Abstract
Introduction
Sagittal jaw growth is influenced during puberty by a ratio of androgens and estrogens. The CYP19A1 (formerly CYP19) gene encodes the cytochrome P450 enzyme aromatase (estrogen synthetase), which converts testosterone to estrogen. Genetic variations including single nucleotide polymorphisms might regulate CYP19A1 gene expression or the function of the aromatase protein and thus influence sagittal jaw growth.
Methods
The annual sagittal jaw growth in 92 pubertal orthodontic patients was determined by using pretreatment and posttreatment cephalometric radiographs. Single nucleotide polymorphisms rs2470144 and rs2445761 were genotyped and haplotypes constructed. Associations between genotypes or haplotypes and the annual sagittal growth were estimated by using JMP (version 9.0; SAS Institute, Cary, NC).
Results
Two single nucleotide polymorphisms were significantly associated with average differences in annual sagittal jaw growth in boys. Haplotype analysis demonstrated that haplotypes Trs2470144Trs2445761 and Crs2470144Trs2445761 had significant effects on annual sagittal maxillary growth and on mandibular growth in boys. No association was found in girls.
Conclusions
A quantitative trait locus that influences male pubertal sagittal jaw growth might exist in the CYP19A1 gene, and single nucleotide polymorphisms rs2470144 and rs2445761 might be inside this quantitative trait locus or be linked to it.
Growth and development of the maxilla and mandible are determined by genetic and environmental factors.1 Identification of these factors and mechanisms would help diagnosis, prediction, and treatment for skeletal variations. Single nucleotide polymorphisms and the haplotypes defined by common single nucleotide polymorphisms can be genotyped to determine normal and variable craniofacial phenotypes.2 Studies have found that single nucleotide polymorphisms (P561T, C422F, and I526L) in the growth hormone receptor gene are associated with mandibular ramus height in Japanese, Korean, and Chinese populations, and the P561T polymorphism has an inhibitory effect on mandibular growth in young children.3–7 Several single nucleotide polymorphisms have been found to be involved in mandibular prognathism.8–10 A polymorphism of the noggin gene (SNP rs1348322) was present in 4 families with mandibular micrognathia.11
Estrogen is a key hormone for skeletal growth, maturation, and maintaining bone mass.12 An increase in serum estrogen promotes the pubertal growth spurt by (1) decreasing osteoclast formation and activity; (2) increasing osteoblast formation, differentiation, proliferation, and function; and (3) stimulating chondrogenesis.13–15
Studies showing estrogen receptors in areas of the jaws including the condyles and palate support the importance of estrogen for jaw mass and growth.16–19 Aromatase (estrogen synthetase) catalyzes the final and rate-limiting step in the conversion of C19 androgens (androstenedione and testosterone) to C18 estrogens (estrone and estradiol); this makes it a key enzyme for estrogen biosynthesis in vivo.20 The CYP19A1 gene is about 123 kb in length, is located at chromosome 15q21.2, and encodes aromatase.21,22
Several functional CYP19A1 single nucleotide polymorphisms have been described. Yang et al23 showed that a CYP19A1 gene polymorphism in intron 1 (SNP rs730154) was significantly associated with adult height variation. Another single nucleotide polymorphism, rs2470144, lies near the exon/promoter I.1 of the CYP19A1, the activity of which is the basis for strikingly elevated levels of circulating estrogen in pregnant women.24,25 Single nucleotide polymorphism rs2445761 lies adjacent to the CYP19A1 promoter 2a. The single nucleotide polymorphisms rs2470144 and rs2445761 have already been found to be associated with variations in the onset of menarche.21 These findings suggest that rs2470144, and rs2445761 per se, or the functional loci in linkage disequilibrium with them, might be involved in regulating the transcription and expression of CYP19A1. Linkage disequilibrium refers to the nonrandom associations among neighboring alleles. This means that a variation or DNA marker might serve as a marker for other genetic variations in the DNA that is close to the marker, as defined by ethnic-specific linkage disequilibrium “blocks” of DNA. Skeletal sexual dimorphisms, such as bone mass in men, and characteristics of the human face, such as the growth of the cheekbones, mandible, and chin, could be significantly affected by estrogen and testosterone.20,26
To better predict and take advantage of the extent of pubertal jaw growth, it is important to understand the inherent genetic factors that influence it. In this study, we investigated the association between pubertal sagittal jaw growth and CYP19A1 rs2470144 and rs2445761 single nucleotide polymorphisms and haplotypes.
MATERIAL AND METHODS
The study was approved by the board of ethics of Sichuan University in China. Participation was voluntary, and informed-consent documents were signed by all participants before they entered this study.
Ninety-two subjects were chosen from approximately 1000 posttreatment patients in the department of orthodontics from the West China Hospital of Stomatology in Chengdu from 2006 to 2009. The inclusion criteria were (1) adolescent patients who started fixed orthodontic treatment in cervical vertebral maturation stage 3 and finished it in stage 4 or 5; (2) Class I skeletal relationship (0° < ANB < 5°); and (3) availability of pretreatment and posttreatment lateral cephalometric radiographs taken by the same digital cephalostat. Exclusion criteria were (1) use of a functional appliance or headgear; (2) use of Class II or Class III elastics for more than 3 months; and (3) incomplete treatment information or blurred radiographs. Cervical skeletal age was determined on lateral cephalometric radiographs based on the cervical vertebral maturation method.27 The ANB angle was ascertained by Winceph software (version 7.0; Rise, Sendai, Japan). Determinations of cervical stage maturation and skeletal relationship were initially done by 2 orthodontists (S.H. and Y.G.) separately, and discordant findings were then resolved by the 2 observers together.
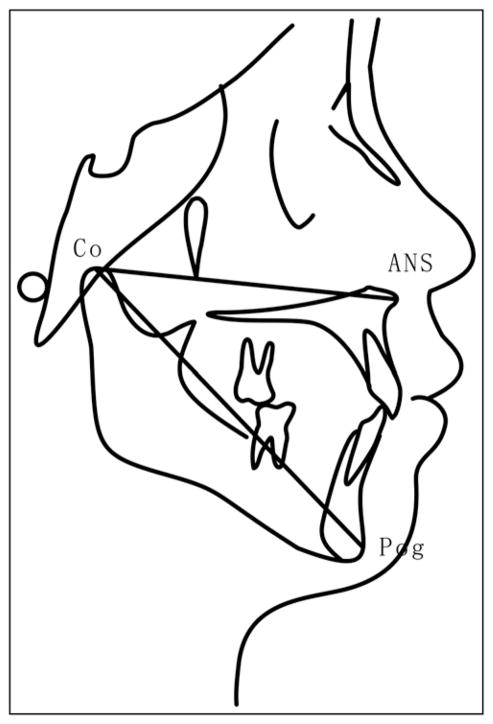
All lateral cephalometric radiographs were numbered with the patient's name hidden. Maxillary and mandibular sagittal lengths (condylion to anterior nasal spine condylion to hard-tissue pogonion, respectively; Fig 1) were measured 3 times independently by the 2 investigators using the Winceph software, with the mean value used for analysis. Annual sagittal growth was calculated for each subject during the observation (orthodontic treatment) period.
Fig 1.
Measurements for maxillary and mandible sagittal length: Co-ANS, Condylion to anterior nasal spine; Co-Pog, condylion to hard-tissue pogonion.
Buccal swabs were collected for DNA extraction and genotyping by scraping firmly against the inside of each cheek 10 times with a sterile cotton-tipped stick. Each specimen was coded with a sample number. Genomic DNA was isolated by using a buccal swab DNA kit (Bioteke, Beijing, China) and stored at −80°C.
Real-time polymerase chain reaction (PCR) was performed with the iq5 Real-time Quantitative PCR instrument (Bio-Rad, Hercules, Calif) by using TaqMan single nucleotide polymorphism assays (Applied Biosystems, Forster City, Calif) to complete the genotyping. The probe sequences designed by Applied Biosystems were rs2445761:TAAATAGTAGAACTTGTGGGATCAA[C/T]GA-TAAACGGACATGGAACTGTTTTA and rs2470144:AGGC CAGCAAGGCCAGGGCCACTGA[C/T]GGAGGGAAATTT-TACAAGGTAAACA. Each reaction mixture contained 10 μL 1x TaqMan Universal PCR Master mix, 0.5 μL 1x Taq-Man SNP kit (probe/primer mix), 2 μL DNA obtained, and 7.5 μL DNase-free water in a final volume of 20 μL. Standard amplification conditions were 95°C for 10 minutes, 40 cycles of 92°C for 15 seconds, and 60°C for 40 seconds; 2 negative controls with sterile water as templates were used in each reaction plate. Allelic discrimination results were according to fluorescent signals from reporters VIC and FAM. The expected genotyping results were CC, CT, and TT for both single nucleotide polymorphisms.
Statistical analysis
Interobserver agreement of the determinations of cervical stage and skeletal relationship was assessed by using Cohen's equation: kappa = PA − PE/1 − PE, where PA is the actual interobserver agreement rate and PE is the expected intraobserver agreement rate. The calculated kappa values were 0.918 and 0.907, respectively. U-tests of the 2 kappa values showed no significant difference (P < 0.01), indicating high consistency of the determinations.
The reproducibility of the cephalometric measurements was assessed by repeating them 1 month later in 40 randomly selected radiographs and calculated by using Dahlberg's equation28 for method error: , where is the difference between duplicated measurements and N is the number of repeated measurements. The errors for maxillary and mandibular length measurements were low, 0.10 and 0.16 mm, and 0.14 and 0.13 mm, on the pretreatment and posttreatment radiographs, respectively.
The chi-square test was used for the Hardy-Weinberg equilibrium of the genotype frequencies for each single nucleotide polymorphism. Linear regressions for the average increases in mandibular and maxillary sagittal length for each genotype in both sexes was performed with Prism 5 (GraphPad Software, San Diego, Calif). Linkage disequilibrium analysis was performed with SHEsis software.29 Haplotype construction was done with the phase 2.1 program.30 The associations between single nucleotide polymorphism genotypes or haplotypes and annual sagittal jaw growth in both sexes were analyzed by the least-squares method conducted by JMP (version 9.0; SAS Institute, Cary, NC) according to the following linear model: Yij = μ + Gi (Hi) + Pj + eij, where Yij is the individual observation value, μ is the overall population mean, Gi is the effect of genotype, Hi is the effect of haplotype, Pj is the effect of initial jaw length, and eij is the random residual effect. A P value less than 0.05 was considered statistically significant.
RESULTS
For the genotype distributions and linkage disequilibrium analysis, the sample consisted of 42 boys and 50 girls. Frequencies of rs2470144 genotypes were 12 CC (28.57%), 20 CT (47.62%), and 10 TT (23.81%) in the boys, and 14 CC (28%), 24 CT (48%), and 12 TT (24%) in the girls. The frequencies of rs2445761 genotypes were 8 CC (19.05%), 24 CT (57.14%), and 10 TT (23.81%) in the boys, and 10 CC (20%), 24 CT (48%), and 16 TT (32%) in the girls.
The genotype frequencies at the 2 single nucleotide polymorphisms in the sample population were both in the Hardy-Weinberg equilibrium (χ2 = 0.1597; P = 0.689 > 0.05 for rs2470144; and χ2 = 0.2433; P = 0.622 > 0.05 for rs2445761). Linkage disequilibrium analysis showed the 2 single nucleotide polymorphisms were in strong linkage disequilibrium (D′ = 0.833; γ2 = 0.534). Haplotypes were constructed with estimated frequencies as shown in Table I. A summary of ages, observation times, and pretreatment jaw lengths in the genotype or haplotype groups is given in Table II.
Table I.
Haplotypes constructed by single nucleotide polymorphisms rs2470144 and rs2445761
| rs2470144 | rs2445761 | Estimated frequency | n* | |
|---|---|---|---|---|
| Haplotype 1 | T | T | 0.4417 | 82 |
| Haplotype 2 | C | C | 0.4199 | 78 |
| Haplotype 3 | C | T | 0.1018 | 18 |
| Haplotype 4 | T | C | 0.0366 | 6 |
Number of each haplotype in the total sample.
Table II.
Summary of age, observation time, and pretreatment jaw lengths by genotype and haplotype groups
| Male
|
Female
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype or haplotype | Age (y) | Observation time (y) | Pre-Mx (mm) | Pre-Mn (mm) | Age (y) | Observation time (y) | Pre-Mx (mm) | Pre-Mn (mm) |
| rs2470144 | ||||||||
|
| ||||||||
| CC | 12.28 (0.59) | 2.54 (0.55) | 79.0 (2.5) | 97.5 (3.6) | 11.12 (0.75) | 1.95 (0.47) | 77.6 (5.0) | 97.1 (4.8) |
|
| ||||||||
| CT | 11.71 (0.93) | 2.26 (0.61) | 78.8 (2.7) | 99.1 (4.0) | 11.51 (0.91) | 1.96 (0.35) | 76.9 (3.5) | 96.6 (2.8) |
|
| ||||||||
| TT | 12.12 (1.51) | 2.31 (0.57) | 77.3 (3.7) | 97.3 (3.8) | 11.46 (0.58) | 2.07 (0.30) | 75.9 (3.6) | 96.1 (4.1) |
|
| ||||||||
| rs2445761 | ||||||||
|
| ||||||||
| CC | 11.93 (0.56) | 2.55 (0.39) | 79.6 (3.5) | 98.8 (4.2) | 10.93 (0.73) | 1.83 (0.39) | 78.5 (4.1) | 97.5 (4.3) |
|
| ||||||||
| CT | 11.81 (0.92) | 2.30 (0.61) | 78.8 (2.3) | 98.8 (3.8) | 11.43 (0.90) | 2.00 (0.41) | 76.6 (4.3) | 96.7 (3.5) |
|
| ||||||||
| TT | 11.84 (1.59) | 2.32 (0.64) | 76.9 (3.2) | 96.3 (3.3) | 11.60 (0.59) | 2.06 (0.28) | 76.2 (3.3) | 96.1 (3.7) |
|
| ||||||||
| Haplotype 1 | ||||||||
|
| ||||||||
| 0 | 12.16 (0.6 4) | 2.52 (0.49) | 79.0 (2.8) | 98.1 (3.7) | 11.12 (0.75) | 1.95 (0.47) | 77.6 (5.0) | 97.1 (4.9) |
|
| ||||||||
| 1 | 11.64 (0.95) | 2.25 (0.63) | 78.7 (2.5) | 99.0 (4.0) | 11.51 (0.91) | 1.96 (0.35) | 76.9 (3.5) | 96.6 (2.8) |
|
| ||||||||
| 2 | 11.61 (1.69) | 2.24 (0.62) | 76.9 (3.6) | 96.5 (3.6) | 11.46 (0.58) | 2.07 (0.30) | 75.9 (3.6) | 96.1 (4.1) |
|
| ||||||||
| Haplotype 2 | ||||||||
|
| ||||||||
| 0 | 11.73 (1.46) | 2.36 (0.59) | 77.2 (3.4) | 97.0 (3.5) | 11.60 (0.59) | 2.05 (0.28) | 76.2 (3.3) | 96.1 (3.7) |
|
| ||||||||
| 1 | 11.85 (0.90) | 2.31 (0.60) | 78.9 (2.4) | 98.8 (3.8) | 11.43 (0.90) | 2.00 (0.41) | 76.6 (4.3) | 96.7 (3.5) |
|
| ||||||||
| 2 | 12.07 (0.43) | 2.63 (0.48) | 80.0 (3.5) | 97.6 (4.8) | 10.93 (0.73) | 1.83 (0.39) | 78.5 (4.2) | 97.5 (4.3) |
|
| ||||||||
| Haplotype 3 | ||||||||
|
| ||||||||
| 0 | 11.64 (1.07) | 2.30 (0.57) | 78.6 (3.2) | 98.6 (4.0) | 11.31 (0.82) | 1.96 (0.36) | 76.9 (3.9) | 96.8 (3.5) |
|
| ||||||||
| 1 | 12.44 (0.67) | 2.52 (0.61) | 78.2 (1.9) | 97.1 (3.0) | 11.81 (0.55) | 2.13 (0.42) | 76.3 (4.7) | 96.1 (4.8) |
|
| ||||||||
| Haplotype 4 | ||||||||
|
| ||||||||
| 0 | 11.84 (1.08) | 2.34 (0.60) | 78.4 (2.8) | 98.0 (3.8) | 11.39 (0.80) | 1.99 (0.37) | 76.8 (4.0) | 96.7 (3.7) |
|
| ||||||||
| 1 | 12.34 (0.47) | 2.38 (0.53) | 79.4 (5.1) | 99.4 (5.7) | – | – | – | – |
|
| ||||||||
| 2 | 11.25 (0.35) | 2.58 (0.00) | 79.0 (4.8) | 100.5 (3.4) | – | – | – | – |
Haplotype 1, Trs2470144Trs2445761; haplotype 2, Crs2470144Crs2445761; haplotype 3, Crs2470144Trs2445761; haplotype 4, Trs2470144Crs2445761. 0, 1, and 2 represent 0, 1, and 2 of this haplotype, respectively.
Pre-Mx, Pretreatment maxillary length; Pre-Mn, pretreatment mandible length; values are presented as means (and standard deviations).
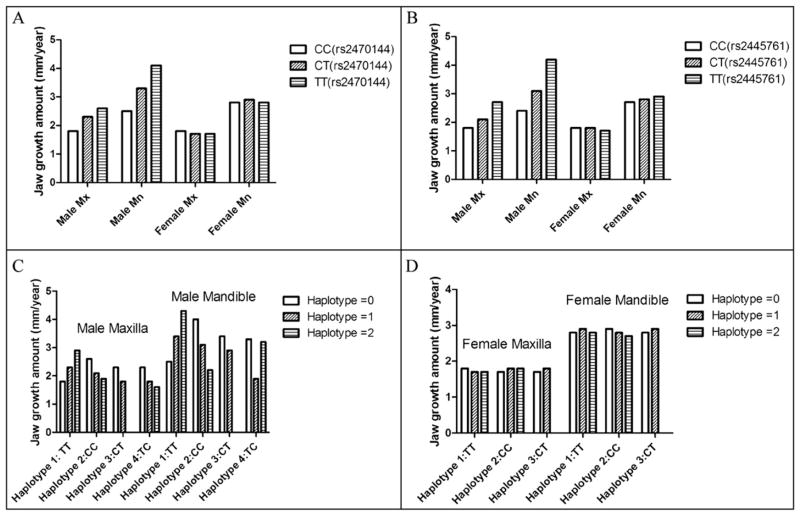
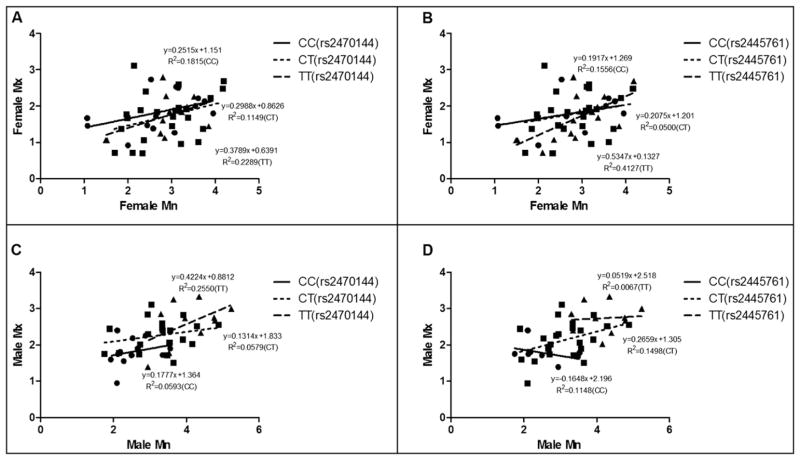
Single nucleotide polymorphism association analysis (Table III) showed that boys with the rs2470144 genotype CC had significantly smaller average annual maxilla growth than those with genotypes CT and TT. For the mandible, the boys with the CC genotype had the smallest amount of average growth, and the boys with TT had the greatest average growth (Table III; Fig 2, A). Boys with rs2445761 genotypes CC and CT had significantly smaller average annual maxilla growth than those with genotype TT, and boys with genotype CC had the smallest average mandibular growth amount, and those carrying TT had the greatest (Table III; Fig 2, B). No significant difference was found in genotypic groups in the girls (Table III; Fig 2, A and B). Linear regression for the different genotypes is presented in Figure 3.
Table III.
Association of single nucleotide polymorphism genotypes with annual jaw growth
| Boys
|
Girls
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Annual Mx growth (mm) | P value | Annual Md growth (mm) | P value | Annual Mx growth (mm) | P value | Annual Md growth (mm) | P value |
| SNP 2470144 | 0.0026* | < 0.0001* | 0.7378 | 0.9481 | ||||
|
| ||||||||
| CC | 1.8 (0.1)a | 2.5 (0.2)a | 1.9 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| CT | 2.3 (0.1)b | 3.3 (0.2)b | 1.7 (0.1)a | 2.9 (0.2)a | ||||
|
| ||||||||
| TT | 2.5 (0.1)b | 4.1 (0.2)c | 1.7 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| SNP 2445761 | 0.0017* | < 0.0001* | 0.7448 | 0.9760 | ||||
|
| ||||||||
| CC | 1.8 (0.2)a | 2.4 (0.2)a | 1.8 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| CT | 2.2 (0.1)a | 3.1 (0.1)b | 1.8 (0.1)a | 2.8 (0.2)a | ||||
|
| ||||||||
| TT | 2.7 (0.1)b | 4.2 (0.2)c | 1.7 (0.2)a | 2.8 (0.2)a | ||||
Values of annual maxillary (Mx) and mandibular (Md) growth are presented as least square means (and standard errors). Values with no common letter (a, b, or c) for the same single nucleotide polymorphism are significantly different at P < 0.05.
SNP, Single nucleotide polymorphism.
P < 0.05 indicates that genotypes of this single nucleotide polymorphism have a significant effect on annual jaw growth.
Fig 2.
Average yearly increases of maxillary and mandibular length during treatment according to A, different rs2470144 genotypes in male and female subjects; B, different rs2445761 genotypes in male and female subjects; C, different haplotypes constructed with rs2470144 and rs2445761 in male subjects; D, different haplotypes constructed with rs2470144 and rs2445761 in female subjects.
Fig 3.
Linear regression for average jaw growth amount (millimeters per year) of mandibular (Mn) and maxillary (Mx) length during treatment time according to A, different rs2470144 genotypes in female subjects; B, different rs2445761 genotypes in female subjects; C, different rs2470144 genotypes in male subjects; D, different rs2445761 genotypes in male subjects.
The haplotype association test (Table IV) showed significant differences with respect to average annual maxillary and mandibular sagittal growth among boys with 2, 1, or 0 haplotype Trs2470144Trs2445761 units. Average annual mandibular growth was significantly different among boys with different numbers of haplotype Crs2470144Crs2445761 units. Boys with haplotype Crs2470144Trs2445761 had less average annual maxillary growth than those without it (Fig 2, C). No significant difference was found in the girls (Table IV; Fig 2, D).
Table IV.
Association of different haplotypes with annual jaw growth
| Boys
|
Girls
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype | Annual Mx growth (mm) | P value | Annual Md growth (mm) | P value | Annual Mx growth (mm) | P value | Annual Md growth (mm) | P value |
| Haplotype 1 | < 0.0001* | < 0.0001* | 0.7378 | 0.9481 | ||||
|
| ||||||||
| 0 | 1.8 (0.1)a | 2.5 (0.2)a | 1.9 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| 1 | 2.3 (0.1)b | 3.4 (0.2)b | 1.7 (0.1)a | 2.9 (0.2)a | ||||
|
| ||||||||
| 2 | 2.8 (0.1)c | 4.3 (0.2)c | 1.7 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| Haplotype 2 | 0.1203 | 0.0002* | 0.7448 | 0.9760 | ||||
|
| ||||||||
| 0 | 2.5 (0.1)a | 4.0 (0.2)a | 1.7 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| 1 | 2.1 (0.1)a | 3.1 (0.1)b | 1.8 (0.1)a | 2.8 (0.2)a | ||||
|
| ||||||||
| 2 | 2.0 (0.3)a | 2.2 (0.4)c | 1.8 (0.2)a | 2.8 (0.2)a | ||||
|
| ||||||||
| Haplotype 3 | 0.0039* | 0.0837 | 0.9833 | 0.8258 | ||||
|
| ||||||||
| 0 | 2.3 (0.1)a | 3.4 (0.2)a | 1.8 (0.1)a | 2.8 (0.1)a | ||||
|
| ||||||||
| 1 | 1.8 (0.1)b | 2.8 (0.3)a | 1.7 (0.2)a | 2.9 (0.3)a | ||||
|
| ||||||||
| Haplotype 4 | 0.1155 | 0.1259 | – | – | ||||
|
| ||||||||
| 0 | 2.3 (0.1)a | 3.3 (0.1)a | 1.8 (0.1) | 2.8 (0.1)a | ||||
|
| ||||||||
| 1 | 1.8 (0.4)a | 2.0 (0.6)b | – | – | ||||
|
| ||||||||
| 2 | 1.6 (0.3)a | 3.3 (0.6)ab | – | – | ||||
Haplotype 1, Trs2470144Trs2445761; haplotype 2, Crs2470144Crs2445761; haplotype 3, Crs2470144Trs2445761; haplotype 4, Trs2470144Crs2445761. 0, 1, and 2 represent 0, 1, and 2 of this haplotype, respectively. Values of annual maxillary (Mx) and mandibular (Md) growth are presented as least square means (and standard errors). Values with no common letter (a, b, or c) for the same haplotype are significantly different at P < 0.05.
P < 0.05 indicates that genotypes of this single nucleotide polymorphism have a significant effect on annual jaw growth.
DISCUSSION
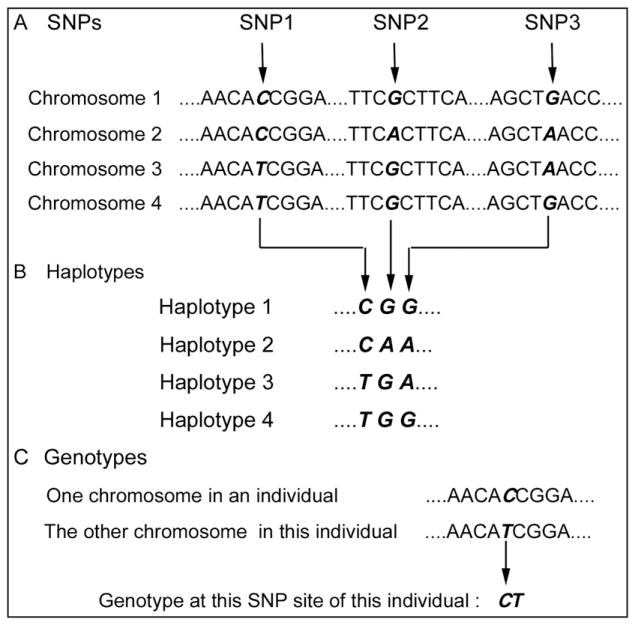
The simplest form of genetic polymorphism is the substitution of one nucleotide for another, termed a single nucleotide polymorphism (Fig 4, A). Single nucleotide polymorphisms are the most common type of DNA sequence variation. They are stable and distributed throughout the genome. A stretch of DNA with a distinctive pattern of single nucleotide polymorphisms at a given location of a chromosome is called a haplotype (Fig 4, B). The pair of alleles at the site of 1 single nucleotide polymorphism is called the genotype of this single nucleotide polymorphism (Fig 4, C).31 Single nucleotide polymorphism association analysis and haplotype association are both useful ways for investigation of associations between genetic markers and putative trait loci, whereas the utility of haplotypes has some advantages: eg, significantly improving the power and robustness of finding potential genes and locating quantitative trait loci.32,33
Fig 4.
A, Single nucleotide polymorphisms are single nucleotide variations among populations; B, a haplotype is a stretch of DNA with a distinctive pattern of single nucleotide polymorphisms at a given location of a chromosome; C, a genotype is the pair of alleles at the site of 1 single nucleotide polymorphism in a subject's paired chromosomes.
In this study, we chose subjects with Class I relationships, since the growth of skeletal Class III relationships is thought to be a polygenic or a single gene trait with variable expressivity and incomplete penetrance with an interaction between genetic and environmental factors.34,35 Although associations have been found between specific polymorphisms and mandibular micrognathia,11 the genetic influence on Class II malocclusion is variable.1 It could be presumed that these skeletal patterns are influenced by a series of more complicated genetic and environmental factors. To better understand variations that result in Class III or Class II relationships, it is important to understand the underlying genetic influence on variations that result in skeletal Class I populations. In this study, we focused on the association of a genetic marker for estrogen synthesis with sagittal jaw growth in a Class I sample.
The annual sagittal growth of the jaws was observed during a skeletal development stage in which the peak in mandibular growth occurs within 1 year after cervical vertebral stage 3 and ends 1 year before cervical stage 5.27 Subjects were excluded if they received treatment that was expected to have an orthopedic effect on jaw growth, such as functional appliance, headgear, or long-time intermaxillary elastics. Because the cephalometric measurement for maxillary growth, condylion to anterior nasal spine, included a mandibular landmark, we also investigated using another measurement, pterygomaxillary fissure to anterior nasal spine, to prevent errors due to variations in the mandible. There was no difference in the conclusions when using pterygomaxillary fissure to anterior nasal spine and condylion to anterior nasal spine (data not shown). Therefore, we report the measurement condylion to anterior nasal spine as it was used in a previous study.36 Since the subjects were all skeletal Class I with a jaw growth relationship considered to be normal at the beginning of treatment, the growth calculated in this study likely represents the natural jaw growth in children with normal skeletal relationships during the pubertal growth spurt.
Single nucleotide polymorphism association analysis showed that genotypes of the 2 single nucleotide polymorphisms were significantly associated with annual sagittal growth of both jaws in boys, but not in girls (Table III). Linear regression of maxillary sagittal growth and mandibular sagittal growth for different genotypes of both single nucleotide polymorphisms show greater differences in the slope and the R2 values in boys than in girls (Fig 3). This demonstrates that there is more individual variation in mandibular and maxillary jaw growth “coordination” in boys than in girls, and might indicate the variable interaction of other factors beside the CYP19A1 genotype. These findings are consistent with those in a similar preliminary study with white subjects, suggesting that the effect of CYP19A1 variation on sagittal jaw growth is seen across ethnic groups.36
The sex difference based on genotype was confirmed by haplotype analysis that demonstrated an association with jaw growth in the boys (Table IV). Haplotypes Trs2470144Trs2445761 and Crs2470144Trs2445761 had highly significant effects on annual sagittal maxillary growth and on mandibular growth in boys.
These associations might be explained by the physical locations or the physiologic functions of the 2 single nucleotide polymorphisms. Single nucleotide polymorphism rs2470144 is near the exon/promoter I.1 of the CYP19A1 and is reported to influence circulating levels of estrogen in pregnant women.24,25 Single nucleotide polymorphism rs2445761 lies adjacent to the CYP19A1 promoter 2a. Both single nucleotide polymorphisms are associated with variations in the onset of menarche in pubertal girls, also suggesting the capacity to influence estrogen levels in vivo.21 Our study suggests that a quantitative trait locus that influences maxillary and mandibular growth might exist in the CYP19A1 gene, and single nucleotide polymorphisms rs2470144 and rs2445761 might be inside this quantitative trait locus or linked to it.
Estrogen and testosterone affect many physical features, including facial ones. Estrogen not only initiates pubertal growth in both sexes, but also limits longitudinal bone growth by progressively inducing closure of the epiphyseal growth plate at the end of puberty.12,20 Low doses of estrogen stimulate the pubertal growth spurt and prolong puberty, whereas higher concentrations might inhibit linear growth and promote growth plate closure.20 The biphasic effect of estrogen in both sexes is in contrast with the longitudinal growth-stimulating effect of androgens in boys.37 Clinical data to support the effect of the inhibition of estrogen activity and an increase in the effective testosterone-to-estrogen ratio come from the administration of aromatase inhibitors, resulting in an increased final height in growing boys with short stature.38,39 There is also evidence that, in pubertal boys, a high testosterone-estrogen ratio is supposed to facilitate the growth of cheekbones, mandible, chin, and bones of the eyebrow ridges, and the lengthening of the lower face.26,40
From our results, it can be hypothesized that genotypes and haplotypes associated with greater growth—eg, geno-type TT and haplotype Trs2470144Trs2445761—might have a suppressive effect on aromatase synthesis, resulting in an increased testosterone-estrogen ratio, which causes greater annual sagittal jaw growth in boys. Those correlated with lower growth, such as genotype CC or haplotype Crs2470144Trs2445761, might have a promoting effect on aromatase synthesis, resulting in a decreased testosterone-estrogen ratio, which causes less annual growth. The reason for no difference in jaw growth among girls with different genotypes or haplotypes could indicate a different effect of estrogens and androgens on their sagittal growth. The mechanisms of androgen and estrogen effects in the sexes are not identical and need further investigation.
Because the regulation of bone growth involves a complex interplay among hormones, mechanical stimuli, and locally produced mediators, further studies are still needed to clarify the connections between various genotypes or haplotypes affecting aromatase synthesis and sagittal growth of the jaws.41 In addition, a markedly larger sample size would facilitate genome-wide association studies to discover other genes that can influence facial growth.
CONCLUSIONS
We found that genotypes rs2470144 and rs2445761, and haplotypes constructed from them, have significant associations with average annual sagittal jaw growth in pubertal male orthodontic patients. Quantitative trait loci that influence maxillary and mandibular growth might exist in the gene CYP19A1.
Acknowledgments
Supported by number 2010FZ0087 funds from the Science and Technology Department of Sichuan Province, China.
Footnotes
The authors report no commercial, proprietary, or financial interest in the products or companies described in this article.
References
- 1.Hartsfield JK., Jr . Genetics and orthodontics. In: Graber LW, Vanarsdall RL, Vig KWL, editors. Orthodontics: current principles and techniques. St Louis: Elsevier Mosby; 2011. pp. 139–56. [Google Scholar]
- 2.Coussens AK, van Daal A. Linkage disequilibrium analysis identifies an FGFR1 haplotype-tag SNP associated with normal variation in craniofacial shape. Genomics. 2005;85:563–73. doi: 10.1016/j.ygeno.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Kang EH, Yamaguchi T, Tajima A, Nakajima T, Tomoyasu Y, Watanabe M, et al. Association of the growth hormone receptor gene polymorphisms with mandibular height in a Korean population. Arch Oral Biol. 2009;54:556–62. doi: 10.1016/j.archoralbio.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Tomoyasu Y, Yamaguchi T, Tajima A, Nakajima T, Inoue I, Maki K. Further evidence for an association between mandibular height and the growth hormone receptor gene in a Japanese population. Am J Orthod Dentofacial Orthop. 2009;136:536–41. doi: 10.1016/j.ajodo.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi T, Maki K, Shibasaki Y. Growth hormone receptor gene variant and mandibular height in the normal Japanese population. Am J Orthod Dentofacial Orthop. 2001;119:650–3. doi: 10.1067/mod.2001.114536. [DOI] [PubMed] [Google Scholar]
- 6.Zhou J, Lu Y, Gao XH, Chen YC, Lu JJ, Bai YX, et al. The growth hormone receptor gene is associated with mandibular height in a Chinese population. J Dent Res. 2005;84:1052–6. doi: 10.1177/154405910508401116. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki Y, Satoh K, Hayasaki H, Fukumoto S, Fujiwara T, Nonaka K. The P561T polymorphism of the growth hormone receptor gene has an inhibitory effect on mandibular growth in young children. Eur J Orthod. 2009;31:536–41. doi: 10.1093/ejo/cjp017. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Li X, Zhang F, Chen F. The identification of a novel locus for mandibular prognathism in the Han Chinese population. J Dent Res. 2011;90:53–7. doi: 10.1177/0022034510382546. [DOI] [PubMed] [Google Scholar]
- 9.Tassopoulou-Fishell M, Deeley K, Harvey EM, Sciote J, Vieira AR. Genetic variation in Myosin 1H contributes to mandibular prognathism. Am J Orthod Dentofacial Orthop. 2012;141:51–9. doi: 10.1016/j.ajodo.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang JY, Park EK, Ryoo HM, Shin HI, Kim TH, Jang JS, et al. Polymorphisms in the Matrilin-1 gene and risk of mandibular prognathism in Koreans. J Dent Res. 2010;89:1203–7. doi: 10.1177/0022034510375962. [DOI] [PubMed] [Google Scholar]
- 11.Gutierrez SJ, Gomez M, Rey JA, Ochoa M, Gutierrez SM, Prieto JC. Polymorphisms of the noggin gene and mandibular micrognathia: a first approximation. Acta Odontol Latinoam. 2010;23:13–9. [PubMed] [Google Scholar]
- 12.Riggs BL, Khosla S, Melton LJ., 3rd Sex steroids and the construction and conservation of the adult skeleton. Endocr Rev. 2002;23:279–302. doi: 10.1210/edrv.23.3.0465. [DOI] [PubMed] [Google Scholar]
- 13.Hughes DE, Dai A, Tiffee JC, Li HH, Mundy GR, Boyce BF. Estrogen promotes apoptosis of murine osteoclasts mediated by TGF-beta. Nat Med. 1996;2:1132–6. doi: 10.1038/nm1096-1132. [DOI] [PubMed] [Google Scholar]
- 14.Majeska RJ, Ryaby JT, Einhorn TA. Direct modulation of osteoblastic activity with estrogen. J Bone Joint Surg Am. 1994;76:713–21. doi: 10.2106/00004623-199405000-00013. [DOI] [PubMed] [Google Scholar]
- 15.Qu Q, Perala-Heape M, Kapanen A, Dahllund J, Salo J, Vaananen HK, et al. Estrogen enhances differentiation of osteo-blasts in mouse bone marrow culture. Bone. 1998;22:201–9. doi: 10.1016/s8756-3282(97)00276-7. [DOI] [PubMed] [Google Scholar]
- 16.Sheridan PJ, Aufdemorte TB, Holt GR, Gates GA. Cartilage of the baboon contains estrogen receptors. Rheumatol Int. 1985;5:279–81. doi: 10.1007/BF00541356. [DOI] [PubMed] [Google Scholar]
- 17.Aufdemorte TB, Van Sickels JE, Dolwick MF, Sheridan PJ, Holt GR, Aragon SB, et al. Estrogen receptors in the temporomandibular joint of the baboon (Papio cynocephalus): an autoradiographic study. Oral Surg Oral Med Oral Pathol. 1986;61:307–14. doi: 10.1016/0030-4220(86)90407-x. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Ejiri S, Nakajima M, Kohno S, Ozawa H. Changes of cancellous bone mass in rat mandibular condyle following ovariectomy. Bone. 1999;25:339–47. doi: 10.1016/s8756-3282(99)00179-9. [DOI] [PubMed] [Google Scholar]
- 19.Ejiri S, Tanaka M, Watanabe N, Anwar RB, Yamashita E, Yamada K, et al. Estrogen deficiency and its effect on the jaw bones. J Bone Miner Metab. 2008;26:409–15. doi: 10.1007/s00774-008-0870-4. [DOI] [PubMed] [Google Scholar]
- 20.Vanderschueren D, Vandenput L, Boonen S, Lindberg MK, Bouillon R, Ohlsson C. Androgens and bone. Endocr Rev. 2004;25:389–425. doi: 10.1210/er.2003-0003. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y, Xiong DH, Yang TL, Guo YF, Recker RR, Deng HW. Polymorphisms of estrogen-biosynthesis genes CYP17 and CYP19 may influence age at menarche: a genetic association study in Caucasian females. Hum Mol Genet. 2006;15:2401–8. doi: 10.1093/hmg/ddl155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haiman CA, Stram DO, Pike MC, Kolonel LN, Burtt NP, Altshuler D, et al. A comprehensive haplotype analysis of CYP19 and breast cancer risk: the multiethnic cohort. Hum Mol Genet. 2003;12:2679–92. doi: 10.1093/hmg/ddg294. [DOI] [PubMed] [Google Scholar]
- 23.Yang TL, Xiong DH, Guo Y, Recker RR, Deng HW. Association analyses of CYP19 gene polymorphisms with height variation in a large sample of Caucasian nuclear families. Hum Genet. 2006;120:119–25. doi: 10.1007/s00439-006-0199-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahendroo MS, Means GD, Mendelson CR, Simpson ER. Tissue-specific expression of human P-450AROM. The promoter responsible for expression in adipose tissue is different from that utilized in placenta. J Biol Chem. 1991;266:11276–81. [PubMed] [Google Scholar]
- 25.Means GD, Mahendroo MS, Corbin CJ, Mathis JM, Powell FE, Mendelson CR, et al. Structural analysis of the gene encoding human aromatase cytochrome P-450, the enzyme responsible for estrogen biosynthesis. J Biol Chem. 1989;264:19385–91. [PubMed] [Google Scholar]
- 26.Schaefer K, Fink B, Mitteroecker P, Neave N, Bookstein FL. Visualizing facial shape regression upon 2nd to 4th digit ratio and testosterone. Coll Antropol. 2005;29:415–9. [PubMed] [Google Scholar]
- 27.Baccetti T, Franchi L, McNamara JA. The cervical vertebral maturation (CVM) method for the assessment of optimal treatment timing in dentofacial orthopedics. Semin Orthod. 2005;11:119–29. [Google Scholar]
- 28.Dahlberg G. Statistical methods for medical and biological students. New York: Interscience Publications; 1940. [Google Scholar]
- 29.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–8. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 30.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561–6. doi: 10.1007/s100380200086. [DOI] [PubMed] [Google Scholar]
- 32.Akey J, Jin L, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? Eur J Hum Genet. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- 33.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–23. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 34.Xue F, Wong RW, Rabie AB. Genes, genetics, and Class III maloc-clusion. Orthod Craniofac Res. 2010;13:69–74. doi: 10.1111/j.1601-6343.2010.01485.x. [DOI] [PubMed] [Google Scholar]
- 35.Cruz RM, Krieger H, Ferreira R, Mah J, Hartsfield J, Jr, Oliveira S. Major gene and multifactorial inheritance of mandibular prognathism. Am J Med Genet A. 2008;146A:71–7. doi: 10.1002/ajmg.a.32062. [DOI] [PubMed] [Google Scholar]
- 36.Hartsfield JK, Jr, Zhou J, Chen S. The importance of analyzing specific genetic factors in facial growth for diagnosis and treatment planning. In: McNamara JA Jr, Kapila SD, editors. Surgical enhancement of orthodontic treatment. Ann Arbor, Mich: University of Michigan; 2010. pp. 267–81. [Google Scholar]
- 37.Juul A. The effects of oestrogens on linear bone growth. Hum Re-prod Update. 2001;7:303–13. doi: 10.1093/humupd/7.3.303. [DOI] [PubMed] [Google Scholar]
- 38.Hero M, Makitie O, Kroger H, Nousiainen E, Toiviainen-Salo S, Dunkel L. Impact of aromatase inhibitor therapy on bone turnover, cortical bone growth and vertebral morphology in pre- and peripubertal boys with idiopathic short stature. Horm Res. 2009;71:290–7. doi: 10.1159/000208803. [DOI] [PubMed] [Google Scholar]
- 39.Dunkel L. Update on the role of aromatase inhibitors in growth disorders. Horm Res. 2009;71(Suppl 1):57–63. doi: 10.1159/000178040. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer K. Female appearance: facial and bodily attractiveness as shape. Psychol Sci. 2006;48:187–204. [Google Scholar]
- 41.Ohlsson C, Bengtsson BA, Isaksson OG, Andreassen TT, Slootweg MC. Growth hormone and bone. Endocr Rev. 1998;19:55–79. doi: 10.1210/edrv.19.1.0324. [DOI] [PubMed] [Google Scholar]