Abstract
Adaptive phenotypic evolution is shaped by natural selection on multiple organismal traits as well as by genetic correlations among traits. Genetic correlations can arise through pleiotropy and can bias the production of phenotypic variation to certain combinations of traits. This phenomenon is referred to as developmental bias or constraint. Developmental bias may accelerate or constrain phenotypic evolution, depending on whether selection acts parallel or in opposition to genetic correlations among traits. We discuss examples from floral evolution where genetic correlations among floral traits contribute to rapid, coordinated evolution in multiple floral organ phenotypes and suggest future research directions that will explore the relationship between the genetic basis of adaptation and the pre-existing structure of genetic correlations. On the other hand, natural selection may act perpendicular to a strong genetic correlation, for example when two traits are encoded by a subset of the same genes and natural selection favors change in one trait and stability in the second trait. In such cases, adaptation is constrained by the availability of genetic variation that can influence the focal trait with minimal pleiotropic effects. Examples from plant diversification suggest that the origin of certain adaptations depends on the prior evolution of a gene copy with reduced pleiotropic effects, generated through the process of gene duplication followed by subfunctionalization or neofunctionalization. A history of gene duplication in some developmental pathways appears to have allowed particular flowering plant linages to have repeatedly evolved adaptations that might otherwise have been developmentally constrained.
Keywords: Developmental constraint, bias, pleiotropy, floral morphology, repeated evolution, gene duplication
Organisms are complex systems of developmentally and functionally related traits (characters), and natural selection acts on more than one trait at a time. For example, selection may favor evolutionary change in one or more traits, while at the same time favor stability in others. The response to selection on multiple traits will depend on the structure of underlying genetic and developmental pathways, which can limit the production of variation in certain phenotypic directions compared to others. This phenomenon is generally known as developmental bias, developmental constraint, or developmental drive (Maynard Smith et al. 1985; Arnold 1992; Arthur 2001) and is predicted to shape evolutionary trajectories of complex phenotypes (Wagner and Altenberg 1996).
Flowers, in particular, are complexes of multiple morphological and physiological traits. While floral evolution has produced a spectacularly diverse array of phenotypes, individual floral traits are functionally and developmentally interrelated. Therefore, flowers are an attractive system in which to investigate evolutionary change in complex phenotypes, the interaction between natural selection and developmental bias, and the emergence of repeated patterns in evolution.
Developmental bias and pleiotropy in phenotypic evolution
The concept of developmental bias represents an important link between the fields of developmental biology, quantitative genetics, and the evolution of development (Maynard Smith 1985; Futuyma 2010; Losos 2011). This is because a population's response to selection will be influenced by the structure of genetic correlations among the phenotypic traits under selection (Lande and Arnold 1983). Genetic correlations are correlated patterns of variation among phenotypic traits across individuals in a population. If two or more traits are uncorrelated, traits can independently respond to selective pressures. However, if traits are correlated, selection acting on one trait can produce a corresponding effect on a second trait. An important source of genetic correlation is pleiotropy, where mutational variation in individual genes affects multiple traits, thereby generating the observed phenotypic correlation.
As an example, consider a developmental pathway that influences two traits. Mutations to this pathway that increase the magnitude of one trait also increase the magnitude of the second trait. The two traits will display a positive genetic correlation across individuals in a population due to pleiotropy. The evolutionary consequences of this genetic correlation will depend on how natural selection acts on these two traits. If natural selection happens to favor phenotypic change in both traits that is parallel to the genetic correlation (i.e., selection favors increased magnitude in both traits or a decrease in both traits), adaptive evolution towards the fitness optimum will be relatively rapid compared to when selection favors a phenotypic change in these traits that is perpendicular to the pattern of genetic correlation (Fig. 1; Lande and Arnold 1983; Maynard Smith 1985; Schluter 1996; Conner 2012). In other words, depending on the alignment between natural selection and genetic correlations among traits, certain novel phenotypes may be more accessible (selected phenotype is more quickly attained by a population) than others.
Fig 1.
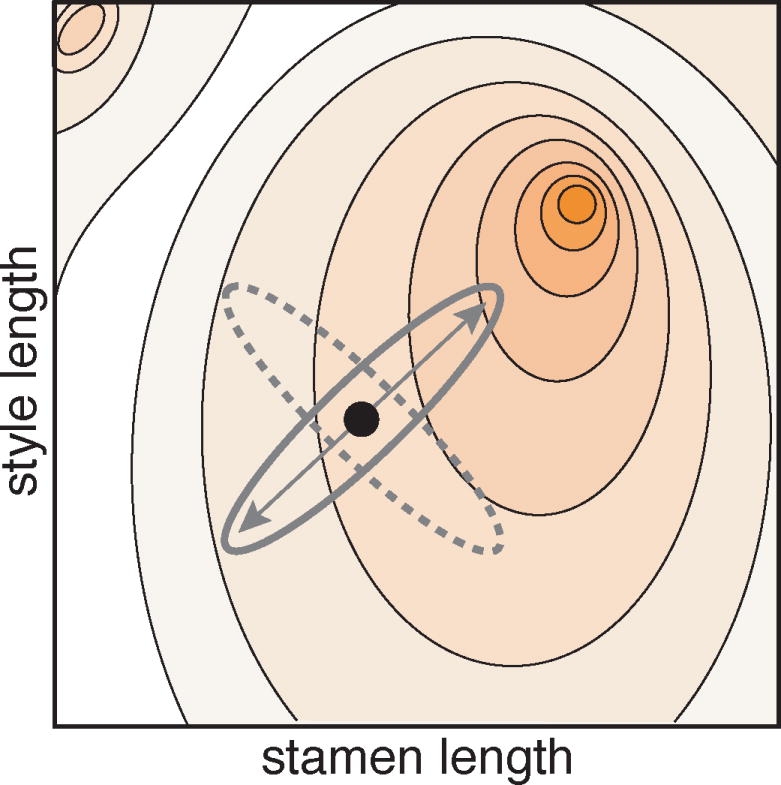
The effect of trait correlation on accessibility of adaptive phenotypes. The background depicts the contours of a fitness landscape that displays fitness (Z axis, represented using contour lines) associated with different combinations of stamen filament length (X axis) and style length (Y axis). The adaptive peak is denoted by the darkest orange point. The black dot represents the current population mean for stamen filament and style lengths. If the two traits are correlated in a direction parallel to the direction of natural selection (solid ellipse) then evolution towards the fitness peak is more easily accessible than a situation in which the two traits are correlated perpendicular to the direction of natural selection (dotted ellipse).
Some genes participate in the development of multiple essential organismal phenotypes. When selection favors adaptive change in one trait but exerts strong stabilizing selection on other traits, selection acts perpendicular to a genetic correlation. Adaptation in these cases can be constrained by the availability of trait-specific mutations – those that can generate the favored change in one aspect of phenotype with minimal pleiotropic effects (Futuyma 2010). The opportunity for such trait-specific mutations can arise through processes of genome evolution. One example is gene duplication followed by the evolution of a narrowed expression domain in one or more gene copies. This process can generate a gene paralog with reduced developmental scope and subsequent mutations to this paralog may enable adaptive evolution of the focal trait with minimal pleiotropic effects (Carroll 2005).
Other mechanisms besides pleiotropy can generate and maintain genetic correlations between traits within a population. For example, genetic correlation can result from tight genetic linkage between loci that affect different traits. In this case, genetic variation at each of the trait-specific loci produces corresponding phenotypic variation. A genetic correlation between the traits is generated because recombination events between the tightly linked loci are rare. Alternatively, strong selection for certain combinations of traits (correlational selection) can generate linkage disequilibrium between unlinked loci affecting different traits, yielding genetic correlations among traits (Lynch and Walsh 1998). Genetic correlations between unlinked loci are unlikely to contribute to developmental bias since they are the result of selection and can be altered by a change in the direction of natural selection (Sinervo and Svensson 2002). Yet consistent correlational selection over long periods of time may alter the genetic and developmental architecture underlying traits – favoring developmental integration of functionally related traits that experience correlational selection, and parcellation of functionally unrelated traits into separate developmental modules (Wagner and Altenberg 1996).
Here we discuss how the evolution of certain novel phenotypes may be accessible or may be constrained due to developmental bias, using examples from floral evolution.
Genetic correlations among floral traits that may facilitate adaptation
A flower is a complex set of organs that facilitate reproduction through pollen movement by abiotic and biotic factors. Animal-pollinated flowers additionally have adaptations that attract and reward pollinators. Floral traits are functionally integrated in order to ensure successful pollination and reproduction; therefore floral adaptation requires coordinated change in multiple morphological and physiological traits. Despite this required complexity for adaptive floral evolution, closely related species often exhibit strikingly divergent floral phenotypes (Stebbins 1970, 1974). This observation generates a paradox–how does coordinated phenotypic change in multiple traits happen on a short evolutionary timescale? This paradox may be resolved in part by the occurrence of developmental biases that facilitate correlated change in floral morphological traits.
Genetic correlations among floral traits are pervasive (Ashman and Majetic 2006) and a material source of genetic correlations among floral traits is pleiotropy (Conner 2002; Smith 2016). Pleiotropy among floral traits derives in part from the developmental homology of different floral organs. The flower is comprised of serially homologous organs: sepals, petals, stamens, and carpels, which share developmental control (Bowman et al. 1991). Genetic correlations also may be generated between floral organs by tight linkage or by consistent correlational selection, for example by pollinators (Conner et al. 2009).
Two examples of evolutionary change in floral phenotype that have occurred frequently during angiosperm evolution are shifts from flowers adapted for outcrossing to flowers adapted to self fertilization (a selfing syndrome), and adaptation to a novel pollinator. Both types of evolutionary change occur on a short evolutionary timescale, with closely related species (e.g., outcrossing versus selfing, or bee-pollinated versus hummingbird-pollinated) differing in multiple floral traits (Stebbins 1970, 1974).
Evolution of selfing syndromes
Transition in mating system from predominantly outcrossing to highly selfing is considered to be one of the most common evolutionary transitions in flowering plants (Stebbins 1974). Shifts to selfing can be selectively favored for reproductive assurance when potential mates or pollinators are scarce, for example in marginal habitats. If a self-incompatibility mechanism exists in the outcrossing species, the evolution of selfing requires the disintegration of this system. Shifts to selfing also frequently involve the adaptive evolution of a selfing syndrome: small flowers (with reduced petal area, shorter stamens, and shorter styles), reduced herkogamy (spatial separation between stigma and anthers; this may often result simply from reduction in flower size), reduced scent and nectar production, and reduced pollen-to-ovule ratio.
The yellow monkeyflower Mimulus guttatus is variable for life history and flower size (Wu et al. 2008). Multiple lineages derived from this species, or a M. guttatus-like ancestor, have transitioned to selfing and display small flowers with reduced pollen production (Ritland and Ritland 1989). Quantitative genetic studies have examined the genetic basis of flower size variation in the M. guttatus species complex using crosses among populations or species (Fenster and Ritland 1994; Fishman et al. 2002; Hall et al. 2006). These studies have identified strong positive genetic correlations among floral dimensions in segregating F2 populations (Table 1). Two of these studies also mapped floral trait QTL in F2 populations (Fishman et al. 2002; Hall et al. 2006). In both studies, QTLs underlying different floral dimensions mapped to exactly or nearly exactly the same location in the genome.
Table 1.
Genetic correlations among floral traits associated with the evolution of selfing syndrome estimated from F2 hybrid populations of Mimulus. AL: stamen filament length, CL: corolla length, CW: corolla width, SL: style length.
| Cross | CW–CL | CW–AL | CW–SL | CL–AL | CL–SL | AL–SL | Reference |
|---|---|---|---|---|---|---|---|
| M. guttatus x M. micranthus | 0.96 | 0.90 | 0.79 | 1.01 | 0.98 | 0.97 | Fenster and Ritland 1994 |
| M. nasutus x M. micranthus | 0.98 | 0.65 | 0.86 | 0.92 | 0.85 | 0.99 | Fenster and Ritland 1994 |
| M. guttatus x M. laciniatus | 0.89 | 0.83 | 0.87 | 0.97 | 0.99 | 0.93 | Fenster and Ritland 1994 |
| M. nasutus x M. laciniatus | 1.10 | 0.82 | 0.80 | 0.88 | 0.82 | 0.82 | Fenster and Ritland 1994 |
| M. guttatus x M. nasutus | 0.73 | 0.67 | 0.82 | 0.99 | 1.08 | 0.96 | Fenster and Ritland 1994 |
| M. micranthus x M. laciniatus | 0.79 | 0.74 | 0.62 | 0.85 | 0.63 | 0.84 | Fenster and Ritland 1994 |
| M. guttatus x M. nasutus | 0.97 | 0.96 | 0.93 | 0.95 | 0.90 | 0.93 | Fishman et al. 2002 |
| M. guttatus (perennial) x M. guttatus (annual) | 0.59 | 0.567 | 0.671 | 0.751 | 0.816 | 0.814 | Hall et al. 2006 |
| M. lewisii x M. parishii | 0.72 | 0.52 | 0.60 | 0.58 | 0.83 | 0.59 | Fishman et al. 2014 |
These data suggest that the types of genetic variants that fix during transitions to a selfing syndome reside at pleiotropic loci that control the size of multiple floral organs. Variation at these loci generates genetic correlations in floral organ size (e.g., between petal and stamen size). During transitions to selfing, selection favors correlated changes in floral organ size which are in alignment with these genetic correlations (Fig. 1). Under this scenario, developmental bias will accelerate adaptive evolution towards the selfing syndrome in the M. guttatus species complex, and may help explain its repeated evolution.
This developmental bias may not be a feature restricted to the M. guttatus complex; other study systems show correlations among floral dimensions that suggest such pleiotropy. The pink monkeyflower, M. lewisii, is distantly related to M. guttatus. It is an outcrosser and its close relative, M. parishii, exhibits a selfing syndrome. Fishman et al. (2014) examined the genetic architecture of floral syndrome divergence between these two species and found that floral traits are strongly and positively correlated, again due to largely coincident QTL for different floral dimensions (Table 1). This suggests that the floral organ sizes are developmentally coupled to a similar degree across multiple Mimulus lineages. Moreover, putatively pleiotropic QTL for flower size associated with evolutionary shifts to selfing were identified in Capsella (Slotte et al. 2012) and Leptosiphon (Goodwillie et al. 2006).
Large positive genetic correlations and coincident QTL among floral dimensions, particularly among lengths of floral organs, are also found in crosses between different populations of Arabidopsis thaliana, Brassica rapa, and wild radish (e.g., Conner and Via 1993; Conner and Sterling 1995; Juenger et al. 2000, 2005; Conner 2002; Brock et al. 2010, 2012; Edwards and Weinig 2011). These intraspecific data suggest that much of the standing genetic variation in floral organ size in these species resides at pleiotropic loci, and that there may be many genomic regions that each affect multiple floral organs.
The hypothesis that flower organ size variation is largely due to genetic variation at pleiotropic loci is consistent with our understanding of the developmental control of floral organ size in A. thaliana. Studies of A. thaliana mutants have identified genes that influence the size or shape of floral organs and the majority of identified genes that control floral organ size act pleiotropically on multiple floral organs (reviewed by Krizek and Anderson 2013). For example, a developmental pathway that includes ARGOS, AINTEGUMENTA (ANT), ARGOS-LIKE (ARL), ORGAN SIZE RELATED 1 (OSR1), and ORGAN SIZE RELATED 2 (OSR2) integrates multiple hormonal signals and controls plant organ size through both cell proliferation and cell expansion (Elliott et al. 1996; Mizukami and Fischer 2000; Hu et al. 2003, 2006; Feng et al. 2011; Qin et al. 2014). This pathway is expressed in multiple floral organs and loss-of-function mutations have pleiotropic effects across these floral organs. Therefore, there are likely many genetic loci that each can produce correlated changes to floral organ size, consistent with the QTL results discussed above.
The evolutionary consequence of this genetic architecture is that concomitant shifts in the size of different floral organs (e.g., both petals and reproductive organs) may be easily accessible to plant populations when favored by selection since pleiotropic variation allows adaptation in multiple traits simultaneously. This architecture may explain why the evolution of the selfing syndrome has occurred so frequently during plant evolution.
Evolution of pollination syndromes
While strong positive correlations among floral dimensions may facilitate rapid evolutionary transitions to a selfing syndrome, not all adaptive shifts in floral evolution can be obtained through a modification of overall flower size; for example, adaptive shifts in pollination syndrome. Pollination syndromes are suites of floral traits that attract and facilitate pollination by particular animal (or abiotic) pollinators (Faegri and Van der Pijl 1979). For example, flowers that display a bee pollination syndrome have short wide flowers with included reproductive organs that are generally colorful (but rarely red) and produce small amounts of nectar. Flowers with a hummingbird pollination syndrome have bright red tubular flowers with exserted reproductive organs and produce large amounts of nectar. The moth pollination syndrome is characterized by very long and narrow floral tubes that are white and produce a sweet fragrance. Although adaptation to a novel pollinator requires coordinated change in multiple morphological and physiological traits, pollination syndrome evolution is highly labile in some plant groups (Thomson and Wilson 2008).
Studies in Ipomopsis (Nakazato et al. 2013), Mimulus (Bradshaw et al. 1995, 1998; Fishman et al. 2013), Nicotiana (Bissell and Diggle 2008, 2010), Penstemon (Wessinger et al. 2014), and Petunia (Hermann et al. 2013, 2015) have used quantitative genetic analyses to examine the genetic architecture of pollination syndrome divergence between closely related species that display alternative pollination syndromes. In each of these study systems, the pollination syndrome transition involves a coordinated change in corolla tube, stamen, and style lengths (Fig. 2) that together result in a proper fit between flowers and pollinators for efficient pollen transfer. For example shifts from bee or fly to hummingbird adaptation (e.g., Ipomopsis, Mimulus, Penstemon) involve evolutionary increases to corolla tube, stamen and style lengths. Transitions between hummingbird and moth pollination syndromes (e.g., Nicotiana, Petunia) also require coordinated evolution in floral organ lengths. Longer corolla tubes, stamens, and styles are associated with moth pollination compared to hummingbird pollination. Similar to the evolution of selfing syndrome, coordinated changes associated with pollination syndrome evolution may be facilitated by genetic correlations among floral organ length traits. Quantitative genetic studies in these systems suggest that pleiotropic loci may underlie genetic correlations and that developmental bias may facilitate pollination syndrome evolution.
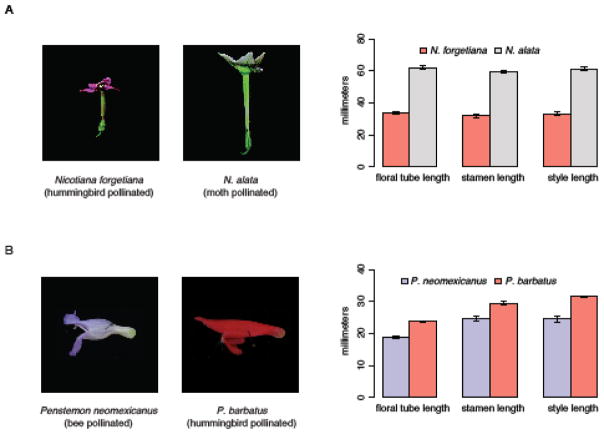
Fig 2.
Two examples of correlated shifts in floral organ length that accompany pollination syndrome evolution. Photographs of parental species showing divergent pollination syndromes and graphical depiction of mean floral organ length in parental species. (A) Nicotiana: images adapted from Bissell and Diggle (2008), data from Bissell and Diggle (2010). (B) Penstemon: images and data from Wessinger et al. (2014). Error bars indicate standard deviations.
Four of these study systems (Ipomopsis, Nicotiana, Penstemon, and Petunia) estimated correlations among floral traits in hybrid populations. In each case, the lengths of floral organs are strongly positively correlated (Table 2). To our knowledge, correlations among floral traits have not yet been reported in the Mimulus (M. cardinalis x M. lewisii) system. Additionally, QTL analyses in hybrid populations were performed in four of these study systems (Ipomopsis, Mimulus, Penstemon, and Petunia). In each case, identified QTLs underlying variation in corolla tube, stamen, and style lengths tended to coincide, with variation in usually at least two, and frequently all three of these traits mapping to the same genomic location. These QTL results suggest that pleiotropic QTL may explain variation in these traits. While QTL analysis has not been performed in Nicotiana, correlations among floral traits were estimated in a fourth-generation hybrid population that had experienced four generations of recombination (Bissell and Diggle 2010), thus the strong genetic correlations among floral organ lengths in Nicotiana (Table 2) are expected to reflect pleiotropy or tight genetic linkage. Together, these studies suggest that genetic correlations among the lengths of floral organs are common in species that have experienced a shift in pollination syndrome, and pleiotropic QTLs may underlie these correlations. These genetic correlations may bias phenotypic transitions towards those that involve coordinated change in floral organ lengths, making certain pollination syndrome transitions evolutionarily accessible.
Table 2.
Genetic correlations among floral traits associated with the evolution of pollination syndromes. TW: corolla tube width, TL: corolla tube length, AL: stamen filament length, SL: style length. For Ipomopsis, TW=tube width at throat. For Petunia, TL=length of proximal floral tube and AL=length of lateral stamen pair. For Penstemon, AL=length of lateral stamen pair. For Nicotiana, AL=length of portion of stamen that is adnate to corolla tube.
| Species | TW– TL | TW– AL | TW– SL | TL–AL | TL–SL | AL–SL | Reference |
|---|---|---|---|---|---|---|---|
| Ipomopsis aggregata (hummingbird) x I. guttata | 0.375 | 0.358 | 0.203 | 0.872 | 0.706 | 0.813 | Nakazato et al. 2013 |
| Nicotiana alata (moth) x N. forgetiana (hummingbird) | 0.3021 | 0.3090 | 0.3051 | 0.9098 | 0.9850 | 0.8970 | Bissell and Diggle 2010 |
| Penstemon barbatus (hummingbird) x P. neomexicanus (bee) | −0.25 | −0.20 | −0.19 | 0.77 | 0.37 | 0.38 | Wessinger et al. 2014 |
| Petunia axillaris (moth) x P. exserta (hummingbird) | n/a | n/a | n/a | 0.6618 | 0.5814 | 0.8421 | Hermann et al. 2015 |
Genetic correlations among floral traits that may oppose adaptation
In our discussion of the evolution of selfing, we found that corolla tube width is often positively correlated with floral organ lengths in a variety of plant species. Yet evolution of hummingbird or moth pollination syndromes favors longer floral organ lengths and narrower corolla tube width. Thus evolutionary change in these cases may occur either through trait-specific variation that allow corolla tube width to be modified independently from floral organ lengths or through pleiotropic mutations that produce contrasting effects on width vs. length traits. Three study systems (Ipomopsis, Nicotiana, and Penstemon) report correlations among width and length traits (Table 2).
Both Ipomopsis and Nicotiana report modest positive correlations between corolla width and floral organ lengths. In Ipomopsis, a subset of corolla width QTL overlap with floral length QTLs, producing positively correlated change in corolla width and floral organ lengths; the remaining QTLs are trait-specific, affecting either width or lengths (Nakazato et al. 2013). These trait-specific QTLs may reflect variation that allowed adaptation to circumvent the positive correlation between length and width traits. In Penstemon, corolla tube width was weakly negatively correlated with floral organ lengths, and none of the QTLs for corolla width overlapped with floral organ length QTLs (Wessinger et al. 2014). In this case, the negative correlations likely result from linkage among QTLs in a small F2 mapping population.
These studies suggest that evolutionary change in pollination syndrome may involve genetic variation that has a different pattern of correlations (i.e., strong correlations among length traits but weaker correlations between width and length traits) compared to transitions to selfing syndrome where strong correlations exist among floral dimensions. Altered patterns of genetic correlations ancestrally present in these lineages may have enabled the evolution of floral tube width to be decoupled from correlated evolution of floral organ lengths. Alternatively, they may reflect the pattern of genetic correlation that passed through the filter of selection during adaptation to novel pollinators.
Detecting biases on the genetic variation that contributes to adaptation
The pattern of genetic correlations identified in hybrid populations (generated from crossing a lineage that displays a derived adaptation to a lineage that retains the ancestral phenotypic state) may reflect the genetic correlations that were present before adaptive evolution in the ancestral population. One way to explore the validity of this assumption is to compare the genetic architecture of phenotypic correlations before and after adaptive evolution. Studies in Mimulus guttatus have found that outcrossing populations show the same pattern of genetic correlations (namely, large positive correlations between floral dimensions; e.g., Scoville et al. 2009) as have been identified in hybrid populations derived from crosses between outcrossing and selfing lineages (Table 1). Additionally, in Nicotiana, both the hummingbird-adapted species N. alata and the hawkmoth-adapted species N. forgetiana show the same pattern of genetic correlations (length traits highly correlated, but weaker correlations between length and width traits) as were identified in a population derived from a cross between N. alta and N. forgetiana (Table 2). These observations suggest that the genetic correlations identified through interspecific crosses aimed at identifying the genetic architecture of adaptive evolution reflect ancestral genetic correlations. Hence these correlations may have facilitated the adaptive shift in each case.
Alternatively, genetic variation that underlies adaptation may not reflect the dominant pattern of genetic correlation that segregated within the ancestral population. Evidence of this comes from selection experiments in wild radish. Conner et al. (2011) applied selection over nine generations perpendicular to a strong genetic correlation (r2=0.85 in the ancestral population) between corolla tube length and filament length in wild radish and observed an immediate and steady response to selection. While most of the genetic variation in corolla tube and filament lengths segregating within wild radish affects both traits and contributes to the strong genetic correlation between these two traits, clearly there is also genetic variation that is free from the constraint of correlation. This trait-specific variation can separately influence floral organs, and may be the variation upon which selection acted.
These different possibilities lead to the following predictions. When selection acts parallel to existing genetic correlations, we expect adaptation to proceed by favoring correlated genetic variation (i.e., genetic variation that affects multiple traits in a direction parallel to selection). This is the basis for our central prediction that genetic correlations facilitate adaptation when selection acts parallel to the correlation. Conversely, when selection acts perpendicular to existing genetic correlations, we expect adaptation to proceed by favoring only genetic variation that affects traits independently.
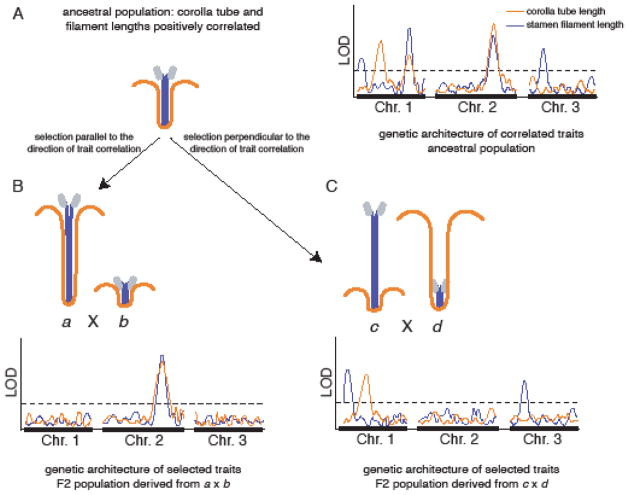
These predictions can be tested using quantitative genetic analyses. We illustrate these analyses using a hypothetical example similar to the wild radish study system of Conner et al. (2011), starting with a focal population from which selection lines are derived. The first step is to estimate QTLs for two correlated traits segregating within an ancestral population, for example using a replicated F2 mapping design (Kelly 2009; Scoville et al. 2009). In a system similar to wild radish, presumably QTLs for corolla tube length and stamen length will colocalize to several genomic locations. These overlapping QTLs reflect loci that contribute to the genetic correlation between traits. Additional QTLs would be non-overlapping, and contribute to trait-specific phenotypic variation (Fig. 3A).
Fig. 3.
Example of potential results from experiments that compare genetic architectures of adaptation to artificial selection when selection is applied parallel vs. perpendicular to a pre-existing positive genetic correlation between two traits, corolla tube length and stamen filament length. (A) Ancestral population shows intermediate corolla tube and filament lengths and quantitative genetic analyses suggest that some QTLs explain variation in both traits, while other QTLs are trait-specific. (B) Selection lines a and b result from artificial selection applied parallel to the genetic correlation; these lines are crossed and quantitative genetic analysis on the hybrid population reveals that overlapping QTLs that explain variation in both corolla tube and filament length underlie the response to selection. (C) Selection lines c and d result from artificial selection applied perpendicular to the genetic correlation; these lines are crossed and quantitative genetic analysis on the hybrid population reveals that trait-specific QTLs underlie the response to selection.
The next step is to derive four selection lines from this ancestral population by applying artificial selection over several generations. Two of these selection lines are generated by applying selection parallel to the genetic correlation, one line in each direction (e.g., line a: long corolla tube, long stamen, line b: short corolla tube, short stamen). The other two selection lines are generated by selection perpendicular to the correlation (e.g., line c: short corolla tube, long stamen, line d: long corolla tube, short stamen). After selection, the genetic architecture underlying the response to selection in each selection regime (i.e., parallel vs. perpendicular selection) is compared using QTL experiments. In particular, the two parallel selected lines (a and b) are crossed and the two perpendicular selected lines (c and d) are crossed to generate two mapping populations. QTL analyses are performed on each population. We predict that different loci will respond to selection in the parallel vs. perpendicular selection lines: we expect overlapping QTL to predominantly contribute to the response to selection in parallel selected lines (Fig. 3B) while trait-specific QTL contribute to the response to selection in perpendicular selected lines (Fig. 3C).
Evaluating the contribution of pleiotropy to genetic correlations
The studies described above clearly show that floral traits associated with the evolution of selfing syndrome and of pollination syndrome divergence can be genetically correlated, often parallel to the direction of selection. QTL studies suggest that genetic correlations may result from loci with pleiotropic effects on multiple floral organs. However, overlapping QTLs do not necessarily prove pleiotropic loci are responsible, since F2 populations do not contain enough recombination events to separate tightly linked loci. For example, in Petunia, fine-mapping floral traits underlying pollination syndrome divergence has revealed three tightly linked loci in a 0.1 cM interval that separately encode (1) the presence/absence of floral scent, (2) the presence/absence of UV absorbant pigmentation as well as visible color and (3) the lengths of floral organs (Hermann et al. 2013; Sheehan et al. 2015).
In order to demonstrate pleiotropy, it is necessary to identify the mutational basis for individual QTL through fine-mapping, positional cloning, and functional verification. This is a large effort in model plant species, and even more so in non-model systems. Despite the required effort, determining the underlying genetic variants that explain trait variation at all loci, but most critically coincident QTL for correlated floral traits, will reveal the structure of developmental pathways for flower size and shape and the role of pleiotropy in generating genetic correlations among floral traits.
In advance of this substantial effort, detailed developmental analyses can provide insight into the contribution of pleiotropy to correlated traits. These analyses will not provide direct evidence for pleiotropy, but certain experimental outcomes could rule out pleiotropy. We describe these analyses below using interspecific variation in floral organ lengths and corolla width associated with pollination syndrome divergence as an example.
The developmental basis for both intra- and interspecific variation in floral organ lengths and width results from divergent patterns of cell proliferation, cell expansion, or a combination of these. These divergent patterns should be reflected in the average cell size and cell number along the lengths of stamen filaments and styles, and along both the length and width dimensions of petal tissue. Comparing the patterns of cell proliferation and cell expansion across different floral organs is a first step towards characterizing the genetic basis for adaptive differences in flower size and shape. For example, in Petunia adaptive differences in style length are due to greater cell proliferation rather than greater cell elongation (Hermann et al. 2015). Under the hypothesis that genetic correlations among floral dimensions in Petunia involve pleiotropic genetic variation, we would expect to see that the correlated differences in floral tube length and stamen filament length show the same developmental alteration as is observed for style length (i.e., greater cell proliferation). Cell size and number has not yet been reported for floral tube and stamen filament tissue in Petunia. These developmental comparisons will not necessarily demonstrate pleiotropy, as linked loci might produce similar changes to cell number and/or size in different floral organs. However, if one observed a different developmental basis in different organs (e.g., increased cell size in stamen filaments and increased cell number in petal tissue), this would provide strong evidence for separate organ-specific loci, even under coincident QTLs, ruling out pleiotropy as a contributor to developmental bias.
Circumventing genetic correlations that oppose adaptation: gene duplication
As demonstrated above by work in radish (Conner et al. 2011), even if natural selection acts perpendicular to a strong genetic correlation, as long as the correlation is less than one, there remains genetic variation upon which selection may act. On the other hand, when two or more traits rely on a completely overlapping set of genes and the genetic correlation is equal to one, pleiotropy can constrain these traits from independently responding to natural selection (Futuyma 2010). This absolute constraint can be alleviated by the origin of a genomic region that has a trait-specific influence on phenotype (Wagner and Altenberg 1996). One process that can accomplish this is gene duplication followed by subfunctionalization or neofunctionalization (Carroll 2005) of either regulatory or coding function. This yields a new gene copy with limited scope that can harbor genetic variation that has trait-specific (non-pleiotropic) phenotypic effects. Recent studies in plants suggest that acquiring a gene copy with narrow expression domain can precede the evolution of certain novel phenotypes.
Although not focused on flowers, a recent example that nicely illustrates this process comes from leaf shape evolution. Lineages within Brassicaceae have experienced evolutionary change in leaf complexity. The functional significance of this leaf shape diversity is not clear, but may reflect local adaptation to climate (reviewed in Ferris et al. 2015). In at least two lineages within this plant family, evolutionary change in the degree of leaf dissection has involved mutations to the homeobox gene REDUCED COMPLEXITY (RCO) (Sicard et al. 2014; Vlad et al. 2014). RCO derives from two tandem gene duplication events of a LATE MERISTEM IDENTITY1 (LMI)-type gene in Brassicaceae yielding three homologs: LMI1, RCO-A, and RCO-B (Vlad et al. 2014). LMI1 is a pleiotropic meristem identity regulator with expression in floral tissue, bracts, and leaves (Saddic et al. 2006). Following the initial duplication event, RCO gene copies have diverged in expression domain from LMI1, having evolved a novel, highly specific expression pattern in leaf margins. A shift from dissected to simple leaves in the lineage leading to A. thaliana has involved the loss of both RCO paralogs (Vlad et al. 2014). A shift towards greater leaf dissection in the lineage leading to Capsella rubella has involved a cis-regulatory mutation to RCO-A that increases gene expression (Sicard et al. 2014). Thus, gene duplication followed by the evolution of a narrowed expression domain has resulted in genomic material for mutations that specifically influence leaf complexity, which may contribute to the accessibility of changes in leaf morphology within the Brassicaceae.
Evolutionary change from flowers with petals to flowers that lack petals (apetaly) has occurred repeatedly in Ranunculaceae, accompanying the evolution of wind pollination or shifts towards pollination by generalized pollinators (Zhang et al. 2013). This pattern is associated with evolution in the APETALA3 (AP3) floral organ identity gene family. Ranunculaceae species have three paralogs of AP3, generated through two duplication events at the base of this clade. Two of the paralogs (AP3-1 and AP3-2) are broadly expressed in flowers and also leaves, presumably the ancestral domain of expression. The AP3-3 paralog is highly expressed in petal tissue only, suggesting that this gene copy has reduced pleiotropic effects, allowing genetic control of petal identity to be partially decoupled from the identity of other plant organs. In at least four lineages, the evolution of apetaly involves pseudogenization or loss of AP3-3.
Loss-of-function (LOF) mutations tend to have large mutational target sizes since there are many nucleotide positions that, if mutated, can disrupt gene function. Therefore, adaptations that can occur through LOF mutations to non-pleiotropic genes are likely to be relatively accessible to populations since there is a relatively short waiting time for these mutations. Therefore, given the history of AP3 gene family evolution, it is unsurprising that apetaly repeatedly occurred through LOF mutations to AP3-3.
Adaptive evolutionary change in flower color can often involve LOF mutations to non-pleiotropic gene copies (reviewed by Rausher 2008; Streisfeld and Rausher 2011; Wessinger and Rausher 2012; Sobel and Streisfeld 2013). Evolutionary transitions in anthocyanin pigment intensity, especially shifts to white flowers, tend to involve mutations to R2R3-MYB transcription factors that regulate the flavonoid pathway (Sobel and Streisfeld 2013). The R2R3-MYB transcription factor gene family is very large in most plants and individual gene copies have narrow domains of expression (Ramsay and Glover 2005). Therefore, mutations to these genes can cause floral tissue-specific changes in pigmentation without affecting the production of pigments or other phytoprotective flavonoids in vegetative tissues. Similarly, evolutionary change from blue to red flowers often involves LOF mutations to the enzyme FLAVONOID 3',5'-HYDROXYLASE (F3'5'H), an ancient gene duplicate of FLAVONOID 3'-HYDROXYLASE (F3'H). Comparative physiology suggests that F3'H is broadly expressed in leaves and flowers of most plants (see Wessinger and Rausher 2012), whereas F3'5'H can show a flower-specific pattern of expression (Wessinger and Rausher 2012, 2014), suggesting it is a relatively non-pleiotropic gene. In Penstemon, repeated adaptive transitions to red flowers (associated with transitions to hummingbird pollination) involves LOF mutations to F3'5'H in 12 separate cases (Wessinger and Rausher 2015), likely because this type of mutation provides an easily accessible means of producing the adaptive phenotype with minimal pleiotropic effects.
Conclusions
During the diversification of flowering plants, novel, yet highly integrated, floral morphologies have repeatedly evolved, even on short evolutionary timescales. An emerging hypothesis is that repeated evolutionary shifts are facilitated by genetic correlations among floral morphological characters in the direction of natural selection. We have outlined the evidence for this from comparative genetic studies focused on the evolution of selfing syndrome from outcrossing and evolutionary transitions in floral pollination syndromes. While correlations in the direction of selection can facilitate rapid phenotypic evolution, natural selection may favor independent evolution of different traits that share a developmental basis. In this case, the accessibility of a selectively favored phenotype may depend on variation in the genetic program that is outside pleiotropic loci, or especially in the case of very strong genetic correlations may rely on the previous evolution of genomic preadaptations that provide the genetic material for non-pleiotropic mutations. This evolutionary contingency may help explain why certain lineages appear prone to the repeated evolution of certain novel phenotypes. Future work investigating the genetic architecture and underlying basis of genetic correlations will clarify the role of genetic correlations in facilitating or constraining adaptation. For example, artificial selection experiments combined with quantitative genetic analyses may allow the genetic architecture of adaptation in response to selection that acts parallel vs. perpendicular to existing genetic correlations to be directly compared. In addition, experiments that characterize the genetic and developmental basis for genetic correlations will begin to address the prevalence of pleiotropic genetic variation.
Highlights.
Developmental bias can facilitate or constrain the evolution of novel phenotypes depending on genetic correlations among traits experiencing selection.
Trait correlations aligned with the direction of selection may accelerate the evolution of certain phenotypes, which may produce a pattern of repeated evolution; potential examples are found in the evolution of flowers adapted to self-pollination and in evolutionary shifts among pollination syndromes.
When natural selection on two or more traits opposes correlations generated by pleiotropy, adaptations may be facilitated by gene duplication, followed by specialization of a gene copy, yielding clade-specific patterns of repeated evolution.
Acknowledgments
We thank John Kelly and two anonymous reviewers for helpful comments on the manuscript. This work has been supported by NIH 5F32GM110988-03 to CW and NSF1624043 to CW and LH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Carolyn A. Wessinger, Email: cwessinger@ku.edu.
Lena C. Hileman, Email: lhileman@ku.edu.
References
- Arnold SJ. Constraints on phenotypic evolution. Am Nat. 140:S85–S107. doi: 10.1086/285398. [DOI] [PubMed] [Google Scholar]
- Arthur W. Developmental drive: an important determinant of the direction of phenotypic evolution. Evol Dev. 2001;3:271–278. doi: 10.1046/j.1525-142x.2001.003004271.x. [DOI] [PubMed] [Google Scholar]
- Ashman TL, Majetic CJ. Genetic constraints on floral evolution: a review and evaluation of patterns. Heredity. 2006;96:343–352. doi: 10.1038/sj.hdy.6800815. [DOI] [PubMed] [Google Scholar]
- Bissell EK, Diggle PK. Floral morphology in Nicotiana: architectural and temporal effects on phenotypic integration. Int J Plant Sci. 2008;169:225–240. [Google Scholar]
- Bissell EK, Diggle PK. Modular genetic architecture of floral morphology in Nicotiana: quantitative genetic and comparative phenotypic approaches to floral integration. J Evol Biol. 2010;23:1744–1758. doi: 10.1111/j.1420-9101.2010.02040.x. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Bradshaw HD, Jr, Wilbert SM, Otto KG, Schemske DW. Genetic mapping of floral traits associated with reproductive isolation in monkeyflowers (Mimulus) Nature. 1995;376:762–765. [Google Scholar]
- Bradshaw HD, Jr, Otto KG, Frewen BE, McKay JK, Schemske DW. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus) Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MT, Dechaine JM, Iniguez-Luy FL, Maloof JN, Stinchcombe JR, Weinig C. Floral genetic architecture: an examination of QTL architecture underlying floral (co)variation across environments. Genetics. 2010;186:1451–1465. doi: 10.1534/genetics.110.119982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MT, Kover PX, Weinig C. Natural variation in GA1 associates with floral morphology in Arabidopsis thaliana. New Phytol. 2012;195:58–70. doi: 10.1111/j.1469-8137.2012.04145.x. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner JK. Genetic mechanisms of floral trait correlations in a natural population. Nature. 2002;420:407–410. doi: 10.1038/nature01105. [DOI] [PubMed] [Google Scholar]
- Conner JK. Quantitative genetic approaches to evolutionary constraint: how useful? Evolution. 2012;66:3313–3320. doi: 10.1111/j.1558-5646.2012.01794.x. [DOI] [PubMed] [Google Scholar]
- Conner J, Sterling A. Testing hypotheses of functional relationships: a comparative survey of correlation patterns among floral traits in five insect-pollinated plants. Am J Bot. 1995;82:1399–1406. [Google Scholar]
- Conner J, Via S. Patterns of phenotypic and genetic correlations among morphological and life history traits in wild radish, Raphanus raphanistrum. Evolution. 1993;47:704–711. doi: 10.1111/j.1558-5646.1993.tb02128.x. [DOI] [PubMed] [Google Scholar]
- Conner JK, Sahli HF, Karoly K. Tests of adaptation: functional studies of pollen removal and estimates of natural selection on anther position in wild radish. Ann Bot. 2009;103:1547–1556. doi: 10.1093/aob/mcp071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner JK, Karoly K, Stewart C, Koelling VA, Sahli HF, Shaw FH. Rapid independent trait evolution despite a strong pleiotropic genetic correlation. Am Nat. 2011;178:429–441. doi: 10.1086/661907. [DOI] [PubMed] [Google Scholar]
- Edwards CE, Weinig C. The quantitative-genetic and QTL architecture of trait integration and modularity in Brassica rapa across simulated seasonal settings. Heredity. 2011;106:661–677. doi: 10.1038/hdy.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Ogburn RM. Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. Int J Plant Sci. 2012;173:724–733. [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faegri K, van der Pijl L. The Principles of Pollination Ecology. Pergamon; New York: 1979. [Google Scholar]
- Feng G, Qin Z, Yan J, Zhang X, Hu Y. Arabidopsis ORGAN SIZE RELATED1 regulates organ growth and final organ size in orchestration with ARGOS and ARL. New Phytol. 2011;191:635–646. doi: 10.1111/j.1469-8137.2011.03710.x. [DOI] [PubMed] [Google Scholar]
- Fenster CB, Ritland K. Quantitative genetics of mating system divergence in the yellow monkeyflower species complex. Heredity. 1994;73:422–435. [Google Scholar]
- Ferris KG, Rushton T, Greenlee AB, Toll K, Blackman BK, Willis JH. Leaf shape evolution has a similar genetic architecture in three edaphic specialists within the Mimulus guttatus species complex. Ann Bot. 2015;116:213–223. doi: 10.1093/aob/mcv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Willis JH. Minor quantitative trait loci underlie floral traits associated with mating system divergence in Mimulus. Evolution. 2002;56:2138–2155. doi: 10.1111/j.0014-3820.2002.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF, Hill JP. The genetics of reproductive barriers in Mimulus (monkeyflowers) Evolution. 2013;67:2547–2560. doi: 10.1111/evo.12154. [DOI] [PubMed] [Google Scholar]
- Fishman L, Beardsley PM, Stathos A, Williams CF, Hill JP. The genetic architecture of traits associated with the evolution of self-pollination in Mimulus. New Phytol. 2014;205:907–917. doi: 10.1111/nph.13091. [DOI] [PubMed] [Google Scholar]
- Futuyma DJ. Evolutionary constraint and ecological consequences. Evolution. 2010;64:1865–1884. doi: 10.1111/j.1558-5646.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- Goodwillie C, Ritland C, Ritland K. The genetic basis of floral traits associated with mating system evolution in Leptosiphon (Polemoniaceae): an analysis of quantitative trait loci. Evolution. 2006;60:491–504. [PubMed] [Google Scholar]
- Hall MC, Basten CJ, Willis JH. Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics. 2006;172:1829–1844. doi: 10.1534/genetics.105.051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann K, Klahre U, Moser M, Sheehan H, Mandel T, Kuhlemeier C. Tight genetic linkage of prezygotic barrier loci creates a multifunctional speciation island in Petunia. Curr Biol. 2013;23:873–877. doi: 10.1016/j.cub.2013.03.069. [DOI] [PubMed] [Google Scholar]
- Hermann K, Klahre U, Venail J, Brandenburg A, Kuhlemeier C. The genetics of reproductive organ morphology in two Petunia species with contrasting pollination syndromes. Planta. 2015;241:1241–1254. doi: 10.1007/s00425-015-2251-2. [DOI] [PubMed] [Google Scholar]
- Hu Y, Xie Q, Chua NH. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell. 2003;15:1951–1961. doi: 10.1105/tpc.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Poy HM, Chua NH. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 2006;47:1–9. doi: 10.1111/j.1365-313X.2006.02750.x. [DOI] [PubMed] [Google Scholar]
- Juenger T, Purugganan M, Mackay TFC. Quantitative trait loci for floral morphology in Arabidopsis thaliana. Genetics. 2000;156:1379–1392. doi: 10.1093/genetics/156.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T, Perez-Perez JM, Bernal S, Micol JL. Quantitative trait loci mapping of floral and leaf morphology traits in Arabidopsis thaliana: evidence for modular genetic architecture. Evol Dev. 2005;7:259–271. doi: 10.1111/j.1525-142X.2005.05028.x. [DOI] [PubMed] [Google Scholar]
- Kelly JK. Connecting QTLs to the G-matrix of evolutionary quantitative genetics. Evolution. 2009;63:813–825. doi: 10.1111/j.1558-5646.2008.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Anderson JT. Control of flower size. J Exp Bot. 2013;64:1427–1437. doi: 10.1093/jxb/ert025. [DOI] [PubMed] [Google Scholar]
- Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Losos JB. Convergence, adaptation, and constraint. Evolution. 2011;65:1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer; Sunderland, MA: 1998. [Google Scholar]
- Maynard Smith J, Burian R, Kauffman S, Alberch P, Campbell J, Goodwin B, Lande R, Raup D, Wolpert L. Developmental constraints and evolution. Q Rev Biol. 1985;6:265–287. [Google Scholar]
- Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato T, Rieseberg LH, Wood TE. The genetic basis of speciation in the Giliopsis lineage of Ipomopsis (Polemoniaceae) Heredity. 2013;111:227–237. doi: 10.1038/hdy.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Zhang X, Zhang X, Feng G, Hu Y. The Arabidopsis ORGAN SIZE RELATED 2 is involved in the regulation of cell expansion during organ growth. BMC Plant Biol. 2014;14:349. doi: 10.1186/s12870-014-0349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Rausher MD. Evolutionary transitions in floral color. Int J Plant Sci. 2008;169:7–21. [Google Scholar]
- Ritland C, Ritland K. Variation of sex allocation among eight taxa of the Mimulus guttatus species complex (Scrophulariaceae) Am J Bot. 1989;76:1731–1739. [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development. 2006;133:1673–1682. doi: 10.1242/dev.02331. [DOI] [PubMed] [Google Scholar]
- Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50:1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. [DOI] [PubMed] [Google Scholar]
- Scoville A, Lee YW, Willis JH, Kelly JK. Contribution of chromosomal polymorphisms to the G-matrix of Mimulus guttatus. New Phytol. 2009;183:803–815. doi: 10.1111/j.1469-8137.2009.02947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan H, Moser M, Klahre U, Esfeld K, Dell'Olivo A, Mandel T, Metzger S, Vandenbussche M, Freitas L, Kuhlemeier C. MYB-FL controls gain and loss of floral UV absorbance, a key trait affecting pollinator preference and reproductive isolation. Nat Genet. 2015 doi: 10.1038/ng.3462. Advance online publication. [DOI] [PubMed] [Google Scholar]
- Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol. 2014;24:1880–1886. doi: 10.1016/j.cub.2014.06.061. [DOI] [PubMed] [Google Scholar]
- Sinervo B, Svensson E. Correlational selection and the evolution of genomic architecture. Heredity. 2002;89:329–338. doi: 10.1038/sj.hdy.6800148. [DOI] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Stern D, Andolfatto P, Wright SI. Genetic architecture and adaptive significance of the selfing syndrome in Capsella. Evolution. 2012;66:1360–1374. doi: 10.1111/j.1558-5646.2011.01540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD. Pleiotropy and the evolution of floral integration. New Phytol. 2016;209:80–85. doi: 10.1111/nph.13583. [DOI] [PubMed] [Google Scholar]
- Sobel JM, Streisfeld MA. Flower color as a model system for studies of plant evo-devo. Front Plant Sci. 2013;4:321. doi: 10.3389/fpls.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Adaptive radiation of reproductive characteristics in angiosperms, I: pollination mechanisms. Ann Rev Ecol Syst. 1970;1:307–326. [Google Scholar]
- Stebbins GL. Flowering Plants: Evolution Above the Species Level. Arnold; London: 1974. [Google Scholar]
- Streisfeld MA, Rausher MD. Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution. 2011;65:629–642. doi: 10.1111/j.1558-5646.2010.01165.x. [DOI] [PubMed] [Google Scholar]
- Thomson JD, Wilson P. Explaining evolutionary shifts between bee and hummingbird pollination: convergence, divergence, and directionality. Int J Plant Sci. 2008;169:23–38. [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, Vuolo F, Ioio RD, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, Huijser P, Bailey CD, Tsiantis M. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science. 2014;343:780–783. doi: 10.1126/science.1248384. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Altenberg L. Complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Wessinger CA, Rausher MD. Lessons from flower colour evolution on targets of selection. J Exp Bot. 2012;63:5741–5749. doi: 10.1093/jxb/ers267. [DOI] [PubMed] [Google Scholar]
- Wessinger CA, Rausher MD. Genetic changes associated with flower color evolution in Penstemon barbatus. Evolution. 2014;68:1058–1070. doi: 10.1111/evo.12340. [DOI] [PubMed] [Google Scholar]
- Wessinger CA, Rausher MD. Ecological transition predictably associated with gene degeneration. Mol Biol Evol. 2015;32:347–354. doi: 10.1093/molbev/msu298. [DOI] [PubMed] [Google Scholar]
- Wessinger CA, Hileman LC, Rausher MD. Identification of major quantitative trait loci underlying floral pollination syndrome divergence in Penstemon. Phil Trans R Soc Lond. 2014;369:20130349. doi: 10.1098/rstb.2013.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Lowry DB, Cooley AM, Wright KM, Lee YW, Willis JH. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity. 2008;100:220–230. doi: 10.1038/sj.hdy.6801018. [DOI] [PubMed] [Google Scholar]
- Zhang R, Guo C, Zhang W, Wang P, Lin L, Duan X, Du Q, Zhao L, Shan H, Hodges SA, Kramer EM, Ren Y, Kong H. Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae) Proc Natl Acad Sci USA. 2013;110:5074–5079. doi: 10.1073/pnas.1219690110. [DOI] [PMC free article] [PubMed] [Google Scholar]