Abstract
More than 75,000 man-made chemicals contaminate the environment; many of these have not been tested for toxicities. These chemicals demand quantitative high-throughput screening assays to assess them for causative roles in neurotoxicities, including Parkinson’s disease and other neurodegenerative disorders. To facilitate high throughput screening for cytotoxicity to neurons, three human neuronal cellular models were compared: SH-SY5Y neuroblastoma cells, LUHMES conditionally-immortalized dopaminergic neurons, and Neural Stem Cells (NSC) from human fetal brain. These three cell lines were evaluated for rapidity and degree of differentiation, and sensitivity to 32 known or candidate neurotoxicants. First, expression of neural differentiation genes was assayed during a 7-day differentiation period. Of the three cell lines, LUHMES showed the highest gene expression of neuronal markers after differentiation. Both in the undifferentiated state and after 7 days of neuronal differentiation, LUHMES cells exhibited greater cytotoxic sensitivity to most of 32 suspected or known neurotoxicants than SH-SY5Y or NSCs. LUHMES cells were also unique in being more susceptible to several compounds in the differentiating state than the undifferentiated state; including known neurotoxicants colchicine, methyl-mercury (II), and vincristine. Gene expression results suggest that differentiating LUHMES cells may be susceptible to apoptosis because they express low levels of anti-apoptotic genes BCL2 and BIRC5/survivin; whereas SH-SY5Y cells may be resistant to apoptosis because they express high levels of BCL2, BIRC5/survivin, and BIRC3 genes. Thus, LUHMES cells exhibited favorable characteristics for neuro-cytotoxicity screening: rapid differentiation into neurons exhibiting high level expression neuronal marker genes, and marked sensitivity of LUHMES cells to known neurotoxicants.
Short Abstract
Three human neuronal cell lines were evaluated as high throughput screening models for neuronal cytotoxicity: SH-SY5Y neuroblastoma cells, LUHMES conditionally-immortalized dopaminergic neurons, and Neural Stem Cells. After 7 days of differentiation LUHMES expressed the highest levels of neuronal markers. Differentiated LUHMES cells exhibited greater cytotoxic sensitivity to most of 32 suspected or known neurotoxicants than differentiated SH-SY5Y or NSCs, and greater cytotoxic sensitivity to 11 compounds compared to undifferentiated LUHMES cells.
Introduction
Toxicants are suspected to play roles in a variety of neural- and psychiatric diseases, such as Parkinson’s disease, autism spectrum disorders, and Alzheimer's disease (Caudle et al., 2012), (Rossignol et al., 2014), (Moulton and Yang, 2012, Smirnova et al., 2014); yet most industrial chemicals have not been examined for possible effects on neurons, due to the lack of high-throughput validated models for testing. Neuronal toxicants may act in a variety of ways by disrupting differentiation, function, or growth and survival, of neurons. Indeed, neurons have several characteristics that are known to make them sensitive to disruption, such as extensive microtubule-supported axons, intensive electrical activity demanding specialized ion channels and mitochondrial activity to recharge electrical potential, and synapses that require specialized enzyme activities to make and catabolize neurotransmitters. While assays have been prepared for a variety of specific neuronal disruptions, simple, reproducible screens are needed to quantitatively evaluate large libraries of compounds (Coecke et al., 2006, Tice et al., 2013). For example, the Tox21 Consortium has collected a library of 10,000 chemicals for use in quantitative high-throughput screening (qHTS (Inglese et al., 2006)). A first step toward this goal might be to screen a phenotypically neuronal cell line for cytotoxicity. Hence, this study focuses on cytotoxicity and growth disruption in differentiating neuronal cell lines, with the expectation that these phenotypes will be amenable to qHTS.
The great challenge of in vitro toxicology is to identify a model that satisfactorily mimics action of toxicants in vivo. It is known that actions of many neurotoxicants are not mimicked by in vitro models (Krug et al., 2013, Smirnova et al., 2015). While the acute toxicity assays described in this manuscript will not address developmental toxicity (Krug et al., 2013) nor cellular recovery from toxicants (Smirnova et al., 2015), the detailed characterization of differentiation markers and cytotoxic or apoptotic responses presented may provide a useful basis to select a cell model(s) for these longer-term characterizations. Thus, in vitro cellular models which are validated as fit-for-purpose may be used to gain insight into the action of a toxicant by studying the model’s responses. Such use demands characterization of the model system to determine whether it mimics the toxicant’s in vivo target cell; in this case neurons. Clearly, none of these 3 models will mimic the full multicellular, 3-dimensional, contextual complexity of the brain. Instead, this study tries to reveal the phenotypes of each model using marker genes, as well as the cells’ sensitivity to necrosis or apoptosis.
Whereas necrosis is generally thought to be a passive process producing cell debris that are likely to cause inflammation; apoptosis is an active process in which the cell partially digests itself and summons neighboring cells to phagocytize it to limit inflammation. It is well-established that cells carry the requisite enzymes to carry out apoptosis immediately in response to apoptotic signals, without the need for de novo transcription and translation. Cells do, however, regulate their sensitivity to apoptosis; e.g. increasing sensitivity while growing and decreasing sensitivity during differentiation (Hu and Xuan, 2008). Predisposition to apoptosis is regulated largely via transcriptional and post-transcriptional control of pro- and anti-apoptotic genes such as the BCL- family, BIRC- family, and others. Cancer cells often accrue mutations that reduce sensitivity to apoptotic signals that would kill normal cells (Hu and Xuan, 2008). Thus, when characterizing transformed and non-transformed cytotoxicity models it is useful to gain an understanding of how those models respond to apoptotic signals.
In order to assess neuronal growth and cytotoxicity we evaluate three cell lines as potential models: SH-SY5Y, LUHMES, and Neural Stem Cells derived from human fetal brain (NSC, StemPro®). SK-N-SH cells were derived from a human neuroblastoma; whereas SH-SY5Y cells were selected from this parental cell line SK-N-SH for their ability to differentiate uniformly into neurons. SH-SY5Y cells can be differentiated to grow extensive neurites; expressing characteristics of acetylcholinergic, glutamatergic, and adenosinergic neurons (Pahlman et al., 1984). Both SK-N-SH and SH-SY5Y cells were previously used to screen 1400 environmental contaminant chemicals for cytotoxicity (Huang et al., 2008, Xia et al., 2008). It should be noted that two other cell lines of neural or neuroblast origin were also used in this screening, N2a mouse neuroblastoma cells (Mathews et al., 1976) and HEK293 human neuronal lineage cells from embryonic kidney tissue (Shaw et al., 2002). In this cytotoxicity screen, a number of chemicals were identified that demonstrated selective toxicity to SH-SY5Y cells and/or SK-N-SH cells, HEK293, or N2a cells, relative to the other 9 cell lines used. These candidate neurotoxicants are included in this study to determine whether NSC and LUHMES are also sensitive to these compounds.
LUHMES (Lund human mesencephalic) cells were derived from striatal primary human dopaminergic cells, and conditionally immortalized using a tetracycline-regulated v-myc gene (Lotharius et al., 2005) (Scholz et al., 2011). As such, LUHMES cells can be grown to achieve desired cell numbers, and then v-myc expression switched off to halt growth and trigger rapid and homogenous differentiation to a phenotype characteristic of dopaminergic neurons. LUHMES cells are expected to have avoided the numerous mutations that are characteristic of tumor-derived cell lines.
Neural Stem Cells (NSC) are derived from human fetal stem cells (hESCs). They are committed to the neural lineage, and typically differentiate into a mixture of neurons, oligodendrocytes, and astrocytes, over the course of 2 weeks or more. By manipulating culture conditions, different neural types can be enriched, including neurons that exhibit neurites and neuronal marker proteins, MAP2 and DCX. (Shin and Vemuri, 2010) Due to the limitations of qHTS in 384-well microplates, NSCs were assessed at 7 days of differentiation whereupon neurites were abundant, but differentiation was incomplete.
It has been widely observed that mammalian neurons, as well as other cell types, are generally more resistant to toxicants than their undifferentiated progenitor cells, particularly cells in the S-phase of the mitotic cycle (Gartlon et al., 2006) (Ninomiya et al., 1997). This study examined whether any particular neuronal cell type is more sensitive to neuronal toxicants than others, and whether differentiated cell types differ in their sensitivity in comparison to the undifferentiated cells. Susceptibility to apoptosis and the underlying apoptosis regulatory mechanisms are also investigated. These results will be used to evaluate these cell models for neuro-cytotoxicity qHTS with respect to rapid and homogenous differentiation and susceptibility to known and suspected neuronal toxicants.
Materials and Methods
Chemicals
All chemical compounds listed in Table 2, as well as DMSO, were purchased from Sigma-Aldrich Chemical Co. Chemicals were dissolved in dimethylsulfoxide (DMSO) at a concentration of 10 mM to make stock solutions, and stored at −20°C.
Table 2.
Known-, candidate-neurotoxicants, and reference compounds used in this study.
| Compound | Abbrevi ation |
C.A.S. number |
Category - sensitive cells |
In vitro neural selectivity* |
|---|---|---|---|---|
| Aflatoxin B1 | AFLA | 1162-65-8 | Known, neuro-protoxicant in animals (Ikegwuonu, 1983) |
|
| 6-hydroxydopamine | 6HD | 28094-15-7 | Known, dopaminergic neurons (Ossola et al., 2008) |
|
| MPTP | MPTP | 23007-85-4 | Known, protoxicant dopaminergic neurons (Schildknecht et al., 2009) |
|
| MPP+ | MPP | 911295-24-4 | Known, dopaminergic neurons (Schildknecht et al., 2009) |
|
| Rotenone | ROTE | 83-79-4 | Known, Mitochondrial toxicant (Caudle et al., 2012) |
|
| Paraquat | PQ | 11089-65-9 | Known, Mitochondrial toxicant (Caudle et al., 2012) |
|
| Vincristine sulfate | VIN | 2068-78-2 | Known, Microtubule disruptor (Courtemanche et al., 2015) |
|
| Cytosine arabinoside | ARAC | 147-94-4 | Known, Peripheral neuropathy via oxidative stress (Geller et al., 2001) |
|
| Trimethyltin chloride | TMT | 1066-45-1 | Known- Neuronal necrosis and degeneration (Winship, 1988) (Gunasekar et al., 2001) |
|
| Methyl mercury (II) | MHG | 115-09-3 | Known (Huang et al., 2014) | |
| Doxorubicin | DOX | 25316-40-9 | Known (Mohamed et al., 2011) | HEK293, SH-SY5Y, SK-N- SH (Huang et al., 2008, Xia et al., 2008) |
| Daunorubicin | DAUN | 23541-50-6 | Known (Joshi et al., 1996) | HEK293, SH-SY5Y, SK-N- SH, N2a (Huang et al., 2008, Xia et al., 2008) |
| Chlorambucil | CLM | 305-03-3 | Known (Verschoyle et al., 1994) | SH-SY5Y, (Huang et al., 2008, Xia et al., 2008) |
| Colchicine | CLCH | 64-86-8 | Known (Goldschmidt and Steward 1989; Mundy and Tilson 1990; | HEK293, SH-SY5Y, SK-N- SH, N2a (Huang et al., 2008, Xia et al., 2008) |
| Ziram | ZIR | 137-30-4 | Known (Wang et al., 2011) | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| Genistein | GEN | 446-72-0 | Candidate | Apoptotic in SHSY5Y (Karmakar et al., 2009) |
| Quercetin | QRC | 117-39-5 | Candidate | Toxic or protective in SH- SY5Y (Ossola et al., 2008) |
| DL-Sulforaphane | SFR | 4478-93-7 | Candidate | Apoptotic in SH-SY5Y (Hsu et al., 2013) |
| Adrenosterone | ADR | 382-45-6 | Candidate | N2a (Huang et al., 2008, Xia et al., 2008) |
| Dithiocyanatomethan | DTCM | 6317-18-6 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| Nelfinavir | NLF | 159989-65-8 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| 2-octyl-4-isothiazolin- 3-one |
OTZ | 26530-20-1 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| Hexachlorophene | HCP | 70-30-4 | Candidate | (Huang et al., 2008, Xia et al., 2008) |
| 2-Methyl-1- nitroanthraquinone |
MNA | 129-15-7 | Candidate | HEK293 (Huang et al., 2008, Xia et al., 2008) |
| Malachite green oxalate |
MALG | 2437-29-8 | Candidate | N2a (Huang et al., 2008, Xia et al., 2008) |
| Sodium dichromate | CR2 | 7789-12-0 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| Captan | CAP | 133-06-2 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| 4- (Methylamino)phenol hemisulfate salt |
MAP | 55-55-0 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| Resorcinol diglycidyl ether |
RSR | 101-90-6 | Candidate | SH-SY5Y (Huang et al., 2008, Xia et al., 2008) |
| 3-methyladenine | 3MA | 5142-23-4 | Reference, Autophagy inhibitor (Roux et al., 2014) |
|
| Tunicamycin | TUNC | 11089-65-9 | Reference, Causes ER-stress (Kosuge et al., 2008) |
|
| Digoxin | DGX | 20830-75-5 | Reference, Non-selective toxicant (Huang et al., 2008, Xia et al., 2008) |
In vitro neural selectivity is reported from (Huang et al., 2008, Xia et al., 2008), for any of the neural-derived cell lines: HEK293, SH-SY5Y, SK-N-SH, or N2a, that was 10-fold more sensitive (10-fold lower IC50) to the toxicant than the median of the other 9 cell lines in the study.
Cell culture and differentiation
The three cell lines were cultured until nearly confluent, trypsinized, then seeded at 1/4th the previous density. When cell lines reached 9 passages, they were discarded and replaced from frozen stocks. The three cell line identities were verified by short tandem repeat profiling (WiCell Research Inst. Madison, Wisconsin, USA), and checked for mycoplasma contamination using the MycoAlert™ Mycoplasma Detection Kit (Lonza. SHSY-5Y cells were obtained from ATCC (American Type Culture Collection, Manassas Virginia), and cultured in EMEM/F12, a 1:1 mixture of Eagle’s Minimum Essential (EMEM, ATCC) and F12 (Invitrogen) including 100µg/mL Penicillin and 100 µg/mL streptomycin, as well as 10% fetal bovine serum (FBS; Hyclone, Logan, Utah, USA). SH-SY5Y cell differentiation was induced in culture medium with 3% FBS, by 10 µM all trans-retinoic acid. LUHMES cells were provided by Professor Marcel Leist, University of Konstanz, Germany. LUHMES cells were grown in culture vessels coated with 50µg/ml each poly-L-ornithine and human fibronectin in Advanced DMEM/F12 medium containing 2 mM L-glutamine, 1X N2-supplement and 40 ng/ml bFGF. LUHMES differentiation was induced by 1µg/ml tetracycline and 1 mM cAMP contained in advanced DMEM/F12 medium with 2 mM L-glutamine, 1X N2-supplement, and 2ng/ml GDNF. (Schildknecht, Poltl et al. 2009) StemPro® Human Neural Stem Cells (NSC) were obtained from Invitrogen (now Thermo-Fisher, Waltham, Massachusetts, USA). Per the manufacturer’s manual, NSC cells were cultured adherently in vessels coated with Geltrex and grown in StemPro® NSC SFM defined complete medium consisting of KnockOut® D-MED/F-12 with 2% StemPro Neural Supplement, 2 mM GlutaMax-I Supplement and 20 ng/ml each human recombinant bFGF and EGF. NSCs were treated to induce neuronal differentiation in StemPro® NSC SFM medium without bFGF and EGF. Cell differentiation was started when the cells were at 60% confluence in vessels, and continued for up to seven days.
Cytotoxicity determinations
Cells were distributed in 384 well plates one day before applying toxicant treatment, (10,000 cells per well in 30 µL medium) for cytotoxicity assays. For gene expression assays, 3×105 cells were cultured per well in 12 well plates with 2 mL medium to enable RNA extraction and quantitative reverse-transcriptase PCR (qPCR). Toxicant stock solutions were diluted with culture medium to make 10× working solutions for each concentration and added to the cells at 10% of the volume in each well. Control cells were treated with 1% DMSO to match treatments. Cells in in duplicate 384 well plates were assayed for intracellular [ATP] using the CellTitreGlo® luminescence assay (Promega, Madison Wisconsin, USA), or the Caspase-Glo® 3/7 luminescence assay, 24 hours after toxicant treatment. Luminescence was read in an EnVision plate reader (Perkin Elmer, Waltham Massachusetts, USA). DMSO treated control wells were used to determine 100% viability. Concentration-response was plotted for each toxicant:cell-type combination, and the IC50 was identified by interpolation as the concentration that reduced intracellular [ATP] to 50% of control. Caspase 3/7 data were compared to DMSO control data and the maximum level for each toxicant are reported as multiple above control (Treated activity/control activity −1).
qPCR
Each toxicant was used to treat each of the 6 cell types for 6 hours in 2 mL of medium in 12-well microplates. A single toxicant concentration was used for undifferentiated and differentiated cells from each cell line, at approximately the IC20. Total RNA was isolated from three biological replicate treatments using Qiagen RNEasy Plus Mini kits (Qiagen, Hilden, Germany; Cat#:74106) including the optional DNAse treatment on the column. Total RNA was quantitated in a NanoDrop-8000, and stored in freezer (-80C). cDNA synthesis was carried out from 0.5 µg total RNA per sample using RT2 HT First Strand Kit (Qiagen). Data for 16 differentiation marker genes were collected using TaqMan® quantitative PCR assays (qPCR; Life Technologies). Data were collected for 84 human apoptosis-pathway genes using Apoptosis qPCR arrays (Qiagen Cat#: PAHS-3012Z). qPCR data were first subjected to thresholding: Values >36 and "undetermined" values were changed to 36, the approximate limit of detection of the qPCR assay. Sample data were normalized as follows: The median threshold value (Ct) was calculated for the three biological replicates of all genes on the array. The residual (median Ct of each sample minus average of median Ct's) was subtracted from the Ct values of each gene in that sample. This normalization method was found to yield lower and more consistent residuals than normalization to the mean of the 5 housekeeping genes’ Ct. Standard deviations were calculated from the 3 replicates for each gene. Standard deviations >2 were flagged and outlier data points were excluded from further analysis. The median was used to represent each set of replicates to minimize variance from sporadic failed reactions observed in each array. Gene expression levels were calculated from the threshold PCR cycle (Ct) values as: GE = 10,000[2Ct(GAPDH) − Ct(Gene)] such that the limit of detection occurring at cycle 36 yields an expression value of approximately 1. Gene expression that was undetectable was assigned a value of 1.0, so that a lower-limit fold-increase could be estimated as expression level for treated cells divided by expression level for control cells. Ratios: The Treated/Control Ratio of expression was calculated from the normalized median Ct values as follows: 2(Control Ct − Treated Ct).
Results
Differentiation gene markers
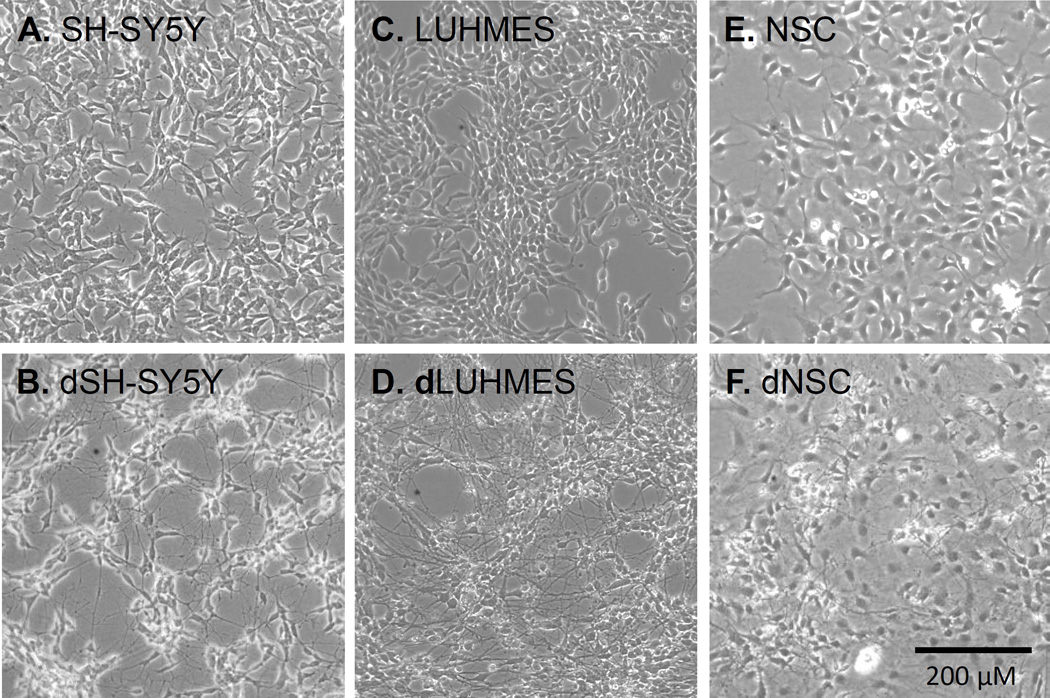
Published differentiation protocols were used for the three cell lines, SH-SY5Y (Pahlman et al., 1984, Cheung et al., 2009), LUHMES (Lotharius et al., 2005), and NSC (Shin and Vemuri, 2010), and cell morphologies were monitored by phase-contrast microscopy as shown in Figure 1. Differentiation protocols were limited to 7 days to facilitate culture in 384-well format required for high-throughput screening. SH-SY5Y cells exhibited short neurites during the growth phase; these neurites became longer and more numerous during differentiation, as described (Pahlman et al., 1984, Cheung et al., 2009). LUHMES cells lacked neurites during the proliferation phase, but exhibited long neurites after about four days (Lotharius et al., 2005). NSCs are committed to the neural lineage, but still multi-potent. NSC also lacked neurites prior to differentiation. A large fraction of these cells formed neurites during the seven days of differentiation, Other neural stem cells acquired markers and morphologies indicative of glia or oligodendrocytes, as described (Shin and Vemuri, 2010). Marker genes were examined to understand the differentiation status of each of these cellular models.
Figure 1. Phase-contrast microscopy images of 6 cell types illustrating the extent of neurite outgrowth in each.
(A) SH-SY5Y cells. (B) SH-SY5Y cells differentiated for 7 days (dSH-SY5Y). (C) LUHMES cells. (D) LUHMES cells differentiated for 7 days (dLUHMES). (E) Neural Stem Cells (NSC). (F) Neural Stem Cells differentiated for 7 days (dNSC).
A panel of gene expression markers was developed using quantitative reverse transcriptase PCR (qPCR) to characterize the 7-day differentiation process of cell lines SH-SY5Y, LUHMES, and NSCs. Among the marker genes selected, GALC and MBP were selected as markers of oligodendrocyte progenitors or oligodendrocytes, and S100B and GFAP as markers of astrocyte progenitors or fully differentiated astrocytes. (Buntinx et al., 2003, Shin and Vemuri, 2010). OLIG2 was assayed as a marker for neurons or oligodendrocyte precursors. The neuronal marker genes included: TUBB3, ENO2, DCX, MAP2, SYN1, and KCNJ6; as well as three genes characteristic of dopaminergic neurons: PARK2, DRD2, and TH. DCX expression may reflect immature, differentiating neurons.(Guichet et al., 2013) Table 1 shows the expression levels of mRNAs for these fifteen genes, normalized to GAPDH mRNA. Note that an expression value of 1 on this scale denotes that a gene’s mRNA was at, or below, the limit of detection by qPCR. SH-SY5Y cells on day 0 exhibited considerable levels of neuronal markers that started low or undetectable in LUHMES cells on Day 0, including ENO2, DCX, MAP2, SYN1, and DRD2 (Table 1). Although in the SH-SY5Y model several neuronal marker genes increased in expression during the seven days of differentiation, including TUBB3, ENO2, MAP2, SYN1, PINK1, DRD2; none of these genes increased more than 3-fold (Table 1; (Zhao et al., 2014), (Guichet et al., 2013), (Gaillard et al., 2009), (Glaab and Schneider, 2015), and (Buntinx et al., 2003)).
Table 1.
Gene expression levels of neuronal marker genes during differentiation of three cell lines.
| SH-SY5Y | LUHMES | NSC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker of Cell: |
Day: | 0 | 1 | 3 | 7 | 0 | 1 | 3 | 7 | 0 | 1 | 3 | 7 |
| N | TUBB3 | 2600 | 2900 | 1100 | 3700 | 2100 | 5600 | 17000 | 22500 | 1400 | 1400 | 2200 | 4600 |
| N | ENO2 | 600 | 800 | 370 | 900 | 21 | 31 | 34 | 1400 | 71 | 57 | 84 | 160 |
| N | MAP2 | 90 | 100 | 46 | 170 | 2 | 10 | 130 | 530 | 300 | 510 | 510 | 590 |
| N | KCNJ6 | 1 | 1 | 1 | 2 | 1 | 1 | 42 | 81 | 15 | 5 | 5 | 7 |
| N | SYN1 | 180 | 250 | 140 | 380 | 1 | 9 | 340 | 630 | 30 | 20 | 31 | 110 |
| N | PINK1 | 95 | 130 | 90 | 200 | 100 | 270 | 240 | 290 | 47 | 60 | 52 | 72 |
| IN | DCX | 480 | 310 | 160 | 260 | 1 | 1 | 92 | 1200 | 36 | 65 | 320 | 920 |
| DN | PARK2 | 20 | 8 | 12 | 21 | 1 | 13 | 11 | 18 | 14 | 10 | 14 | 16 |
| DN | DRD2 | 10 | 15 | 8 | 24 | 1 | 12 | 160 | 350 | 1 | 1 | 3 | 11 |
| DN | TH | 1 | 2 | 1 | 3 | 1 | 1 | 1 | 100 | 1 | 1 | 1 | 1 |
| ON | OLIG2 | 1 | 1 | 1 | 2 | 1 | 1 | 4 | 94 | 2 | 1 | 2 | 5 |
| ON | GALC | 65 | 63 | 28 | 120 | 3 | 9 | 11 | 14 | 1 | 1 | 1 | 1 |
| O | MBP | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 |
| A | GFAP | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| A | S100B | 2 | 3 | 2 | 5 | 1 | 1 | 1 | 2 | 8 | 1 | 1 | 1 |
| -- | GAPDH | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 | 10000 |
Genes are listed in the second column in bold and italics. Each gene is a marker for cell types abbreviated in the first column: N, neurons; IN, immature neurons; DN, dopaminergic neurons; ON, oligodendrocytes and neurons; O, oligodendrocytes; G, glia; A, astrocytes. Cell lines and days of differentiation are listed in the first- and second rows. Gene expression values were determined using qPCR. Gene expression values below the limit of detection were set to 1.0 (Ct > 36 were trimmed to Ct = 36 and set to 1). Numbers represent mRNA abundance 2^(36-Normalized Ct(gene)). >Five-fold and >25-fold increases in expression compared to day 0 are highlighted in pink or red, respectively.
In LUHMES cells, gene expression analysis of neuronal marker genes reveals undifferentiated cells on day 0 with no apparent neuronal character, progressing to differentiating cells on day 7, with large increases in most neuronal marker genes (Table 1). On day 0 the only marker genes detectable in LUHMES cells were: TUBB3, ENO2, PINK1, and very low MAP2. These four genes increased approximately 10-fold for TUBB3, 3-fold for PINK1, 65-fold for ENO2, and 270-fold for MAP2. For genes that were undetectable in undifferentiated LUHMES cells, GALC, PARK2, DCX, OLIG2, KCNJ6, SYN1, DRD2, and TH, we presume that they were expressed at the lower limit of detection in order to calculate the minimum ratio of increase. These estimates yield ratios of increase of 10-fold for GALC, 18-fold for PARK2, and 80-fold or greater for: DCX, OLIG2, KCNJ6, SYN1, DRD2, and TH. These data also identify genes that respond with distinct kinetics during differentiation: early: PARK2, late: ENO2, DCX, OLIG2, and TH; and gradual: TUBB3, MAP2, KCNJ6, SYN1, and DRD2. Although GALC is an abundant marker in oligodendrocytes, GALC is also expressed in neurons (Cahoy et al., 2008). Messenger RNAs for ENO2 and a very low level MAP2 were detected in undifferentiated LUHMES cells, but induced ~70-fold and 250-fold, respectively, in day 7 differentiating LUHMES cells. TUBB3 was expressed at a significant level, 1400–2600 units, in each of the three undifferentiated cell lines, but increased most in differentiating LUHMES cells, ~10-fold. Several genes that are characteristic of dopaminergic neurons were greatly increased in differentiating LUHMES cells, and not at all, or only slightly in other cells, including PARK2 (18-fold), DRD2 (350-fold), and TH (100-fold). It should also be noted that at 7 days of differentiation, mRNA levels of expression of nearly all of these neuronal marker genes were higher for LUHMES cells than for the other two cell models, suggesting a greater rapidity and degree of neuronal character in day 7 LUHMES cells.
Differentiation of NSC resulted in expression of neuronal marker gene-mRNA levels, and ratios, that were generally intermediate between the other two differentiating cell lines. Such genes included TUBB3 (3-fold), DCX (26-fold), MAP2 (2-fold), OLIG2 (2.5-fold), SYN1 (3.6-fold), and DRD2 (11-fold). Upon differentiation, the NSC model forms a mixture of neurons with neurites and synapses, glial cells, and oligodendrocytes. OLIG2 is not a pure neuronal marker gene, but rather specifies oligodendrocyte or neuronal subtype lineages (Meijer et al., 2012). Other markers indicate considerable neuronal character in differentiating NSC cell population, including DCX (26-fold), MAP2 (2-fold), SYN1 (4-fold), and DRD2 (11-fold). These neuronal markers are consistent with the appearance of abundant neurites in these cells. Marker genes for other neural cell types were not expressed at significant levels in any of the three models, including glial cell markers GFAP, astrocyte marker S100B, or oligodendrocyte marker MBP. While a longer differentiation period than seven days would likely allow for further differentiation of NSCs, longer culture periods are irreproducible and so are impractical in the 384-well microplates that are used for screening chemical libraries.
Cytotoxicity determination using 32 toxicants
Cell lines SH-SY5Y, LUHMES, and NSC were compared before and after differentiation (dSH-SY5Y, dLUHMES, and dNSC), for cytotoxic sensitivity to 32 candidate neurotoxicants. Suspected neurotoxicants were selected from a previous study because they showed selective cytotoxicity to SH-SY5Y human neuroblastoma cells (Pahlman et al., 1984), SK-N-SH cells from which SH-SY5Y were derived (Pahlman et al., 1984), HEK293 cells that are derived from neuronal lineage cells within a human embryonic kidney (Shaw et al., 2002), or N2a mouse neuroblastoma cells (Pahlman et al., 1984); relative to the 9 non-neural cell lines in the study (Huang et al., 2008, Xia et al., 2008). Table 2 lists 15 “known” in vivo neurotoxicants’ selected from the literature, 12 “candidate” neurotoxicants that showed selectivity for neural-derived cell lines in vitro (Huang et al., 2008, Xia et al., 2008), and 3 “reference” toxicants with well-characterized effects on cells, such as tunicamycin to provoke ER-stress. These 32 compounds listed in Table 2 were studied further for cytotoxic and apoptotic effects on the SH-SY5Y, LUHMES, and NSC cell lines.
Undifferentiated SH-SY5Y, LUHMES, and NSC cells, or 7-day differentiating cells dSH-SY5Y, dLUHMES, and dNSC, were distributed to 384-well plates for an additional day; then treated with one of 32 known or suspected neurotoxicants at a range of concentrations from 0.78 to 100 µM in 2-fold steps. After 24 hours of exposure, cytotoxicity was assessed by measuring intracellular ATP levels, a sensitive gauge of cell death or reduction in cellular energy levels (Xia et al., 2008). On replicate wells, Caspase 3/7 activity was also measured after 24 hours to assess cell death by apoptosis. IC50 values for [ATP] were derived by interpolation at the point each curve showed 50% depletion of the total intracellular ATP relative to the plot for DMSO controls on that day.
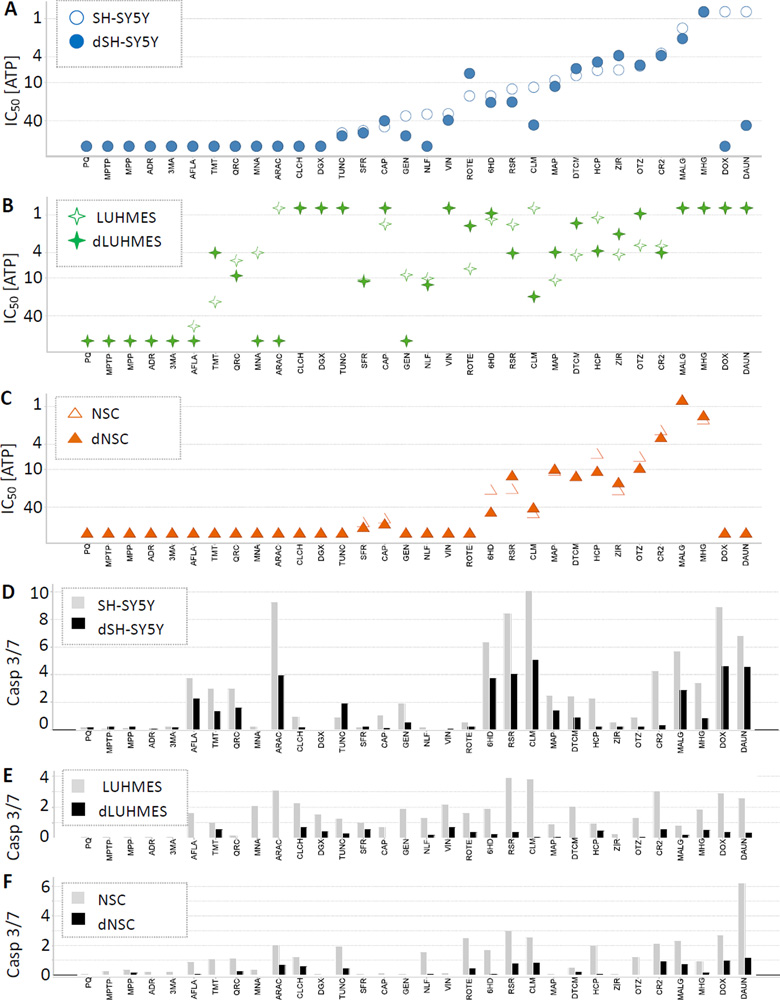
Figure 2 illustrates the relative cytotoxic sensitivities of SH-SY5Y, LUHMES, and NSC, and differentiating cells dSH-SY5Y, dLUHMES, and dNSC, to 32 known or suspected neurotoxicants. Several trends were apparent, such as that differentiating and undifferentiated cells from each cell line were usually similar in sensitivity to a given compound, albeit with important exceptions noted below. In general, when cell lines differed in sensitivity to a toxicant, LUHMES and/or dLUHMES tended to be more sensitive than the other cell lines, including 22 of 32 toxicants: 6HD, AFLA, ARAC, CAP, CLCH, CLM, DGX, DTCM, GEN, HCP, MAP, MNA, NLF, OTZ, QRC, ROTE, RSR, STR, TMT, TUNC, VIN, and ZIR. Several exceptions were observed in which all cell types were similarly sensitive, including: CR2, MALG and MHG. MALG showed a unique pattern, in which LUHMES, dLUHMES, NSC and dNSC exhibited comparable sensitivity to MALG, whereas SH-SY5Y and dSH-SY5Y showed less sensitivity. SH-SY5Y cells and dSH-SY5Y cells were in many cases more sensitive to toxicants than NSC and dNSC cells, including DTCM, GEN, ROTE, TUNC, VIN, and ZIR (Figure 2). There was only one toxicant, MALG, for which both NSC and dNSC were more sensitive than SH-SY5Y cells and dSH-SY5Y cells, but several toxicants for which the sensitivities were roughly comparable, i.e. 6HD, CAP, CR2, HCP, MAP, OTZ, and RSR.
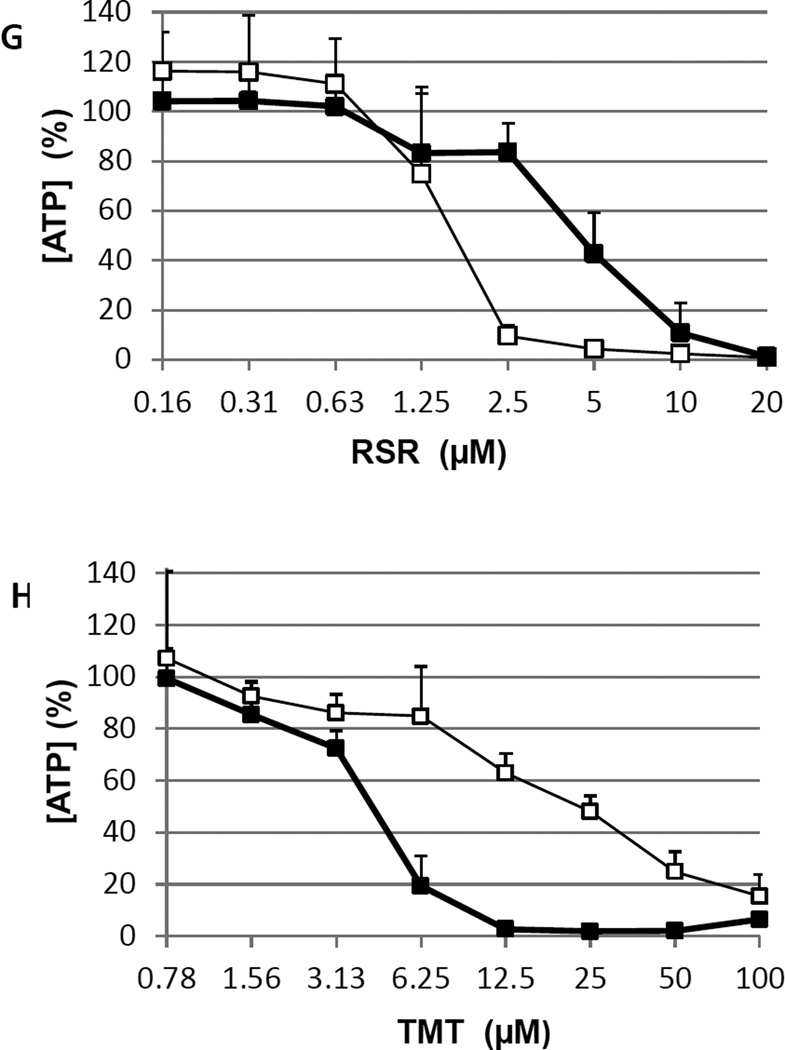
Figure 2. [ATP] IC50’s and Caspase 3/7 Activities for 32 Compounds on 6 Cell Types.
[ATP] IC50’s (A) ○SH-SY5Y, ● dSH-SY5Y, (B) ✧LUHMES, ✦dLUHMES, and (C) △NSC, and ▲dNSC. Caspase 3/7 activities are shown in (D) SH-SY5Y, dSH-SY5Y; (E) LUHMES, dLUHMES; and (F) NSC and dNSC; using light grey bars to represent undifferentiated cells, and black bars to represent differentiating cells. IC50 values above 100 µM or below 0.78 µM are plotted at 100 µM or 0.78 µM in Figure 2 A, B, C. The maximum Caspase 3/7 activity is shown for each compound:cell combination as –fold change above DMSO control values in Figure 2 D, E, F. (G) and (H) are plots of RSR and TMT vs. intracellular [ATP] comparing LUHMES (empty squares and light lines) to dLUHMES (filled squares and heavy lines), including ‘error bars’ showing standard deviation. In these examples, LUHMES is more sensitive than dLUHMES to RSR (G), whereas dLUHMES is more sensitive than LUHMES to TMT (H).
In some cases, undifferentiated and differentiating cells from the same cell line exhibited markedly different toxicant sensitivity. NSC and dNSC showed disparate sensitivities to the fewest compounds, with NSC more sensitive than dNSC to 6HD, HCP, and OTZ (Figure 2). Undifferentiated SH-SY5Y cells exhibited greater sensitivity to 6HD, CLM, DAUN, DOX, GEN, MALG, NLF, RSR, and VIN than dSH-SY5Y cells. It is likely that DNA damaging agents, such as the Topoisomerase II poisons DOX and DAUN, target undifferentiated SH-SY5Y cells, because dsDNA breaks caused by DOX and DAUN would disrupt DNA replication and mitosis. Conversely, dSH-SY5Y cells were more sensitive than SH-SY5Y cells to ROTE and ZIR, and slightly more sensitive to CAP, DTCM, and HCP. Both NSC and dNSC were insensitive to most test compounds up to 100 µM. NSC were more sensitive than dNSC to 6HD, HCP, and OTZ, whereas the converse was true for RSR and ZIR. All three cell lines were more sensitive to ZIR in the differentiating state than when undifferentiated.
LUHMES cells were more sensitive than dLUHMES cells, as expected, to 13 compounds: AFLA, ARAC, CLM, GEN, HCP, MNA, NLF, QRC, ROTE, and RSR (Figure 2 B); as well as DAUN, DIG, and DOX when assayed at lower doses. Surprisingly, differentiating non-growing dLUHMES cells were markedly more sensitive than LUHMES cells to 11 test compounds, including: 6HD, CAP, DTCM, MAP, OTZ, TMT, and ZIR; as well as CLCH, MALG, MHG, and VIN, when assayed at lower doses (Supplementary data). Example [ATP] viability plots are shown for toxicant RSR, in which LUHMES are more sensitive than dLUHMES (Figure 2G); and toxicant OTZ, in which dLUHMES are more sensitive than LUHMES (Figure 2H). In general, undifferentiated cells are observed to be more sensitive to cytotoxicity and apoptosis as observed for a variety of mammalian cell types (Ekwall, 1983). The greater sensitivity of undifferentiated cells to toxicants is often explained by the susceptibility of cells to DNA damage and apoptosis during DNA replication and mitosis. It was surprising, therefore, to find dLUHMES more sensitive than LUHMES to 11 compounds.
Apoptosis - Caspase 3/7 activity
An assay for Caspase 3/7 activity was used to determine whether cell death was apoptotic in nature. Maximal Caspase 3/7 activity from dose-response is shown, for each compound in Figure 2, panels D, E, and F, as fold-increase above DMSO control activity for each cell type. For compound:cell-type combinations where Caspase 3/7 activity was significantly elevated, Caspase 3/7 activity peaked at a chemical concentration approximately one step (3.2-fold) below the IC50 observed with the intracellular [ATP] assay. This is expected, since induction of apoptosis both activates Caspase 3/7 activity, and limits it by degrading the Caspase proteases. In general, differentiating cells showed lower Caspase 3/7 activities than undifferentiated cells (Figure 2 D, E, F). Undifferentiated SH-SY5Y cells expressed higher peak Caspase 3/7 activities than LUHMES or NSC cells treated with the same compounds, with four compounds provoking 8–10-fold increases over controls. There was a strong tendency for compounds that increased Caspase 3/7 activity in an undifferentiated cell type, to also increase Caspase 3/7 activity in the corresponding differentiating cell type. The only exceptions were increased (>1.5-fold above control) Caspase 3/7 activity in LUHMES but not dLUHMES by: AFLA, ARAC, CLM, DTCM, GEN, and MNA; and in NSC but not dNSC by 6HD and HCP. Differentiating dSH-SY5Y cells had increased Caspase 3/7 activity compared to dLUHMES or dNSC cells. This result is concordant with the lower expression of differentiation markers in dSH-SY5Y cells, and the weak expression of neuronal markers in dSH-SY5Y (Table 1), suggesting that dSH-SY5Y cells are only partly differentiated. In SH-SY5Y and dSH-SY5Y cells, every cytotoxic compound increased Caspase 3/7 activity; except NLF with SH-SY5Y, and VIN with both SH-SY5Y and dSH-SY5Y cells (Figure 2D). Peak Caspase 3/7 activities varied widely for SH-SY5Y and dSH-SY5Y cells, with the highest activities observed for CLM, ARAC, RSR, DOX, DAUN, 6HD, and MALG. Based on these Caspase 3/7 assay results, particularly the robust increases that occurred in dSH-SY5Y as compared to other differentiating cell lines, 7 compounds were selected for further study to examine mechanisms of apoptotic sensitivity: 6HD, DOX, DTCM, MALG, MAP, MHG, and RSR.
Apoptosis gene expression, comparing 6 cellular models
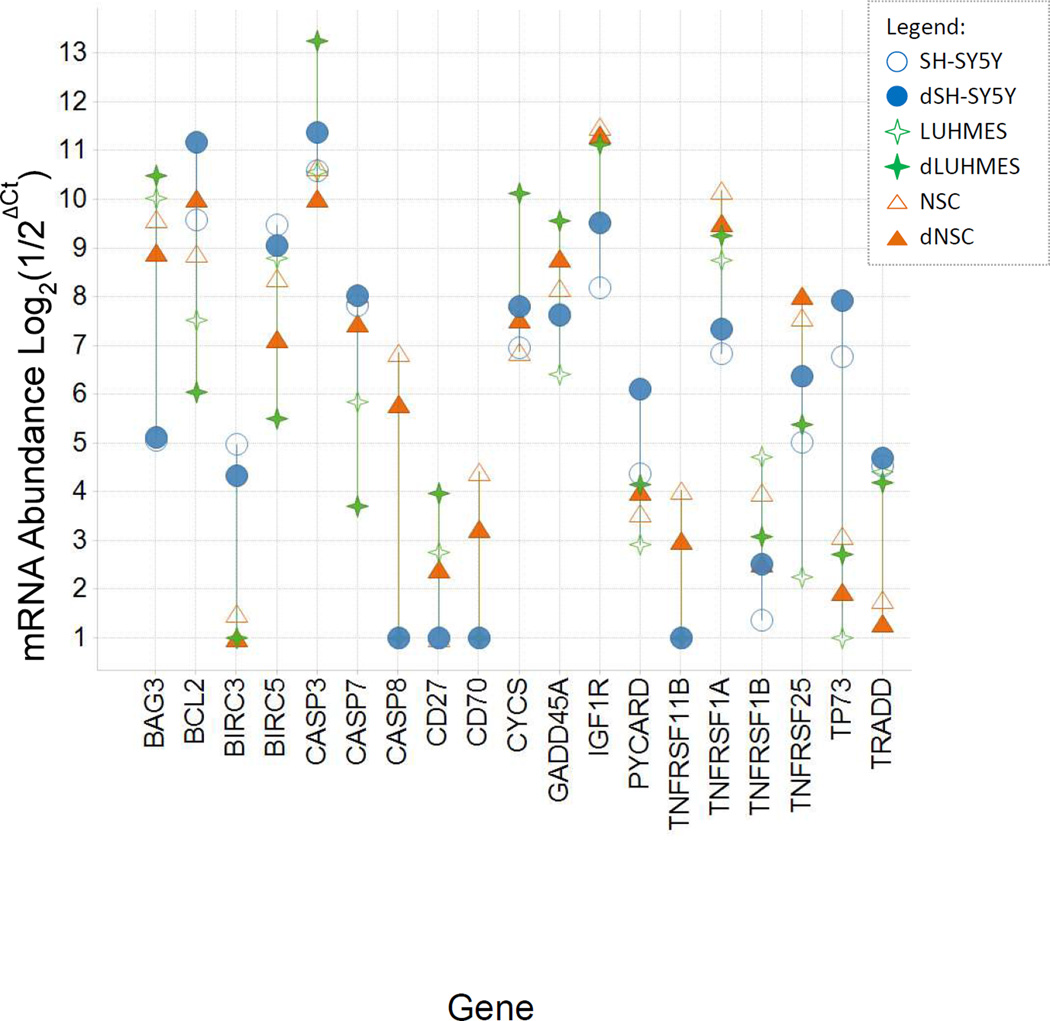
Messenger RNAs from 84 genes that regulate sensitivity to apoptosis were assayed using qPCR. Differences in mRNA abundance for apoptosis regulatory genes may yield insights into differences between the three cell models, and differences between differentiating and undifferentiated cells of the same model. Most of the 84 genes were expressed at similar levels in the various cell lines, however 19 genes showed differences between cell lines of ≥ 16-fold, and are shown in Figure 3. Most of these genes can be classified as either pro- or anti-apoptotic, for roles that shift the balance towards apoptosis (e.g. BAK, FAS, HRK, and TNFRSF1A), or potentiate apoptosis (e.g. Caspase genes, CD70, CYCS, and TNFRSF1B); or as anti-apoptotic (e.g. BAG3, BCL2, BIRCs, NOL3, TNFRSF 1B, TNFRSF25, TP73, and TRADD). A few genes can reportedly play either pro- or anti-apoptotic roles, such as NFκB.
Figure 3. Relative mRNA abundance of apoptosis regulatory genes comparing 6 cell types.
qPCR was used to determined relative abundances of mRNAs. Gene expression values were normalized to GAPDH, and expressed in Log2 units such that a value of 1 on this scale denotes that a gene’s mRNA was at, or below, the limit of detection by qPCR. Genes shown differ in expression by ≥ 16-fold (≥ 24) between cell types.
In characterizing the SH-SY5Y cell model, several anti-apoptotic genes were expressed more highly in both SH-SY5Y and dSH-SY5Y cells than in the other two models, including BCL2 (~2–30-fold), BIRC3 (~10-fold), and TP73 (~16–30-fold). Conversely, anti-apoptotic gene BAG3 was expressed at ~30-fold lower levels in both SH-SY5Y and dSH-SY5Y cells than in the other two models. Apoptosis-potentiating gene FAS was expressed at ~4-fold lower level in dSH-SY5Y, than in undifferentiated SH-SY5Y cells, and 3–20-fold lower levels than in the other two models. Anti-apoptotic gene TNFRSF25 (Tumor Necrosis Factor Receptor 25) was most abundant in dSH-SY5Y cells. Expression of TP73 was abundant in both SH-SY5Y and dSH-SY5Y cells compared to the other cell lines studied. TP73 is a cell stress responsive transcription factor in the TP53 family that serves as a tumor suppressor gene. (Romani et al., 1999)
In dLUHMES cells several survival genes e.g. BCL2, BIRC3, and BIRC5 were expressed at much lower levels in dLUHMES cells than in the other cell types or in undifferentiated LUHMES cells (Figure 3). Several genes that potentiate apoptosis were expressed at the highest levels in dLUHMES cells, including CASP3, CD27, CYCS, and DAPK1. Conversely, CASP7 is expressed at the lowest level in dLUHMES cell type.
NSC and dNSC expressed several apoptotic genes that were not detected in the other cell types. For example, CASP8, CD70, and TNFRSF11B were expressed uniquely by NSC and dNSC (Figure 3). Caspase 8 functions downstream of death receptors for the apoptotic cytokines: TRAIL, TNF, or FASL. (Hopkins-Donaldson et al., 2000) Similarly, CD70 encodes an inflammatory TNF-family cytokine that activates circulating lymphocytes, and TNFRSF11B encodes Osteoprotegerin, a decoy receptor for the inflammatory cytokine TRAIL (Kichev et al., 2014).
Gene expression responses to 7 apoptotic toxicants
The six cell types treated with seven selected apoptotic toxicants were also examined for changes in mRNA expression of 84 apoptosis-relevant genes. The 42 resulting cell:toxicant combinations necessitated selection of a single concentration level for each treatment. Gene expression was assayed at 6 hours to examine short term adaptation to the toxicants, and to avoid effects that followed the peak of Caspase activation which typically occurs in vitro at 12–24 hours (Xia et al., 2008). For each cell line:toxicant pairing a single concentration was selected that caused some decrease in [ATP] but did not kill all the cells, approximately the IC20 at 24 hr. Specifically, SH-SY5Y, dSH-SY5Y, NSC and dNSC were treated for 6 hours with: DOX 0.1 µM, MAP 12.5 µM, DTCM 10 µM, 6HD 2 µM, MALG 1.56 µM, MHG 1.56 µM, and RSR 10 µM. The greater sensitivity of LUHMES and dLUHMES cells to these toxicants necessitated using lower concentrations of toxicants for these cell types. LUHMES and dLUHMES were treated with: DOX .01 µM, MAP 10 µM, DTCM 5 µM, 6HD 1 µM, MALG 0.78 µM, MHG 0.78 µM and RSR 1.56 µM.
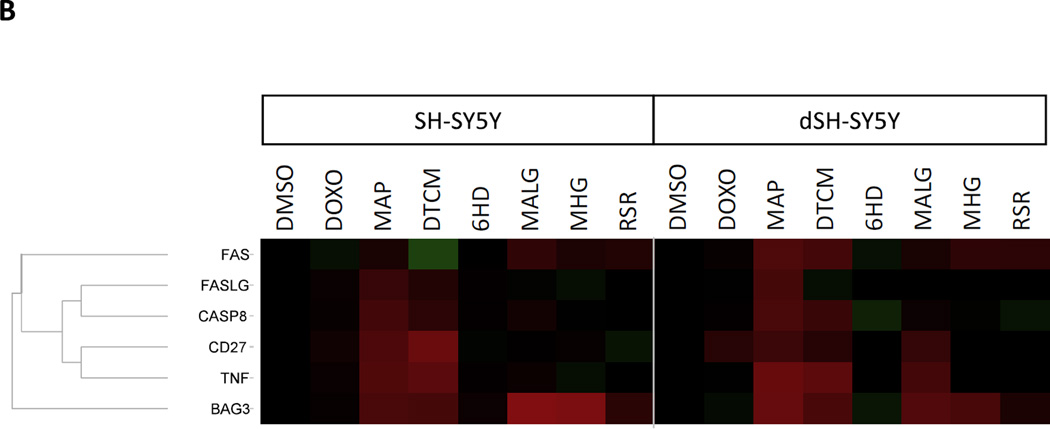
The gene expression responses at 6 hours were examined for changes shared either between cell-types for the same toxicant, or between toxicants for the same cell type, to give insights into how these cell models differ in their responses. Figure 4 illustrates several such patterns, particularly shared between SH-SY5Y and dSH-SY5Y cells in responses to multiple toxicants. One such pattern from Figure 4 was a conserved pattern of gene responses by SH-SY5Y and dSH-SY5Y cells to MAP and to DTCM. These responses included increases in mRNAs for: CASP8, TNF and CD27/TNFRSF27, FASLG and FAS (Figure 4B; except FAS was down-regulated in SHSY5Y by DTCM and FASL mRNA was unchanged in dSH-SY5Y by DTCM).
Figure 4. Apoptosis-Related Gene Expression Profiles of 6 cell types treated with each of 7 toxicants.
SH-SY5Y, dSH-SY5Y, NSC and dNSC were treated for 6 hours with: DOX 0.1 µM, MAP 12.5 µM, DTCM 10 µM, 6HD 2 µM, MALG 1.56 µM, MHG 1.56 µM, and RSR 10 µM. LUHMES and dLUHMES were more sensitive to these toxicants, and so were treated with: DOX .01 µM, MAP 10 µM, DTCM 5 µM, 6HD 1 µM, MALG 0.78 µM, MHG 0.78 µM, and RSR 1.56 µM. Three biological replicates were analyzed by qPCR. The LUHMES samples treated with 1.56 µM RSR were lost due to excessive cytotoxicity. Red represents increased expression and green represents decreased expression of each gene’s mRNA relative to DMSO-treated cells, with colors saturated at 16-fold. (A) Overview of all 6 cell types and 61 apoptosis-related genes. The dendrogram illustrates hierarchical clusters of genes that showed similar expression profiles. Genes were clustered using the UPGMA method and the Euclidian distance metric with Spotfire software (TIBCO Software Co., Palo Alto, CA). The 61genes, listed from top to bottom, were: BIRC5, BIRC2, CIDEB, IGF1R, HRK, CD70, BIRC3, TNFSF10, AIFM1, XIAP, TP73, BCL2L2, FADD, BID, CFLAR, DAPK1, BAG1, CASP6, BNIP3L, CRADD, CYCS, APAF1, BCL10, BFAR, NOD1, BAK1, BNIP2, CASP7, TNFRSF11B, TRADD, TNFRSF1B, BCL2A1, BCL2L10, ABL1, TNFRSF21, TNFRSF1A, TNFRSF10B, BCL2L11, CD40LG, TNFSF8, CASP14, LTBR, CASP4, CASP1, CASP5, CD40, TNFRSF10A, FASLG, TNFRSF9, IL10, CASP8, CIDEA, CASP10, LTA, TNF, PYCARD, FAS, CD27, BIK, GADD45A, and BAG3.
(B) Detail illustrating expression of 6 genes regulated similarly by multiple toxicants in SH-SY5Y and dSH-SY5Y cells.
A second clear pattern was the increased expression of BAG3 in both SH-SY5Y and dSH-SY5Y cells in response to five of the seven apoptotic toxicants (MAP, DTCM, MALG, MHG, and weakly by RSR). The same five apoptotic toxicant treatments also resulted in down-regulation of BIRC3 and BIRC5 genes in SH-SY5Y and dSH-SY5Y cells, except BIRC5 did not respond detectably to RSR or MAP in either cell type, and BIRC3 did not respond detectably to DTCM in dSH-SY5Y nor to RSR in either cell type. Note that in control cells, BAG3 expression was approximately 16-to-32-fold lower in SH-SY5Y and dSH-SY5Y cells than in the other four cell types, whereas BIRC3 and BIRC5/survivin expression was highest in control SH-SY5Y and dSH-SY5Y cells compared to the other four cell types (Figure 4).
These apoptotic-gene expression responses were examined for additional changes shared between cell-types for the same toxicant(s), or between toxicants for the same cell type. Overall, SH-SY5Y and dSH-SY5Y cells shared similar gene expression profiles to several toxicants including the BAG3/ BIRC3/BIRC5, and death receptor pathways, whereas NSC and dNSC showed few similarities in responses to the seven apoptotic toxicants, and LUHMES and dLUHMES shared essentially no similar responses.
Discussion
Differentiation gene markers
This study assessed three cell lines and cells resulting from their respective differentiation protocols, for three responses: efficient expression of neuronal markers, sensitivity to 32 known or candidate neurotoxicants, and transcriptional expression of apoptosis regulatory genes including responses to 7 apoptotic compounds. Morphological observation indicated that most SH-SY5Y cells showed short neurites before differentiation and long neurites after differentiation. Expression of neuronal marker genes was assessed for: TUBB3, ENO2, DCX, MAP2, SYN1, and KCNJ6, to determine neuronal character of each cell type (Table 1). Among these neuronal markers, DCX expression may reflect immature differentiating neurons, and was abundant in all three differentiating cell lines. Morphological observations for SH-SY5Y cells were consistent with the mRNA levels of marker genes, which revealed low-to-moderate expression of neuronal marker genes before differentiation and 2–3-fold increased levels of neuronal marker genes after 7 days of differentiation. These observations of very limited differentiation marker gene expression by SH-SY5Y cells are consistent with other published reports (Cheung et al., 2009, Forster et al., 2016), although it is possible that a better differentiation protocol exists. It should also be noted that SH-SY5Y cells exhibit extensive chromosomal duplications and deletions (Krishna et al., 2014), as expected for a tumor-derived cell line.
NSCs showed no neurites before differentiation, but cells of varied morphologies were visible after 7 days of differentiation, including a minority of cells displaying neurites. Neuronal marker genes in NSCs were moderate before differentiation and generally several-fold higher after 7 days of differentiation. The moderate levels of neuronal markers: TUBB3, ENO2, DCX, MAP2, SYN1, and KCNJ6, expressed at 7 days of differentiation suggest an immature state of NSC-derived neurons (Table 1). These observations are consistent with published observations that NSCs benefit from several weeks of differentiation. (Shin and Vemuri, 2010) Very low expression of oligodendrocyte markers GALC and MBP, and of astrocyte markers S100B and GFAP, further indicates the need for a longer differentiation period for NSCs (Shin and Vemuri, 2010). Interpretation of cytotoxicity to NSCs was complicated by the populations of cells that were differentiating into neurons, glia, and oligodendrocytes. The mixture of cell types from NSCs are advantageous for some neurobiology studies; but for neurotoxicity screening it was difficult to ascertain which cell type(s) were killed by each toxicant. A longer differentiation protocol, 2–4 weeks are recommended (Shin and Vemuri, 2010), would enable additional differentiation of these cell types. However, a differentiation period of several weeks is difficult to replicate from one experiment to the next such that cell types are in the same proportions and same degree of maturity, and so is problematic for qHTS. Refinements to stem cell differentiation protocols are ongoing in many labs and will likely result in more facile neuronal models from NSCs.
LUHMES cells showed no neurites before differentiation, but long neurites extending from nearly all cells after differentiation. Marker gene expression reflected morphology, with virtually no expression of neuronal marker genes before differentiation, yet very abundant expression of neuronal markers after 7 days of differentiation. Following 7 days of differentiation, dLUHMES cells expressed neuronal differentiation marker genes more abundantly than dSH-SY5Y or dNSC, including: TUBB3, ENO2, DCX, MAP2, SYN1, and KCNJ6; as well as four genes characteristic of dopaminergic neurons: PINK1, PARK2, DRD2, and TH. (Guichet et al., 2013) In contrast, undifferentiated LUHMES cells expressed virtually no differentiation markers (Table 1). Overall, LUHMES differentiated rapidly, and dLUHMES cells expressed neuronal markers and dopaminergic markers most abundantly, suggesting a phenotype more characteristic of neurons than the other two cell lines. This panel of marker genes was useful to assess cell lineages, and could be used to develop improved differentiation protocols for these cell lines.
Cytotoxicity determination using 32 toxicants
LUHMES and/or dLUHMES tended to be more sensitive to the toxicants than the other cell lines, including 22 of 32 toxicants. Table 2 indicates that these 32 chemicals tested includes 15 known neurotoxicants, 14 candidate neurotoxicants that showed selective toxicity to 4 neural-derived cell lines compared to 9 non-neural cell lines, and 3 reference compounds. In most cases undifferentiated cells from each cell line were similarly sensitive to a given toxicant, or more sensitive than differentiating cells. However, dLUHMES cells were unique among the differentiating cell lines in being more sensitive than the undifferentiated LUHMES cells to 11 toxicants, including known neurotoxicants colchicine, methyl-mercury (II), and vincristine. (Goldschmidt and Steward, 1989, Huang et al., 2014, Courtemanche et al., 2015) The greater sensitivity of the differentiating cells to so many compounds is unexpected, and may reflect sensitivity of specialized neurons to particular neurotoxicants observed in vivo. The cell culture media used were minimally different for LUHMES and dLUHMES, hence it is possible that the sensitivity of dLUHMES to some compounds depends on the highly differentiated phenotype of dLUHMES cells indicated by abundant expression of markers of neuronal differentiation in Table 1.
The selective toxicity of several toxicants for dLUHMES cells suggests relevance of this model to screen for neuro-cytotoxicity and to further investigate toxic mechanisms. Neurons are thought to be susceptible to particular toxicants due to their formation of long delicate neurites, to high energy demands for neurotransmission, and due to specialized neurotransmitters and synaptic activities. Selective action of CLCH, VIN, MHG, and TMT on microtubules has been invoked to explain toxicity of these tubulin disruptors to neurons; because these neurons elaborate extensive neurites that are structurally dependent on tubulin. (Goldschmidt and Steward, 1989) (Mundy and Tilson, 1990) (Bondy and Hall, 1986, Johansson et al., 2007) For example, ARAC is known to cause peripheral neuropathy through oxidative stress (Geller et al., 2001). Rotenone also causes oxidative stress, in this case by inhibiting mitochondrial electron transport Complex I. This oxidative stress is thought to explain why rotenone exposure is associated with Parkinson’s disease. (Caudle et al., 2012) The nitrogen mustard CLM is known to be neurotoxic in mice (Verschoyle et al., 1994), and showed selective toxicity to SH-SY5Y cells in qHTS for cytotoxicity (Huang et al., 2008, Xia et al., 2008), however no basis for neuronal-selectivity is described in the literature. All three cell lines were more sensitive to ZIR in the differentiating state than when undifferentiated in this study. The selective toxicity of ZIR for differentiating cells of all three cell lines raises the question of whether it is similarly selective in vivo. Indeed, there is suggestive evidence that sodium dimethyldithiocarbamate (ZIR is Zinc dimethyldithiocarbamate) causes motor deficits in mice by inhibiting proteasomal E1 Ligase activity.(Chou et al., 2008) Taken together, these results suggest further study of ZIR pesticide is justified as a possible neurotoxicant. While these cultured cell models do not model chemical distribution to the brain, nor model the precise conditions experienced by neurons in vivo, sensitivity of dLUHMES cells to many of the tested toxicants suggests they may be a useful model to identify and characterize neurotoxicants.
Apoptosis - Caspase 3/7 activity
Caspase 3/7 assays were performed to identify which compounds kill cells via apoptosis. A subset of toxicants stimulated Caspase 3 and/or Caspase 7 activity in some cell types, leading to selection of seven toxicants for study of the underlying gene expression changes. Toxicants that increased Caspase 3/7 activity in an undifferentiated cell type, also tended to increase Caspase 3/7 activity in the corresponding differentiating cell type (Figure 2 D, E, F). The principal exceptions were Caspase 3/7 activity was increased at least 1.5-fold above control in LUHMES but not dLUHMES by: AFLA, ARAC, CLM, DTCM, GEN, and MNA; and in NSC but not dNSC by 6HD and HCP. Nearly all the toxicants caused greater increases in Caspase 3/7 activity in differentiating dSH-SY5Y cells compared to the same toxicants in dLUHMES or dNSC cells (Figure 2D). This may be explained by the lower expression of differentiation markers in dSH-SY5Y cells, in agreement with the weak expression of neuronal markers in dSH-SY5Y (Table 1), suggesting that dSH-SY5Y cells are only nominally different from SH-SY5Y cells.
Apoptosis gene expression, comparing 6 cellular models
Control cells of the six cell types were assayed for mRNA levels of 84 apoptosis-related genes using qPCR (Figure 3). In the SH-SY5Y cell model, anti-apoptotic genes BCL2, BIRC3, and TP73 were expressed more highly in both SH-SY5Y and dSH-SY5Y cells than in the other two models. These BCL2, BIRC3, and TP73 genes may play roles in rendering these tumor-derived cells resistant to apoptosis. Conversely, anti-apoptotic gene BAG3 was expressed at ~30-fold lower levels in both SH-SY5Y and dSH-SY5Y cells than in the other two models, suggesting a apoptosis-susceptible activity. Apoptosis-potentiating gene FAS was expressed at ~4-fold lower level in dSH-SY5Y, than in undifferentiated SH-SY5Y cells, and 3–20-fold lower levels than in the other two models. This observation suggests that weak expression of FAS is a possible mechanism of dSH-SY5Y cells resistance to FAS-Ligand-mediated apoptosis. Expression of TNFRSF25 (Tumor Necrosis Factor Receptor 25), thought to play an anti-apoptotic role via stimulating the NFκB pathway, was most abundant in dSH-SY5Y cells. Expression of TP73 was abundant in both SH-SY5Y and dSH-SY5Y cells compared to the other cell lines studied. TP73 is a cell stress responsive transcription factor in the TP53 family that serves as a tumor suppressor gene. Since TP73 gene is deleted in many neuroblastomas (Romani et al., 1999), it is unclear why it is relatively abundantly expressed in SH-SY5Y. Overall, the high expression of anti-apoptotic genes BCL2, BIRC3, and TP73, and low expression of FAS suggest mechanisms by which SH-SY5Y tumor-derived cells may be resistant to apoptosis.
Gene expression results in cells not treated with toxicants suggest that dLUHMES cells may be susceptible to apoptosis because they express low levels of anti-apoptotic genes BCL2 and BIRC5/survivin relative to the other cell types (Figure 3).
In control cells, NSC and dNSC express several pro-apoptotic genes that were not detected in the other cell types. CASP8, CD70, and TNFRSF11B were expressed uniquely by NSC and dNSC (Figure 3). Caspase 8 functions downstream of death receptors for apoptotic cytokines TRAIL, TNF, or FASL (Hopkins-Donaldson et al., 2000), suggesting that NSC and dNSC may be the only cell lines in this study that may be expected to respond to TRAIL, TNF, or FASL. Similarly, CD70 encodes an inflammatory TNF-family cytokine that activates circulating lymphocytes, and TNFRSF11B encodes Osteoprotegerin, a decoy receptor for the inflammatory cytokine TRAIL (Kichev et al., 2014). Expression of CASP8, CD70, TNFRSF11B uniquely by NSC and dNSC, suggests that this model is capable of responding to inflammatory cytokines in a manner that the other two models cannot, possibly due to the multiple neural lineages that can result from differentiation of NSCs.
Gene expression responses to 7 apoptotic toxicants
This qPCR was performed not only in the six control cell types, but also in each cell type 6 hours following treatment with threshold levels of the 7 apoptotic toxicants. The gene expression responses to apoptotic toxicants at 6 hours weare overall quite diverse; however several patterns were apparent, groups of genes that respond similarly to several toxicants and shared between several cell types. One clear pattern from Figure 4 was a conserved pattern of gene responses shared by SH-SY5Y and dSH-SY5Y cells in response to MAP and DTCM. These responses included increases in mRNAs for: CASP8, TNF and CD27/TNFRSF27, FASLG and FAS (Figure 4B; except FAS is down-regulated in SHSY5Y by DTCM and FASL mRNA is unchanged in dSH-SY5Y by DTCM). These five genes are components of the proapoptotic “death receptor pathway” (Garofalo et al., 2010). The death receptor pathway or “extrinsic apoptotic pathway” may be summarized as the TNF gene product TNFα binding to the TNF Receptor, or FASLG gene product binding to its receptor FAS, signaling to the death receptor pathway apoptotic effector CASPASE 8 to trigger apoptosis. TNFα and FAS-Ligand cytokines are often produced by glial cells, however, work with primary rodent neurons has also demonstrated production of both TNFα (Fernyhough et al., 2005) and FASLG (Medana et al., 2001) by neurons. Alternatively, mitochondrial impairment and cytochrome C release can trigger apoptosis, as demonstrated for dLUHMES in response to ROT and MPP (Krug et al., 2014). These increases in components of the death receptor pathway in SH-SY5Y and dSH-SY5Y may indicate increased activity in this apoptotic pathway. Interestingly, the concerted induction of these death receptor pathway genes was observed in SH-SY5Y and dSH-SY5Y cells, but not in LUHMES, dLUHMES, NSC, or dNSC, even in response to the same toxicants. This suggests that the death receptor pathway induction is selectively enabled in the SH-SY5Y neuroblastoma model.
Another conserved response observed in Figure 4 was the increased expression of BAG3 and concomitant down-regulation of BIRC3 and BIRC5/survivin genes in SH-SY5Y and dSH-SY5Y cells in response to five of the seven apoptotic toxicants (MAP, DTCM, MALG, MHG, and weakly by RSR). Previous work has shown that BAG3 associates with HSP70 to regulate survivin/BIRC5, a protein that prevents apoptosis by interacting with BCL2, and prevents mitotic catastrophe by modulating microtubules during mitosis. Most neuroblastoma cell lines depend on BIRC5/survivin gene product for growth and survival (Lamers et al., 2012). Mechanistically, BAG3:HSP70 act in tumor cells to modulate activity of mitosis regulatory proteins including CDKN1A/p21 and BIRC5/Survivin (Colvin et al., 2014). BIRC3 is a BIRC5-family member which has not been well-characterized. In the current study, treatment of SH-SY5Y and dSH-SY5Y cells with five apoptotic toxicants increased the mRNA for BAG3, and decreased the mRNA for BIRC5. The increase in BAG3 expression suggests that these five toxicants may cause presentation of denatured proteins to HSP70:BAG3 invoking autophagy and aggresome formation (Gamerdinger et al., 2011). Down-regulation of BIRC5/survivin and its close homolog BIRC3 further suggest a cell stress response to protein denaturation or DNA Damage (Gamerdinger et al., 2011, Colvin et al., 2014), however the mechanism of BIRC3 and BIRC5 down-regulation by the toxicants is unclear. Since SH-SY5Y and dSH-SY5Y cells responded to five apoptotic toxicants by increasing BAG3 expression from a low basal level, and decreasing BIRC3 and BIRC5 from high basal levels, we postulate that tumor-derived SH-SY5Y cells have some regulatory mutation that maintains the cells in a growth-permissive state, but that SH-SY5Y cells maintain the capacity to upregulate BAG3 and in turn downregulate BIRC5 in response to cellular stress by protein denaturation or DNA Damage.
The apoptotic-gene expression responses summarized in Figure 4 were examined for additional changes shared between cell-types for the same toxicant(s), or between toxicants for the same cell type. Whereas SH-SY5Y and dSH-SY5Y cells shared gene expression profiles to several toxicants (BAG3/BIRC3/BIRC5, and death receptor pathways discussed above); NSC and dNSC showed few common responses to the seven apoptotic toxicants, and LUHMES and dLUHMES shared essentially no common responses among the seven toxicants. These relationships are consistent with the degrees of similarity in expression of differentiation marker genes shown in Table 1, including high similarity between SH-SY5Y and dSH-SY5Y cells, moderate similarity between NSC and dNSC cells, and low similarity between LUHMES and dLUHMES cells. These results further suggest that differentiating cells in the dLUHMES and dNSC models reacted differently to apoptotic stimuli than the corresponding undifferentiated LUHMES and NSC cells, which is also in agreement with the degrees of differences observed between these cell types prior to toxicant treatment (Figure 3). The contrasting response of dLUHMES and LUHMES to 6HD is instructive. Differentiating dLUHMES cells exhibited slightly less sensitivity to killing by 6HD than LUHMES cells. In contrast, dLUHMES responded to 6HD with increased expression of a series of apoptosis-related genes in a response that was not observed in LUHMES cells. This suggests that dLUHMES may adapt to 6HD in a way that LUHMES do not. Although the response of dLUHMES to 6HD includes potentially pro-apoptotic as well as antiapoptotic gene-responses, increased expression of BCL2A1 and IL10 suggest an adaptation to decrease sensitivity of dLUHMES cells to 6HD.
These results provide a basis to select cell lines for further neuronal toxicity studies based collectively on markers of differentiation, sensitivity to known and suspected neurological toxicants, and expression of genes that regulate sensitivity to apoptosis. NSCs differentiated into multiple cell types: neurons, glia, and oligodendrocytes; providing an opportunity to study interactions between these cell types, although NSCs may require several weeks of differentiation to acquire mature neuronal characteristics. SH-SY5Y cells appeared to be most appropriate for study of neuroblastoma. LUHMES cells expressed neuronal markers most rapidly and abundantly; and differentiated LUHMES cells also appeared to be the most sensitive model overall for assessing potential neuronal cytotoxicity. While no cellular model can replicate all the toxicokinetic and toxicological sensitivities of human neurons, and these differentiation protocols are subject to improvements; 7-day differentiated LUHMES cells emerged as a promising model for qHTS of libraries of possible neurotoxicants.
Supplementary Material
Acknowledgments
The authors are grateful to Dr. Marcel Leist for guidance on LUHMES cells, and John Braisted, Dr. Ruili Huang, and Dr. Yuhong Wang for helpful advice and discussions. These studies were supported by the Tox21 Consortium, and the N.I.H. Intramural Research Program at NCATS.
References
- Bondy SC, Hall DL. The relation of the neurotoxicity of organic tin and lead compounds to neurotubule disaggregation. Neurotoxicology. 1986;7:51–55. [PubMed] [Google Scholar]
- Buntinx M, Vanderlocht J, Hellings N, Vandenabeele F, Lambrichts I, Raus J, Ameloot M, Stinissen P, Steels P. Characterization of three human oligodendroglial cell lines as a model to study oligodendrocyte injury: morphology and oligodendrocyte-specific gene expression. J Neurocytol. 2003;32:25–38. doi: 10.1023/a:1027324230923. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle WM, Guillot TS, Lazo CR, Miller GW. Industrial toxicants and Parkinson's disease. Neurotoxicology. 2012;33:178–188. doi: 10.1016/j.neuro.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YT, Lau WK, Yu MS, Lai CS, Yeung SC, So KF, Chang RC. Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology. 2009;30:127–135. doi: 10.1016/j.neuro.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Chou AP, Maidment N, Klintenberg R, Casida JE, Li S, Fitzmaurice AG, Fernagut PO, Mortazavi F, Chesselet MF, Bronstein JM. Ziram causes dopaminergic cell damage by inhibiting E1 ligase of the proteasome. J Biol Chem. 2008;283:34696–34703. doi: 10.1074/jbc.M802210200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coecke S, Eskes C, Gartlon J, Kinsner A, Price A, van Vliet E, Prieto P, Boveri M, Bremer S, Adler S, Pellizzer C, Wendel A, Hartung T. The value of alternative testing for neurotoxicity in the context of regulatory needs. Environ Toxicol Pharmacol. 2006;21:153–167. doi: 10.1016/j.etap.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Colvin TA, Gabai VL, Gong J, Calderwood SK, Li H, Gummuluru S, Matchuk ON, Smirnova SG, Orlova NV, Zamulaeva IA, Garcia-Marcos M, Li X, Young ZT, Rauch JN, Gestwicki JE, Takayama S, Sherman MY. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014;74:4731–4740. doi: 10.1158/0008-5472.CAN-14-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche H, Magot A, Ollivier Y, Rialland F, Leclair-Visonneau L, Fayet G, Camdessanche JP, Pereon Y. Vincristine-induced neuropathy: Atypical electrophysiological patterns in children. Muscle Nerve. 2015 doi: 10.1002/mus.24647. [DOI] [PubMed] [Google Scholar]
- Ekwall B. Screening of toxic compounds in mammalian cell cultures. Ann N Y Acad Sci. 1983;407:64–77. doi: 10.1111/j.1749-6632.1983.tb47814.x. [DOI] [PubMed] [Google Scholar]
- Fernyhough P, Smith DR, Schapansky J, Van Der Ploeg R, Gardiner NJ, Tweed CW, Kontos A, Freeman L, Purves-Tyson TD, Glazner GW. Activation of nuclear factor-kappaB via endogenous tumor necrosis factor alpha regulates survival of axotomized adult sensory neurons. J Neurosci. 2005;25:1682–1690. doi: 10.1523/JNEUROSCI.3127-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster JI, Koglsberger S, Trefois C, Boyd O, Baumuratov AS, Buck L, Balling R, Antony PM. Characterization of Differentiated SH-SY5Y as Neuronal Screening Model Reveals Increased Oxidative Vulnerability. J Biomol Screen. 2016 doi: 10.1177/1087057115625190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard A, Decressac M, Frappe I, Fernagut PO, Prestoz L, Besnard S, Jaber M. Anatomical and functional reconstruction of the nigrostriatal pathway by intranigral transplants. Neurobiol Dis. 2009;35:477–488. doi: 10.1016/j.nbd.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Condorelli GL, Croce CM, Condorelli G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ. 2010;17:200–208. doi: 10.1038/cdd.2009.105. [DOI] [PubMed] [Google Scholar]
- Gartlon J, Kinsner A, Bal-Price A, Coecke S, Clothier RH. Evaluation of a proposed in vitro test strategy using neuronal and non-neuronal cell systems for detecting neurotoxicity. Toxicol In Vitro. 2006;20:1569–1581. doi: 10.1016/j.tiv.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Geller HM, Cheng KY, Goldsmith NK, Romero AA, Zhang AL, Morris EJ, Grandison L. Oxidative stress mediates neuronal DNA damage and apoptosis in response to cytosine arabinoside. J Neurochem. 2001;78:265–275. doi: 10.1046/j.1471-4159.2001.00395.x. [DOI] [PubMed] [Google Scholar]
- Glaab E, Schneider R. Comparative pathway and network analysis of brain transcriptome changes during adult aging and in Parkinson's disease. Neurobiol Dis. 2015;74:1–13. doi: 10.1016/j.nbd.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Goldschmidt RB, Steward O. Comparison of the neurotoxic effects of colchicine, the vinca alkaloids, and other microtubule poisons. Brain Res. 1989;486:133–140. doi: 10.1016/0006-8993(89)91285-7. [DOI] [PubMed] [Google Scholar]
- Guichet PO, Bieche I, Teigell M, Serguera C, Rothhut B, Rigau V, Scamps F, Ripoll C, Vacher S, Taviaux S, Chevassus H, Duffau H, Mallet J, Susini A, Joubert D, Bauchet L, Hugnot JP. Cell death and neuronal differentiation of glioblastoma stem-like cells induced by neurogenic transcription factors. Glia. 2013;61:225–239. doi: 10.1002/glia.22429. [DOI] [PubMed] [Google Scholar]
- Gunasekar P, Li L, Prabhakaran K, Eybl V, Borowitz JL, Isom GE. Mechanisms of the apoptotic and necrotic actions of trimethyltin in cerebellar granule cells. Toxicol Sci. 2001;64:83–89. doi: 10.1093/toxsci/64.1.83. [DOI] [PubMed] [Google Scholar]
- Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000;60:4315–4319. [PubMed] [Google Scholar]
- Hsu YC, Chang SJ, Wang MY, Chen YL, Huang TY. Growth inhibition and apoptosis of neuroblastoma cells through ROS-independent MEK/ERK activation by sulforaphane. Cell Biochem Biophys. 2013;66:765–774. doi: 10.1007/s12013-013-9522-y. [DOI] [PubMed] [Google Scholar]
- Hu X, Xuan Y. Bypassing cancer drug resistance by activating multiple death pathways--a proposal from the study of circumventing cancer drug resistance by induction of necroptosis. Cancer Lett. 2008;259:127–137. doi: 10.1016/j.canlet.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Huang R, Southall N, Cho MH, Xia M, Inglese J, Austin CP. Characterization of diversity in toxicity mechanism using in vitro cytotoxicity assays in quantitative high throughput screening. Chem Res Toxicol. 2008;21:659–667. doi: 10.1021/tx700365e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Law S, Li D, Yu X, Li B. Mercury poisoning: a case of a complex neuropsychiatric illness. Am J Psychiatry. 2014;171:1253–1256. doi: 10.1176/appi.ajp.2013.12101266. [DOI] [PubMed] [Google Scholar]
- Ikegwuonu FI. The neurotoxicity of aflatoxin B1 in the rat. Toxicology. 1983;28:247–259. doi: 10.1016/0300-483x(83)90121-x. [DOI] [PubMed] [Google Scholar]
- Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Castoldi AF, Onishchenko N, Manzo L, Vahter M, Ceccatelli S. Neurobehavioural and molecular changes induced by methylmercury exposure during development. Neurotox Res. 2007;11:241–260. doi: 10.1007/BF03033570. [DOI] [PubMed] [Google Scholar]
- Joshi P, Vig PJ, Veerisetty V, Cameron JA, Sekhon BS, Desaiah D. Increase in brain nitric oxide synthase activity in daunorubicin-treated rats. Pharmacol Toxicol. 1996;78:99–103. doi: 10.1111/j.1600-0773.1996.tb00188.x. [DOI] [PubMed] [Google Scholar]
- Karmakar S, Choudhury SR, Banik NL, Ray SK. Combination of N-(4-hydroxyphenyl) retinamide and genistein increased apoptosis in neuroblastoma SK-N-BE2 and SH-SY5Y xenografts. Neuroscience. 2009;163:286–295. doi: 10.1016/j.neuroscience.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichev A, Rousset CI, Baburamani AA, Levison SW, Wood TL, Gressens P, Thornton C, Hagberg H. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) signaling and cell death in the immature central nervous system after hypoxia-ischemia and inflammation. J Biol Chem. 2014;289:9430–9439. doi: 10.1074/jbc.M113.512350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuge Y, Imai T, Kawaguchi M, Kihara T, Ishige K, Ito Y. Subregion-specific vulnerability to endoplasmic reticulum stress-induced neurotoxicity in rat hippocampal neurons. Neurochem Int. 2008;52:1204–1211. doi: 10.1016/j.neuint.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Krishna A, Biryukov M, Trefois C, Antony PM, Hussong R, Lin J, Heinaniemi M, Glusman G, Koglsberger S, Boyd O, van den Berg BH, Linke D, Huang D, Wang K, Hood L, Tholey A, Schneider R, Galas DJ, Balling R, May P. Systems genomics evaluation of the SH-SY5Y neuroblastoma cell line as a model for Parkinson's disease. BMC Genomics. 2014;15:1154. doi: 10.1186/1471-2164-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug AK, Balmer NV, Matt F, Schonenberger F, Merhof D, Leist M. Evaluation of a human neurite growth assay as specific screen for developmental neurotoxicants. Arch Toxicol. 2013;87:2215–2231. doi: 10.1007/s00204-013-1072-y. [DOI] [PubMed] [Google Scholar]
- Krug AK, Gutbier S, Zhao L, Poltl D, Kullmann C, Ivanova V, Forster S, Jagtap S, Meiser J, Leparc G, Schildknecht S, Adam M, Hiller K, Farhan H, Brunner T, Hartung T, Sachinidis A, Leist M. Transcriptional and metabolic adaptation of human neurons to the mitochondrial toxicant MPP(+) Cell Death Dis. 2014;5:e1222. doi: 10.1038/cddis.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers F, Schild L, Koster J, Versteeg R, Caron HN, Molenaar JJ. Targeted BIRC5 silencing using YM155 causes cell death in neuroblastoma cells with low ABCB1 expression. Eur J Cancer. 2012;48:763–771. doi: 10.1016/j.ejca.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Lotharius J, Falsig J, van Beek J, Payne S, Dringen R, Brundin P, Leist M. Progressive degeneration of human mesencephalic neuron-derived cells triggered by dopamine-dependent oxidative stress is dependent on the mixed-lineage kinase pathway. J Neurosci. 2005;25:6329–6342. doi: 10.1523/JNEUROSCI.1746-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews RA, Johnson TC, Hudson JE. Synthesis and turnover of plasma-membrane proteins and glycoproteins in a neuroblastoma cell line. Biochem J. 1976;154:57–64. doi: 10.1042/bj1540057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medana I, Li Z, Flugel A, Tschopp J, Wekerle H, Neumann H. Fas ligand (CD95L) protects neurons against perforin-mediated T lymphocyte cytotoxicity. J Immunol. 2001;167:674–681. doi: 10.4049/jimmunol.167.2.674. [DOI] [PubMed] [Google Scholar]
- Meijer DH, Kane MF, Mehta S, Liu H, Harrington E, Taylor CM, Stiles CD, Rowitch DH. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. 2012;13:819–831. doi: 10.1038/nrn3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed RH, Karam RA, Amer MG. Epicatechin attenuates doxorubicin-induced brain toxicity: critical role of TNF-alpha, iNOS and NF-kappaB. Brain Res Bull. 2011;86:22–28. doi: 10.1016/j.brainresbull.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Moulton PV, Yang W. Air pollution, oxidative stress, and Alzheimer's disease. J Environ Public Health. 2012;2012:472751. doi: 10.1155/2012/472751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy WR, Tilson HA. Neurotoxic effects of colchicine. Neurotoxicology. 1990;11:539–547. [PubMed] [Google Scholar]
- Ninomiya Y, Adams R, Morriss-Kay GM, Eto K. Apoptotic cell death in neuronal differentiation of P19 EC cells: cell death follows reentry into S phase. J Cell Physiol. 1997;172:25–35. doi: 10.1002/(SICI)1097-4652(199707)172:1<25::AID-JCP3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Ossola B, Kaarainen TM, Raasmaja A, Mannisto PT. Time-dependent protective and harmful effects of quercetin on 6-OHDA-induced toxicity in neuronal SH-SY5Y cells. Toxicology. 2008;250:1–8. doi: 10.1016/j.tox.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Pahlman S, Ruusala AI, Abrahamsson L, Mattsson ME, Esscher T. Retinoic acid-induced differentiation of cultured human neuroblastoma cells: a comparison with phorbolester-induced differentiation. Cell Differ. 1984;14:135–144. doi: 10.1016/0045-6039(84)90038-1. [DOI] [PubMed] [Google Scholar]
- Romani M, Scaruffi P, Casciano I, Mazzocco K, Lo Cunsolo C, Cavazzana A, Gambini C, Boni L, De Bernardi B, Tonini GP. Stage-independent expression and genetic analysis of tp73 in neuroblastoma. Int J Cancer. 1999;84:365–369. doi: 10.1002/(sici)1097-0215(19990820)84:4<365::aid-ijc6>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Rossignol DA, Genuis SJ, Frye RE. Environmental toxicants and autism spectrum disorders: a systematic review. Transl Psychiatry. 2014;4:e360. doi: 10.1038/tp.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C, Lesueur C, Aligny C, Brasse-Lagnel C, Genty D, Marret S, Laquerriere A, Bekri S, Gonzalez BJ. 3-MA inhibits autophagy and favors long-term integration of grafted Gad67-GFP GABAergic precursors in the developing neocortex by preventing apoptosis. Cell Transplant. 2014;23:1425–1450. doi: 10.3727/096368913X670174. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Poltl D, Nagel DM, Matt F, Scholz D, Lotharius J, Schmieg N, Salvo-Vargas A, Leist M. Requirement of a dopaminergic neuronal phenotype for toxicity of low concentrations of 1-methyl-4-phenylpyridinium to human cells. Toxicol Appl Pharmacol. 2009;241:23–35. doi: 10.1016/j.taap.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Scholz D, Poltl D, Genewsky A, Weng M, Waldmann T, Schildknecht S, Leist M. Rapid, complete and large-scale generation of post-mitotic neurons from the human LUHMES cell line. J Neurochem. 2011;119:957–971. doi: 10.1111/j.1471-4159.2011.07255.x. [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Shin S, Vemuri M. Culture and Differentiation of Human Neural Stem Cells. Springer Protoc Hand. 2010:51–73. [Google Scholar]
- Smirnova L, Harris G, Leist M, Hartung T. Cellular resilience. Altex. 2015;32:247–260. doi: 10.14573/altex.1509271. [DOI] [PubMed] [Google Scholar]
- Smirnova L, Hogberg HT, Leist M, Hartung T. Developmental neurotoxicity - challenges in the 21st century and in vitro opportunities. Altex. 2014;31:129–156. doi: 10.14573/altex.1403271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. 2013;121:756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschoyle RD, Carthew P, Holley JL, Cullis P, Cohen GM. The comparative toxicity of chlorambucil and chlorambucil-spermidine conjugate to BALB/c mice. Cancer Lett. 1994;85:217–222. doi: 10.1016/0304-3835(94)90278-x. [DOI] [PubMed] [Google Scholar]
- Wang A, Costello S, Cockburn M, Zhang X, Bronstein J, Ritz B. Parkinson's disease risk from ambient exposure to pesticides. Eur J Epidemiol. 2011;26:547–555. doi: 10.1007/s10654-011-9574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winship KA. Toxicity of tin and its compounds. Adverse Drug React Acute Poisoning Rev. 1988;7:19–38. [PubMed] [Google Scholar]
- Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, Tice RR, Austin CP. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HB, Ma H, Ha XQ, Zheng P, Li XY, Zhang M, Dong JZ, Yang YS. Salidroside induces rat mesenchymal stem cells to differentiate into dopaminergic neurons. Cell Biol Int. 2014;38:462–471. doi: 10.1002/cbin.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.