Abstract
Purpose
Altered proteasome functions are associated with multiple cardiomyopathies. While the proteasome targets poly-ubiquitinated proteins for destruction, it itself is modifiable by ubiquitination. We aim to identify the exact ubiquitination sites on cardiac proteasomes and examine whether they are also subject to acetylations.
Experimental design
Assembled cardiac 20S proteasome complexes were purified from five human hearts with ischemic cardiomyopathy, then analyzed by high-resolution MS to identify ubiquitination and acetylation sites. We developed a library search strategy that may be used to complement database search in identifying PTM in different samples.
Results
We identified 63 ubiquitinated lysines from intact human cardiac 20S proteasomes. In parallel, 65 acetylated residues were also discovered, 39 of which shared with ubiquitination sites.
Conclusion and clinical relevance
This is the most comprehensive characterization of cardiac proteasome ubiquitination to-date. There are significant overlaps between the discovered ubiquitination and acetylation sites, permitting potential crosstalk in regulating proteasome functions. The information presented here will aid future therapeutic strategies aimed at regulating the functions of cardiac proteasomes.
Keywords: 20S proteasome, acetylation, PTM, spectral library, ubiquitination
Heart failure is commonly associated with proteasome functional insufficiency and protein turnover perturbation [1, 2]. Insufficient proteolysis can lead to further protein damage and disease progression, suggesting the regulations of proteasome activities are important for understanding disease mechanism and identifying therapeutic targets [2, 3]. The core 20S proteasome is a ~750-kDa protein complex formed by two pairs of heptameric rings, each composed of seven protein subunits (α1–7, β1–7). Recent advances in proteomics have discovered that the 20S proteasome is heavily modified by protein PTM including phosphorylation, acetylation, sumoylation, and ubiquitination [4, 5]. Interestingly, although the ubiquitin-proteasome system targets poly-ubiquitinated proteins for degradation, ubiquitination of proteasome subunits may have direct regulatory functions [6, 7], suggesting the ubiquitinated lysines on 20S proteasomes may influence proteasomal activities and disease manifestations.
Multiple proteomics studies have detected numerous ubiquitination sites on 20S proteasome subunits in various cultured cells [8-12], but currently it remains unclear how common ubiquitination is displayed on cardiac 20S proteasomes and which modified residue may regulate cardiac phenotypes. Ubiquitination profiles are known to vary greatly across cell types [13] and to exhibit broad range of occupancies, thus warranting targeted individual examination of modification profile in clinically relevant samples. Furthermore, it is essential to distinguish whether the detected ubiquitination occurs on mature proteasome complexes, since whole-cell profiling conflates modifications on functioning complexes, free subunits, defected ribosomal products, and protein aggregates.
To identify the ubiquitination sites present on cardiac 20S proteasomes, we purified functional 20S proteasomes from human heart samples from the individual heart explants of five ischemic cardiomyopathy patients through a multi-step centrifugation and chromatography method [11, 14]. Targeted purification yields a large amount of target proteasomal subunits for deep profiling of protein modifications, and circumvents the documented sequence bias of anti-diglycyl-lysine or anti-acetyl-lysine enrichment strategies [13, 15, 16]. Profiling assembled complexes further allows differentiation between modifications on assembled complexes and on free subunits, with the latter potentially ubiquitinated for degradation independent of functional regulations. We then analyzed the samples with a customized workflow as follows. The purified proteasomes were analyzed with a high-resolution Orbitrap mass spectrometer in totally ten replicate experiments (5 biological replicates with 2 technical replicates each). Following trypsin digestion, ubiquitination leaves behind a characteristic mass shift of 114.0429 Da that allows modified peptides to be discerned from the accurate mass and MS/MS spectra. We conducted database search using ProLuCID and instructed the algorithm to accommodate for variable shifts of 114.0429 or 42.0106 Da on lysine, as well as 79.9663 Da on serine/threonine/tyrosine as a differential (see details in Supplemental Methods). We used a conservative filter to remove potential noises of shotgun data, requiring the modified peptide to be confidently detected in at least two individual human samples, from at least five individual spectra (Figure 1A). The filtered ubiquitinated spectra were manually inspected (Supplemental Data), and where possible, we attempted to cross-validate the spectra using a custom PTM spectral library search algorithm we developed, which works with our in-house spectral library search engine at COPaKB [17]. The algorithm evaluates signal correlation gains between experimental and library spectra after PTM mass shifts, and constitutes an orthogonal means than database search to evaluate PTM-peptide spectrum matches. The use of spectra library search for PTM is uncommon at present due to immature spectral libraries and methods, but we envision it will gain in utility to support and even supersede database search as technologies mature and similar methods to the one described here become available (see details in Supplemental Methods).
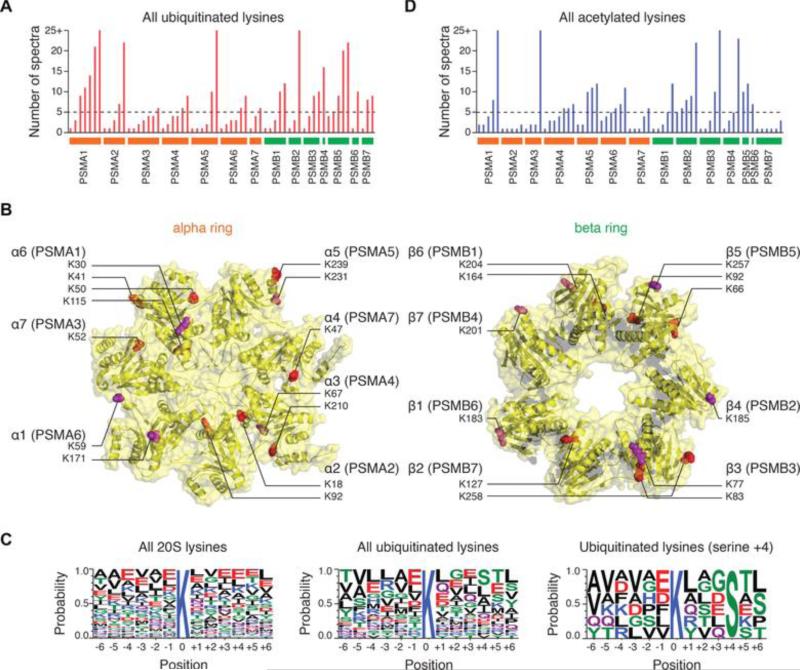
Figure 1. Ubiquitination of cardiac 20S proteasomes.
(A) The number of identified ubiquitinated lysine in each 20S proteasome subunit is shown. The dashed line represents the spectrum count cutoff for the stringency filter. A modification site is only accepted if identified in ≥ 2 subjects and ≥ 5 independent spectra. (B) The location of the identified lysine ubiquitination sites in 20S proteasome (Protein Data Bank structure 1IRU) alpha and beta subunits. Red-colored residues represent ubiquitination sites; purple residues are ubiquitinated and acetylated. (C) Compared to the aligned sequences around all 20S proteasome lysine positions (left), the ubiquitinated lysines (middle) show a slight enrichment of serine in the +4 position (right). (D) The number of identified acetylated lysine in each 20S proteasome subunit is shown.
Using this approach, we identified 2,125 unique peptide sequences belonging to cardiac 20S proteasomes, with average subunit sequence coverage of >80%. Over 90% of all identified spectra belonged to 20S proteasomes. We uncovered a total of 63 lysine modification sites (Supplemental Table 1), out of which 26 passed the stringency filter (identified in 2 subjects and at least 5 individual spectra) (Figure 1A). The 26 sites are located throughout all 14 subunits of the core 20S proteasome (Table 1). The ubiquitinated peptide spectra occupy ~1 – 20% of the spectral count of their corresponding residues (Supplemental Methods). To our best knowledge, 22 of these sites were previously never observed in cardiac tissues, 11 were previously not reported in the literature, and none were examined in clinically relevant samples. Overall, the α ring was more heavily ubiquitinated than the β ring despite their comparable total spectral counts (Figure 1B). This is consistent with our meta-analysis of large-scale studies, and the higher accessibility of the α ring to PTM enzymes. PSMA1 (α6 subunit) in particular appeared to be the most ubiquitinated.
Table 1.
Confidently identified ubiquitination sites in intact human cardiac 20S proteasomes
| Gene name | Subunit name | Residue position | Example sequence of modified peptide | Acetylated? | Known ubiquitination? | Known in cardiac tissue? |
|---|---|---|---|---|---|---|
| PSMA1 | α6 | 30 | IHQIEYAMEAVK(Ub)QGSATVGLK | Yes | [11], [13] | |
| 41 | SK(Ub)THAVLVALKR | Unreported | ||||
| 50 | SKTHAVLVALK(Ub)R | Unreported | ||||
| 115 | LVSLIGSK(Ub)TQIPTQR | [8], [11], [13] | Yes | |||
| PSMA2 | α2 | 18 | ERGYSFSLTTFSPSGK(Ub)LVQIEYALAAVAGGAPSVGIK | Unreported | ||
| 92 | K(Ub)LAQQYYLVYQEPIPTAQLVQR | [11-13], [16] | ||||
| PSMA3 | α7 | 52 | CKDGVVFGVEK(Ub)LVLSK | Unreported | ||
| PSMA4 | α3 | 67 | IYK(Ub)LNEDMACSVAGITSDANVLTNELR | Yes | Unreported | |
| 210 | LSAEK(Ub)VEIATLTR | [11], [13], [16] | Yes | |||
| PSMA5 | α5 | 231 | LNATNIELATVQPGQNFHMFTK(Ub)EELEEVIKD | Yes | [11] | |
| 239 | LNATNIELATVQPGQNFHMFTKEELEEVIK(Ub)D | [11], [9] | ||||
| PSMA6 | α1 | 59 | VPDK(Ub)LLDSSTVTHLFK | Yes | [10], [16] | |
| 171 | ATAAGVK(Ub)QTESTSFLEK | Yes | [11], [13], [16] | |||
| PSMA7 | α4 | 47 | GRDIVVLGVEK(Ub)K | Unreported | ||
| PSMB1 | β6 | 164 | SFK(Ub)AGGSASAMLQPLLDNQVGFK | [11], [13] | ||
| 204 | LVK(Ub)DVFISAAERDVYTGDALR | Yes | [11], [13] | |||
| PSMB2 | β4 | 185 | IDK(Ub)NGIHDLDNISFPK | Yes | [11], [16] | |
| PSMB3 | β3 | 77 | LNLYELK(Ub)EGR | Yes | [11-13], [16] | Yes |
| 83 | IK(Ub)PYTLMSMVANLLYEKR | Unreported | ||||
| PSMB4 | β7 | 201 | EVLEK(Ub)QPVLSQTEAR | Yes | [9], [11], [13], [16] | Yes |
| 66 | TTTLAFK(Ub)FR | Unreported | ||||
| PSMB5 | β5 | 92 | ATAGAYIASQTVKK(Ub) | Unreported | ||
| 257 | VSSDNVADLHEK(Ub)YSGSTP | Yes | [11] | |||
| PSMB6 | β1 | 183 | GMTK(Ub)EECLQFTANALALAMER | Yes | Unreported | |
| PSMB7 | β2 | 127 | MLK(Ub)QMLFR | [11], [16] | ||
| 258 | TTAVLTEK(Ub)ITPLEIEVLEETVQTMDTS | Unreported |
Proteasome sequences are remarkably conserved throughout eukaryotes, making it moot to distinguish the significance of modified residues from homology. Similarly, the small number of residues does not facilitate the deduction of common sequence motifs. Nevertheless, we did observe a suggestive enrichment of serines in the +4 position from ubiquitinated lysines over all lysines in 20S proteasome (3.9-fold, P < 0.005) and in the whole proteome (3.5-fold, P < 0.05) (Figure 1C). The sites with serine at the +4 position do not show strong sequence homology, suggesting the serine residue alone may act in target recognition, although the motif was not observed from large-scale ubiquitination profiling data. Previous study reported an enrichment of a K*XL motif [9] or negatively charged residues [11] around ubiquitination sites, but the existence of ubiquitination motifs have been disputed [12]. We did not observe any reported motifs in our dataset, possibly because the idiosyncrasy of proteasome sequences renders it impervious to proteome-wide generalizations.
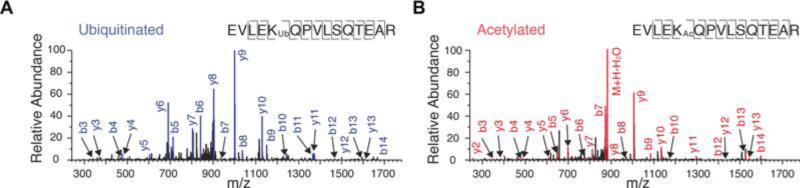
Parallel to ubiquitination, proteasome lysines can also be modified by acetylation [4]. It has been suggested that acetylation and ubiquitination can crosstalk by competing for common lysine residues [16, 18]. Our recent investigation showed that acetylation directly elevates proteasome functions in the mouse heart [19], a phenomenon that can potentially be obstructed by ubiquitination should it occur at a regulatory acetylation site. To test whether lysine acetylation and ubiquitination sites occur preferentially at identical lysines, we identified 65 unique lysine acetylation sites from the same experiments (Supplemental Table 2), out of which 27 passed stringency filters (Figure 1D). In total, 39 ubiquitinated sites were also acetylation targets (61%), although only 11 stringent sites were shared (41%), suggesting some overlap may be due to noise. This figure is nevertheless higher than reported proteome-wide averages (20–30%) [12, 16]. Figure 2 shows an example site (PSMB4 K201) that is subject to both modifications. Overall, an ubiquitinated lysine is statistically much more likely to be also an acetylation site over any modifiable lysine on the human 20S proteasome sequences (found ubiquitinated or acetylated in any human cell sample) (Χ2 = 11.39; df = 1; P = 7.0 × 10−4). This enrichment became more pronounced when all lysines on the 20S proteasome were considered (P < 1.0 × 10−4). The significant overlap of ubiquitination and acetylation sites is suggestive of co-regulation on specific residues through competitive occupancy between the two modifications.
Figure 2. Common ubiquitination and acetylation sites on 20S proteasomes.
Peptide spectrum matches showing PSMB4 K201 can be (A) ubiquitinated or (B) acetylated on cardiac 20S proteasomes.
In summary, we provided the first systematic characterization of confident ubiquitination sites on human cardiac 20S proteasomes. We identified ubiquitinated lysines on all 14 core 20S proteasome subunits from assembled proteasome complexes, including sites that share with acetylation targets. To our knowledge, 22 ubiquitination sites were first reported in cardiac tissues here. Future studies are needed to differentiate poly-ubiquitin linkages at these sites from mono-ubiquitination, or the possibility of ubiquitin-like modifiers (e.g., NEDD8 and ISG15) on 20S proteasomes (Supplemental Figure S1), which would leave behind identical trypsin proteolytic residues [11, 20]. The information presented here is essential toward understanding the functions of these modification sites on the proteasomes, e.g., by developing modification-specific MRM transitions for the modification sites, it will be possible to perform target quantification of their occupancy in health and disease [21, 22]. Finally, the general approach described will be useful for acquiring tissue-specific modification profiles of other protein complexes. The PTM data are integrated into our knowledgebase COPaKB (http://www.heartproteome.org) [17] to support future investigations.
Supplementary Material
Clinical Relevance.
Proteasome insufficiency contributes to protein aggregation and promotes the progression of cardiac hypertrophy and failure. Interventions targeting proteasome modifications and functions are being explored as potential therapeutic avenues. To discover the extent of ubiquitination on human cardiac proteasomes, we profiled lysine ubiquitination and acetylation in five human clinical samples of ischemic cardiomyopathy hearts using high-resolution MS. These data are the first step toward understanding how cardiac 20S proteasomes can be regulated by multiple PTMs in health and disease.
Acknowledgments
This work was supported by the NIH awards HL-R37-63901 and HHSN268201000035C, and the Laubisch endowment to P. Ping; AHA fellowships 13POST14700031 to M.P. Lam; 12PRE11610024 to E. Lau.
Footnotes
Non-standard abbreviations: None.
The authors have declared no conflict of interest.
References
- 1.Lam MP, Wang D, Lau E, Liem D, et al. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J Clin Invest. 2014 doi: 10.1172/JCI73787. Accepted; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Robbins J. Proteasomal and lysosomal protein degradation and heart disease. J Mol Cell Cardiol. 2013 doi: 10.1016/j.yjmcc.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Predmore JM, Wang P, Davis F, Bartolone S, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scruggs SB, Zong NC, Wang D, Stefani E, Ping P. Post-translational modification of cardiac proteasomes: functional delineation enabled by proteomics. Am J Physiol Heart Circ Physiol. 2012;303:H9–18. doi: 10.1152/ajpheart.00189.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Z, Scruggs SB, Gilda JE, Ping P, Gomes AV. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. J Mol Cell Cardiol. 2013 doi: 10.1016/j.yjmcc.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ventadour S, Jarzaguet M, Wing SS, Chambon C, et al. A new method of purification of proteasome substrates reveals polyubiquitination of 20 S proteasome subunits. J Biol Chem. 2007;282:5302–5309. doi: 10.1074/jbc.M610005200. [DOI] [PubMed] [Google Scholar]
- 7.Isasa M, Katz EJ, Kim W, Yugo V, et al. Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol Cell. 2010;38:733–745. doi: 10.1016/j.molcel.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics. 2007;7:868–874. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- 11.Kim W, Bennett EJ, Huttlin EL, Guo A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10:M110, 003590. doi: 10.1074/mcp.M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner SA, Beli P, Weinert BT, Scholz C, et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics. 2012;11:1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau E, Wang D, Zhang J, Yu H, et al. Substrate- and isoform-specific proteome stability in normal and stressed cardiac mitochondria. Circ Res. 2012;110:1174–1178. doi: 10.1161/CIRCRESAHA.112.268359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriksen P, Wagner SA, Weinert BT, Sharma S, et al. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol Cell Proteomics. 2012;11:1510–1522. doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner SA, Beli P, Weinert BT, Nielsen ML, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111, 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zong N, Li H, Li H, Lam MP, et al. Integration of Cardiac Proteome Biology and Medicine by a Specialized Knowledgebase. Circ Res. 2013;113:1043–1053. doi: 10.1161/CIRCRESAHA.113.301151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Fang C, Zong NC, Liem DA, et al. Regulation of acetylation restores proteolytic function of diseased myocardium in mouse and human. Mol Cell Proteomics. 2013;12:3793–3802. doi: 10.1074/mcp.M113.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam MP, Lau E, Scruggs SB, Wang D, et al. Site-specific quantitative analysis of cardiac mitochondrial protein phosphorylation. J Proteomics. 2012 doi: 10.1016/j.jprot.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam MP, Scruggs SB, Kim TY, Zong C, et al. An MRM-based workflow for quantifying cardiac mitochondrial protein phosphorylation in murine and human tissue. J Proteomics. 2012;75:4602–4609. doi: 10.1016/j.jprot.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.