetitive aspect found for 1, a TEM study was carried out to investigate the effect of 1 on DNA structure. Remarkably, treatment with 1 led to condensation of pBR322 plasmid DNA (Figure S5). This result is tentatively attributed to the high overall positive charge (12+) and multiple DNA intercalation sites of 1. Thus, the data clearly indicate that 1 interacts with DNA in a non-covalent manner, resulting in both DNA intercalation and condensation.
Figure 2.
DNA binding: Scatchard analysis of DNA-binding mode of 1 (A) and 2 (B) (r is the ratio of bound EB to total nucleotide concentration; c is the concentration of EB; Rf is the ratio of the compounds to total nucleotide concentration).
Cellular response
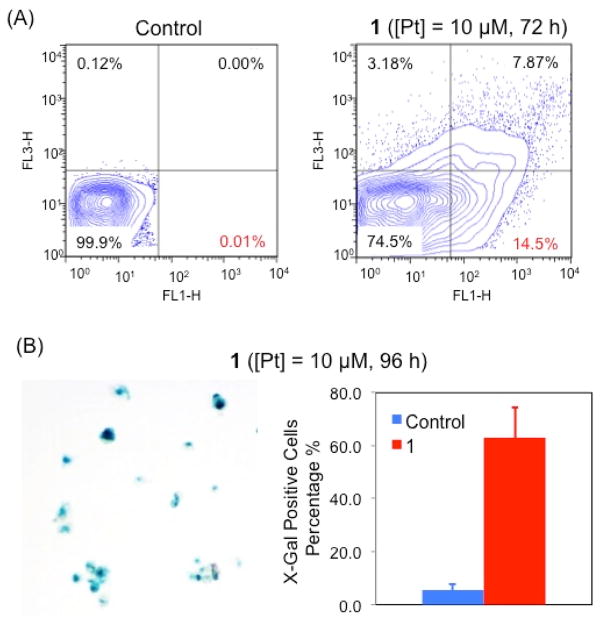
Apoptosis and senescence represent two major cellular responses to DNA damage. Apoptosis was studied using the Annexin V/PI assay. In healthy cells, phosphatidylserine (PS) is located on the cytoplasmic surface of the cell membrane, but in cells undergoing apoptosis, PS is translocated from the inner to the outer side of the plasma membrane, exposing PS to the external cellular environment where it can be detected by annexin V conjugates. Combining Annexin V and PI, both early and late stage apoptosis can be detected. By using the dual staining Annexin V/PI approach, the occurrence of apoptosis was studied in A2780 cells treated with 1 ([Pt] = 10 μM for 72 h) (Figure 3A). This platinum cage induces significant populations of cells to under early- and late-stage apoptosis (14.5% and 7.9%, respectively).
Figure 3.
Cellular response to DNA damage: (A) Annexin V/PI flow cytometric analysis of the apopotic events of A2780 cells treated with 1 ([Pt] = 10 μM); (B) X-gal staining studies of the senescent events of A2780 cells treated with 1 ([Pt] = 10 μM)..
Senescence, which causes permanent cell cycle arrest, is an another consequence of DNA damage.36–37 Upon treatment with 1 ([Pt] = 10 μM), the proliferation of A2780 cells was inhibited, as shown in Figure S6. Most of the treated cells were alive, but they were much larger in size compared to untreated cells (Figure S6). Cell cycle analysis using flow cytometry showed A2780 cells dosed with 1 (([Pt] = 10 μM) arrested at the G1 phase (Figure S7), consistent with senescence. To confirm the occurrence of senescence, X-gal (a marker of senescence) staining studies were carried out using 1-treated A2780 cells. As shown in Figure 4B, 60% of the treated group were deemed X-gal positive (or undergoing senescence), significantly higher than the control group. Collectively our cell-based studies strongly support that 1 triggers both apoptosis and senescence owing to DNA damage.
CONCLUSIONS
We present a detailed analysis of the cytotoxic mechanism of a Pt6L4 cage, 1. The platinum cage exhibits micromolar potency against a panel of human cancer cells (comparable to that of cisplatin) and no cross-resistance with cisplatin. The Pt6L4 cage is taken up in large amounts by cancer cells, accumulating in the nucleus and inducing DNA damage. RNAi-based signature assays suggest that DNA damage most likely results from non-covalent interactions. Biophysical analysis confirmed that 1 interacts with DNA in a non-covalent manner; these studies showed unambiguously that 1 is able to intercalate between DNA base pairs and induce DNA condensation. Strikingly, 1-mediated DNA damage leads to both apoptosis and senescence. Overall, this study provides much needed information on the biological properties of a supramolecular ensemble, which could pave the way for further pre-clinical studies with novel supramolecular inorganic constructs.
EXPERIMENTAL SECTION
General information
The compounds Pt(bpy)Cl2 and 2 were prepared according to previous reports.27,35 All reagents were purchased from Strem, Aldrich, or Alfa and used without further purification under normal atmospheric conditions. Calf-thymus DNA was purchased from Invitrogen, pBR322 plamid DNA from New England Biolabs and deuterated solvents were purchased from Cambridge Isotope Laboratory (Andover, MA). 1H NMR and DOSY NMR spectra were recorded on a Bruker AVANCE-400 NMR spectrometer with a Spectro Spin superconducting magnet in the Massachusetts Institute of Technology Department of Chemistry Instrumentation Facility (MIT DCIF). Chemical shifts in 1H NMR spectra were referenced to solvent signals (1H NMR: DMSO at δ = 2.50 ppm). Electrospray ionization mass spectrometry (ESI-MS) spectra were obtained on an Agilent Technologies 1100 Series liquid chromatography/MS instrument. UV-vis spectra were recorded on a Cary 50 spectrophotometer. UV-vis experiments were performed in septum-capped UV-vis cells (Starna). Fluorescence spectra were obtained on a Quanta Master 4 L-format scanning spectrofluorimeter (Photon Technology International) at 25 or 37 °C. GFAAS measurements were obtained on a Perkin Elmer AAnalyst 600 spectrometer. Distilled water was purified by passage through a Millipore Milli-Q Biocel water purification system (18.2 MΩ) with a 0.22 μm filter.
Synthesis and characterization of the Pt6L4 cage (1)
To a mixture of [Pt(bpy)Cl2] (124 mg, 0.29 mmol) and silver nitrate (100 mg, 0.59 mmol) was added 3 mL water, and the suspension was heated at 80 °C for 3 h with protection from light. The resulting suspension was filtered, and the clear aqueous solution was added to the ligand (61 mg, 0.20 mmol). The suspension with a final volume of 10 mL was heated at 100 °C for 40 min, and 50 μL 70 % HNO3 and 1-adamantane carboxylic acid (35 mg, 0.20 mmol) was added. The mixture was further heated at 80 °C overnight, and then was cooled down to R.T.. 1-adamantane carboxylic acid was removed by extraction with CHCl3 (4 × 10 mL), and the aqueous solution was evaporated to dryness at 55 °C under reduced pressure. The solid residue was redissolved in 10 mL water, and the mixture was filtered via a 0.2 μm syringe filter. To the clear solution was added 40 μL of 70% HNO3 slowly with stirring at 0 °C. Upon addition of HNO3, a white precipitate formed immediately. The product was collected by centrifugation, washed with acetone, and dried in vacuum. Yield: 130 mg, 39 %. 1H NMR (400 MHz, D2O) δ 9.42 (d, J = 6.8 Hz, 24H), 8.88 (d, J = 6.8 Hz, 24H), 8.48 (d, J = 8.4 Hz, 12H), 8.41 (t, J = 7.6 Hz, 12H), 7.93 (d, J = 5.6 Hz, 12H), 7.60 (t, J = 6.4 Hz, 12H); DOSY NMR (400 MHz, DMSO-d6, R.T.): Ddiff = 7.8 × 10−11 m2/s and r = 1.4 nm.
RNAi signatures29–33
Eμ-Mycp19arf-/- lymphoma cells were infected with GFP-tagged shRNAs such that 15–25% of the population were GFP positive. An eighth of a million cells in 250 μL B-cell media (BCM) were then seeded into 24-well plates. For wells that would remain untreated as a control, only 1/16th of a million cells were seeded. Next, 250 μL of medium containing the active agent was added to the cells. After 24 h, 300 μL of cells from untreated wells were removed and replaced by 300 μL fresh BCM. All wells then received 500 μL of BCM before being placed in the incubator for another 24 h. At 48 h, cells transduced with the control vector, MLS, were checked for viability via FACS on a FACScan instrument (BD Biosciences) using propidium iodide as a live/dead marker. Next, a 700 μL aliquot of untreated were removed from the wells and replaced with 700 μL of fresh media, followed by an additional 1 mL of fresh media. Wells for which the compound had killed 80–90% of cells (LD80-90) were then diluted further by adding 1 mL of BCM. Finally, at 72 h, all wells for which an LD80-90 was achieved, as well as the untreated samples, were analyzed via FACS to determine GFP% enrichment. Linkage ratios (LR) and p-values were generated as described previously.29–33 All FACS was conducted using a FACScan (BD Biosciences).
Immunoblotting analysis
A2780 cells (5 × 105 cells) were incubated with the Pt complex ([Pt] = 0–10 μM) for 72 h at 37 °C. Cells were washed with PBS, scraped into SDS-PAGE loading buffer (64 mM Tris-HCl (pH6.8)/9.6% glycerol/2% SDS/5% β-mercaptoethanol/0.01% Bromophenol Blue), and incubated at 95 °C for 10 min. Whole cell lysates were resolved by 4–20 % sodium dodecylsulphate polyacylamide gel electrophoresis (SDS-PAGE; 200 V for 25 min) followed by electro transfer to a poly-vinylidene difluoride membrane, PVDF (350 mA for 1 h). Membranes were blocked in 5% (w/v) non-fat milk in PBST (PBS/0.1% Tween 20) and incubated with the appropriate primary antibodies (Cell Signalling Technology and Santa Cruz). After incubation with horseradish peroxidase-conjugated secondary antibodies (Cell Signalling Technology), immune complexes were detected with the ECL detection reagent (BioRad) and analyzed by using an Alpha Innotech ChemiImagerTM 5500 fitted with a chemiluminescence filter.
Scatchard analysis
For all fluorescence measurements, the excitation wavelength was 530 nm, and the emission was monitored between 540 and 740 nm. The buffer used in these studies was PBS with pH = 7.4. For the Scatchard analysis, 1 (Rf = 0.01–0.06) or 2 (Rf = 0–8) was incubated with 4 μM of calf thymus DNA for 1 min at room temperature before adding ethidium bromide. The ethidium bromide concentration varied from 1 to 8 μM. A plot of r/cf versus r is described by r/cf = K(n - r), where cf is the concentration of unbound ethidium bromide and r is the ratio of bound ethidium bromide to total nucleotide concentration [DNA]. The plot therefore provides the intrinsic binding constant (K) for ethidium bromide (slope) and the maximum value of r (n, the intercept of the abscissa).
TEM study
pBR322 DNA (24 μM) was mixed with or without the Pt cage ([Pt] = 18 μM) in aqueous solution. The aqueous sample (3.5 μL) was adsorbed onto glow discharged carbon-coated TEM grids for 4 min and then wiped away, followed by staining using 3.5 μL 2 % aqueous uranyl acetate for 45 sec. Imaging was performed using an JEOL 1400 operated at 80 kV.
Supplementary Material
Scheme 1.
The platinum compounds studied in this project.
Acknowledgments
This work was supported by National Cancer Institute Grant CA34992 and the Kathy and Curt Marble Cancer Research Fund from the Frontier Research Program of the Koch Institute at MIT. Spectroscopic instrumentation at the MIT DCIF is maintained with funding from NIH Grant 1S10RR13886-01.
Footnotes
ASSOCIATED CONTENT
Experimental details regarding the characterization of 1, cell culture, the MTT assay, Pt cellular distribution, Pt content on DNA, TEM images, and cell cycle analysis are presented in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rosenber B, Vancamp L, Trosko JE, Mansour VH. Nature. 1969;222:385. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 2.Canetta R, Rozencweig M, Carter SK. Cancer Treat Rev. 1985;12:125. doi: 10.1016/0305-7372(85)90027-1. [DOI] [PubMed] [Google Scholar]
- 3.Misset JL, Bleiberg H, Sutherland W, Bekradda M, Cvitkovic E. Critical Rev Onco Hema. 2000;35:75. doi: 10.1016/s1040-8428(00)00070-6. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Lippard SJ. Nat Rev Drug Discov. 2005;4:307. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 5.Kelland L. Nat Rev Cancer. 2007;7:573. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 6.Wheate NJ, Walker S, Craig GE, Oun R. Dalton Transc. 2010;39:8113. doi: 10.1039/c0dt00292e. [DOI] [PubMed] [Google Scholar]
- 7.Cook TR, Zheng YR, Stang PJ. Chem Rev. 2012;113:734. doi: 10.1021/cr3002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarty R, Mukherjee PS, Stang PJ. Chem Rev. 2011;111:6810. doi: 10.1021/cr200077m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caulder DL, Raymond KN. Acc Chem Res. 1999;32:975. [Google Scholar]
- 10.Fujita M, Ogura K. Coord Chem Rev. 1996;148:249. [Google Scholar]
- 11.Cook TR, Vajpayee V, Lee MH, Stang PJ, Chi KW. Acc Chem Res. 2013;46:2464. doi: 10.1021/ar400010v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malina J, Hannon MJ, Brabec V. Chem - Eur J. 2015 doi: 10.1002/chem.201501307. ASAP. [DOI] [PubMed] [Google Scholar]
- 13.Phongtongpasuk S, Paulus S, Schnabl J, Sigel RKO, Spingler B, Hannon MJ, Freisinger E. Angew Chem, Int Ed. 2013;52:11513. doi: 10.1002/anie.201305079. [DOI] [PubMed] [Google Scholar]
- 14.Terenzi A, Ducani C, Blanco V, Zerzankova L, Westendorf AF, Peinador C, Quintela JM, Bednarski PJ, Barone G, Hannon MJ. Chem - Eur J. 2012;18:10983. doi: 10.1002/chem.201201519. [DOI] [PubMed] [Google Scholar]
- 15.Ducani C, Leczkowska A, Hodges NJ, Hannon MJ. Angew Chem, Int Ed. 2010;49:8942. doi: 10.1002/anie.201004471. [DOI] [PubMed] [Google Scholar]
- 16.Boer DR, Kerckhoffs JMCA, Parajo Y, Pascu M, Uson I, Lincoln P, Hannon MJ, Coll M. Angew Chem, Int Ed. 2010;49:2336. doi: 10.1002/anie.200906742. [DOI] [PubMed] [Google Scholar]
- 17.Mishra A, Chang Lee S, Kaushik N, Cook TR, Choi EH, Kumar Kaushik N, Stang PJ, Chi K-W. Chem - Eur J. 2014;20:14410. doi: 10.1002/chem.201403372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grishagin IV, Pollock JB, Kushal S, Cook TR, Stang PJ, Olenyuk BZ. Proc Natl Acad Sci U S A. 2014;111:18448. doi: 10.1073/pnas.1418712111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vajpayee V, Lee Sm, Park JW, Dubey A, Kim H, Cook TR, Stang PJ, Chi K-W. Organometallics. 2013;32:1563. doi: 10.1021/om301174s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubey A, Min JW, Koo HJ, Kim H, Cook TR, Kang SC, Stang PJ, Chi KW. Chem - Eur J. 2013;19:11622. doi: 10.1002/chem.201300870. [DOI] [PubMed] [Google Scholar]
- 21.Freudenreich J, Dalvit C, Suss-Fink G, Therrien B. Organometallics. 2013;32:3018. [Google Scholar]
- 22.Schmitt F, Freudenreich J, Barry NPE, Juillerat-Jeanneret L, Suss-Fink G, Therrien B. J Am Chem Soc. 2012;134:754. doi: 10.1021/ja207784t. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt F, Freudenreich J, Barry NPE, Juillerat-Jeanneret L, Suss-Fink G, Therrien B. J Am Chem Soc. 2012;134:754. doi: 10.1021/ja207784t. [DOI] [PubMed] [Google Scholar]
- 24.Pitto-Barry A, Zava O, Dyson PJ, Deschenaux R, Therrien B. Inorg Chem. 2012;51:7119. doi: 10.1021/ic202739d. [DOI] [PubMed] [Google Scholar]
- 25.Zheng YR, Suntharalingam K, Johnstone TC, Lippard SJ. Chem Sci. 2015;6:1189. doi: 10.1039/c4sc01892c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis JEM, Gavey EL, Cameron SA, Crowley JD. Chem Sci. 2012;3:778. [Google Scholar]
- 27.Kusukawa T, Fujita M. J Am Chem Soc. 2002;124:13576. doi: 10.1021/ja020712k. [DOI] [PubMed] [Google Scholar]
- 28.Megyes T, Jude H, Grosz T, Bako I, Radnai T, Tarkanyi G, Palinkas G, Stang PJ. J Am Chem Soc. 2005;127:10731. doi: 10.1021/ja0523690. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Pritchard JR, Williams RT, Lauffen-burger DA, Hemann MT. Nat Chem Biol. 2011;7:92. doi: 10.1038/nchembio.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard JR, Bruno PM, Gilbert LA, Capron KL, Lauffenburger DA, Hemann MT. Proc Natl Acad Sci U S A. 2013;110:E170. doi: 10.1073/pnas.1210419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard JR, Bruno PM, Hemann MT, Lauffen-burger DA. Mol BioSyst. 2013;9:1604. doi: 10.1039/c2mb25459j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suntharalingam K, Lin W, Johnstone TC, Bruno PM, Zheng YR, Hemann MT, Lippard S. J J Am Chem Soc. 2014;136:14413. doi: 10.1021/ja508808v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suntharalingam K, Awuah SG, Bruno PM, Johnstone TC, Wang F, Lin W, Zheng YR, Page JE, Hemann MT, Lippard SJ. J Am Chem Soc. 2015;137:2967. doi: 10.1021/ja511978y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe-Grant M, Wu KC, Bauer WR, Lippard SJ. Biochemistry. 1976;15:4339. doi: 10.1021/bi00664a031. [DOI] [PubMed] [Google Scholar]
- 35.Cusumano M, Di Pietro ML, Giannetto A. Inorg Chem. 2006;45:230. doi: 10.1021/ic050880o. [DOI] [PubMed] [Google Scholar]
- 36.Rodier F, Campisi J. J Cell Biol. 2011;192:547. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornsby PJ. J Clinical Oncology. 2007;25:1852. doi: 10.1200/JCO.2006.10.3101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.