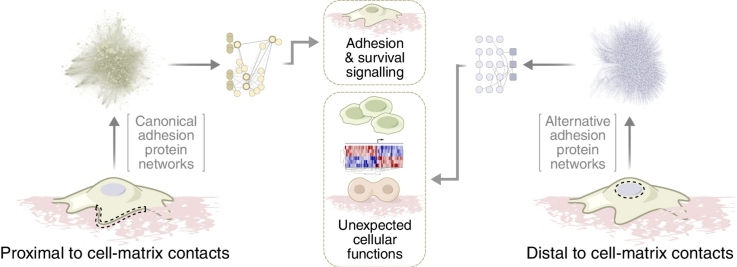
Graphical abstract
Highlights
-
•
Cell adhesion to the extracellular matrix (ECM) is vital for multicellular life.
-
•
Complexes of structural and signalling proteins link the ECM to the cytoskeleton.
-
•
Adhesion signalling networks are complex, diverse, dynamic and tightly regulated.
-
•
Some adhesion proteins perform unexpected functions away from cell-matrix contacts.
-
•
Nuclear roles for adhesion proteins have been revealed in diseases such as cancer.
Abstract
Cell adhesion to the extracellular matrix is generally mediated by integrin receptors, which bind to intracellular adhesion proteins that form multi-molecular scaffolding and signalling complexes. The networks of proteins, and their interactions, are dynamic, mechanosensitive and extremely complex. Recent efforts to characterise adhesions using a variety of technologies, including imaging, proteomics and bioinformatics, have provided new insights into their composition, organisation and how they are regulated, and have also begun to reveal unexpected roles for so-called adhesion proteins in other cellular compartments (for example, the nucleus or centrosomes) in diseases such as cancer. We believe this is opening a new chapter on understanding the wider functions of adhesion proteins, both proximal and distal to cell-matrix contacts.
Current Opinion in Cell Biology 2016, 39:93–100
This review comes from a themed issue on Cell regulation
Edited by Manuela Baccarini and Ivan Dikic
For a complete overview see the Issue and the Editorial
Available online 27th February 2016
http://dx.doi.org/10.1016/j.ceb.2016.02.013
0955-0674/© 2016 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
The extracellular matrix (ECM) forms an essential part of the cellular microenvironment; adhesion of cells to the ECM is critical for much of metazoan development, and its perturbation contributes to disease. The composition of ECM is highly diverse, containing proteins, glycoproteins and proteoglycans that interact to form a complex milieu [1]. It provides a structural support for cells to enable tissue formation and mechanosensing, and it binds soluble ligands and cell-surface receptors to trigger and coordinate cellular signalling [2]. Cells also use cell-surface adhesion receptors to sense the topology and stiffness of the pericellular ECM [3]. Mechanical information is transmitted via receptor-associated proteins to, and from, the actin cytoskeleton. Thus, adhesion receptors integrate and process biochemical and biophysical cues to control many aspects of cell behaviour, including differentiation, proliferation and migration.
The proteins that mediate adhesion signalling have been studied for decades. Recently, progress has been made in cataloguing the components of adhesions in various cell types, revealing that adhesion signalling is complex and diverse, both in terms of the number of components and the interrelations between them in signalling networks. Furthermore, the spatial restriction of this signalling is thought to drive emergent properties of multicellular systems in a way that is not yet fully understood [4]. Working out how cell adhesion systems function at a holistic network level is currently under intense scrutiny.
Here, we review recent progress in the elucidation of adhesion protein networks that mediate cell adhesion and provide the downstream effector signalling mechanisms. We also highlight new studies that have uncovered wider roles for adhesion protein signalling downstream of — and distal from — cell-ECM receptors. These studies suggest important new roles for adhesion proteins in diverse cellular locales.
Adhesion signalling complexes: defining the players
The best-characterised family of cell-surface ECM receptors is the integrins, members of which interact with a range of ligands in the extracellular milieu [5]. Upon ligand binding, intracellular adhesion proteins are recruited to clustered integrin heterodimers at the plasma membrane, forming adhesion complexes [6, 7]. These consist of signalling and structural proteins that connect integrins to the actin cytoskeleton, the sum of which has been termed the ‘adhesome’ [8]. The latest literature-curated adhesome database contained 232 proteins derived from studies using multiple cell types and experimental conditions [9••].
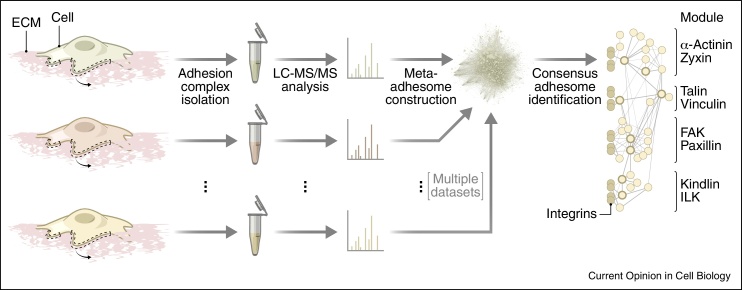
Until recently, the comprehensive, global analysis of adhesomes was restricted by the challenges of purifying the labile, membrane- and cytoskeleton-linked adhesion complexes. The development of biochemical methodologies to isolate integrin-associated proteins, coupled with advances in proteomics and informatics, has largely overcome the earlier major challenges, thus enabling the characterisation of adhesion complexes by quantitative mass spectrometry [10, 11, 12, 13, 14]. Computational integration of multiple adhesion-site proteomes yielded an experimentally defined ‘meta-adhesome’, from which a core set of 60 frequently identified proteins — a ‘consensus’ adhesome — was identified [15••] (Figure 1).
Figure 1.
Definition of a consensus adhesome. Adhesion complexes induced by the integrin ligand fibronectin were stabilised and purified (curly arrows) and their proteomes were characterised by quantitative mass spectrometry (LC–MS/MS) in multiple studies using different cell types. Integration of these datasets generated a meta-adhesome, from which a core consensus adhesome was established [15••]. Network nodes (circles) represent interacting proteins; thick node borders indicate proteins that define the axes of emergent consensus adhesome modules (labelled, right).
It was clear from the first mass spectrometric analyses of isolated adhesion complexes that the number of proteins in these assemblies was greater than previously appreciated [16, 17, 18]. This showed that integrin-mediated adhesions are sites of considerable molecular complexity and diversity, and it is likely that they are sophisticated signalling hubs with physical and functional links to the cytoskeleton and to other organelles and cellular processes. Moreover, adhesion complexes induced by different extracellular ligands, or recruited to different integrin receptors, contain both common and condition-specific subsets of proteins [16, 19, 20]. Therefore, understanding the precise and context-dependent relationships between multiple adhesion proteins, and the mechanisms by which they control cell behaviour, have become important future priorities.
Adhesion signalling close to integrins: mechanosensing the microenvironment
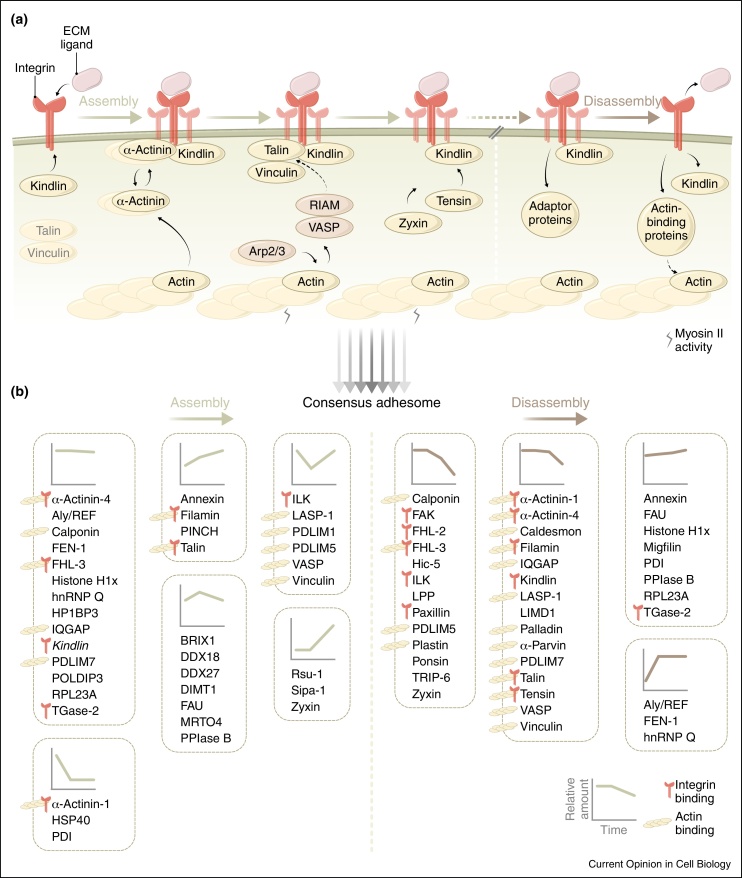
The assembly and disassembly of adhesion complexes are tightly and dynamically regulated. However, the precise interactions of adhesion proteins are poorly defined in both space and time. A recent fluorescence correlation microscopic analysis of tagged adhesion proteins led to a model of hierarchical protein recruitment to integrins at early (nascent) adhesions [21•]. This proposed initial binding of kindlin-2 to α5β1 integrin, a role for α-actinin in nucleation of adhesions and subsequent association of talin and vinculin in response to myosin II activation (Figure 2). Talin forms a complex with vinculin before it associates with integrin [21•], as appears to be the case for several other adhesome components [22•]. Active myosin II generates mechanical forces that can change the conformation of proteins, including talin [23]. In filopodial and lamellipodial protrusions, talin links integrin to RIAM, which can promote actin polymerisation [24]. Focal adhesion kinase (FAK) may also accumulate at adhesion sites at the front of cells before paxillin [25], while some molecules, such as zyxin and tensin, are generally absent from nascent adhesions [26]. However, the temporal sequence of events may be cell and context specific. Proteomic quantification of assembly and disassembly of isolated adhesion complexes has revealed distinct temporal profiles of protein recruitment [15••]. These proteomic studies support the early recruitment of α-actinin and the later appearance of zyxin at adhesion sites (Figure 2). Moreover, adaptor proteins are apparently lost from adhesion complexes more rapidly than actin-binding proteins during disassembly, suggesting a relatively late disruption of the integrin-actin connection during adhesion turnover [15••].
Figure 2.
Temporal dynamics of adhesion complex composition. (a) Hierarchical recruitment of adhesion proteins to integrins (assembly, left), as determined by fluorescence microscopy studies. Some proteins may exist as pre-formed complexes in the cytoplasm (such as talin and vinculin, grey font). α-Actinin aggregates are transiently incorporated into developing adhesions and link integrins to the actin cytoskeleton. In membrane protrusions, RIAM binds Ena/VASP (brown nodes) and talin to link integrins to actin. Recruitment of talin to β1 integrin tails and maturation of adhesions requires myosin II activity, as indicated. Loss of adhesion proteins during disassembly (right), as suggested by proteomic experiments, also appears to occur hierarchically. (b) Assembly (left) and disassembly (right) dynamics of consensus adhesome proteins. Line profiles for each cluster show trends of protein abundance over time, as quantified by mass spectrometry [15••]. Integrin-binding and actin-binding proteins are indicated. Kindlin in the assembly dataset is kindlin-3 (italics), whereas kindlin-2 is the family member in the consensus adhesome.
Despite the remarkable consistency of very early adhesion assembly, regardless of ligand density, rigidity or intracellular tension [27, 28•], the stability and growth of nascent adhesions are regulated by physical links to the cytoskeleton and are influenced by actin-associated proteins (for example, formins, septins and synaptopodins [29, 30, 31]). Microtubules also influence adhesion complex composition and dynamics [32•, 33, 34, 35, 36], with their targeting to adhesion sites being regulated by integrin activation state [37•].
The interactions of vinculin with talin and actin probably form the major mechanosensory module that controls adhesion site composition, organisation and stability [38, 39•, 40, 41], with a role also for FAK [42, 43, 44]. Proteomics experiments have identified many proteins, including a number with LIM domains, that are preferentially incorporated into more mature adhesions under myosin II-generated tension [15••, 17, 18], so increasing the repertoire of likely proteins involved in mechanotransduction [45, 46]. Particular integrins (for example, α5β1 and αV) selectively recruit proteins that mediate differential responses to force, indicating that receptor-specific and multiple modes of rigidity sensing exist [20]. The key mechanisms by which adhesion complex components, and their molecular interactions, enable cells to sense their microenvironment remain to be fully defined in the future.
Unexpected adhesion proteins: old functions for new proteins
Bioinformatic interrogation of datasets of adhesion proteomes has identified a substantial number of proteins that were not previously recognised as adhesion-linked proteins, and these have a broad range of predicted cellular functions [12, 15••, 37•]. Among the ‘non-canonical’ adhesion components, translation regulators are frequently detected in adhesion proteomes, supporting a long-held view that local protein translation occurs at adhesion sites [47]. Proteins involved in cytokinesis have also been discovered in adhesion complexes (e.g. RCC2 [16], CDK1 [48•]), strengthening the links between integrins and cell division [49]. There may also be transient ‘moonlighting’ roles for these proteins in integrin-mediated adhesion that have not yet been explored [50].
Adhesion protein roles far from integrins: new functions for old proteins
Classical adhesion proteins appear to perform unanticipated functions at non-adhesion cellular sites. For example, the integrin-binding protein kindlin-1 has been found at centrosomes, where it ensures correct assembly of mitotic spindles [51]. FAK, in association with paxillin, is also required for correct spindle orientation [52•]. It therefore seems that a number of proteins may link cell adhesion status with spindle assembly and so tight control of cell division.
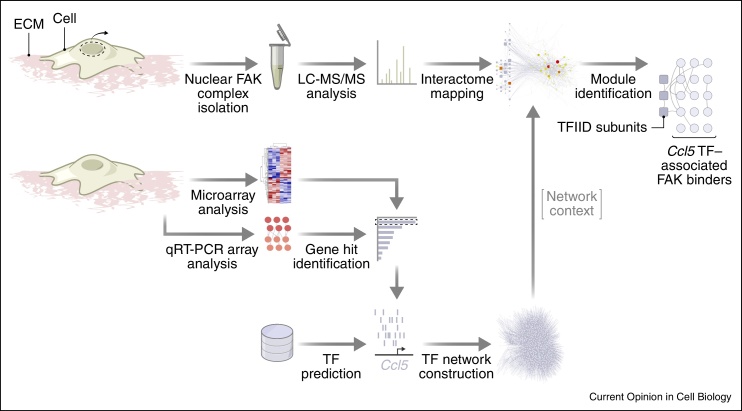
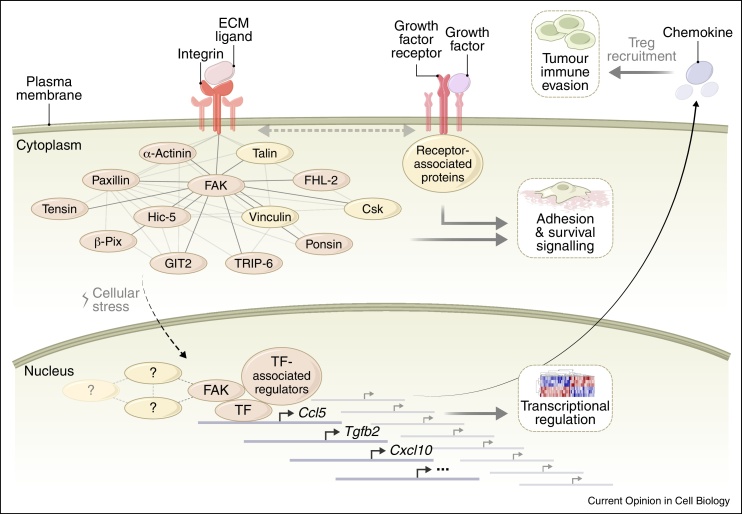
FAK is also targeted to the nucleus (i) when normal cells receive cellular stress, where it can promote cell survival and regulate VCAM-1 expression [53, 54, 55], or alter heterochromatin organisation and promote muscle differentiation [56], and (ii) when it is overexpressed in squamous cell carcinoma (SCC) cells, thereby controlling the tumour microenvironment and immune evasion [57••]. Although FAK is predominantly visualised at focal adhesions by fluorescence microscopy, biochemical fractionation of SCC cells that express high levels of FAK revealed abundant nuclear FAK that we now know causes profoundly important effects on transcription [57••]. Indeed, FAK is associated with chromatin, and an integrative proteomic, bioinformatic and network approach discovered that FAK binds to components of the basal transcription machinery and upstream regulators of sequence-specific transcription factors that control chemokine production, exemplified by the FAK-regulated chemokine Ccl5 [57••] (Figure 3). While the detailed mechanisms by which FAK controls nuclear transcription of genes in a selective manner are still to be worked out, that study concluded that FAK scaffolds selective regulators of chemokine transcription, many in a kinase-dependent manner, in turn leading to regulatory T cell recruitment and immune evasion [57••] (Figure 4). The intermolecular interaction of the FAT and FERM domains of FAK is likely to be disrupted to reveal its nuclear localisation sequence [58, 59•], and nuclear FAK may remain monomeric, supporting its nuclear scaffolding role (and potentially hindering its observation in the nucleus by immunofluorescence). Importantly, there is no detectable nuclear FAK in normal skin keratinocytes [57••], implying that nuclear accumulation of FAK is linked to the cancer phenotype. This raises the possibility that nuclear functions of FAK, and potentially other adhesion proteins, could be specific to pathological states, such as cancer, providing potential therapeutic opportunities.
Figure 3.
Integrative analysis of non-canonical FAK function. FAK complexes were isolated from purified SCC cell nuclei (curly arrow) and their interactomes were characterised by quantitative mass spectrometry (LC–MS/MS). The discovered protein interaction network was contextualised by mapping onto a network neighbourhood of transcription factors (TFs). Selected TFs were predicted to bind promoters of genes regulated by FAK (e.g. Ccl5), as identified by microarray and quantitative reverse transcription polymerase chain reaction (qRT-PCR) array analyses. This identified predicted TFs of FAK-regulated genes (e.g. transcription factor II D (TFIID) subunits) and their upstream regulators that interact with nuclear FAK [57••].
Figure 4.
Role of FAK in the interplay between the microenvironment, adhesion receptor complexes and the nucleus. The network of FAK interaction partners from the consensus adhesome, and their predicted interactions (light grey lines; FAK interactions, dark grey lines), is shown associated with integrins at the plasma membrane. Synergistic integrin and growth factor receptor signalling regulates cell adhesion, migration and proliferation. Several FAK-interacting partners have been reported in the nucleus (brown nodes). Upon cellular stress, FAK localises to the nucleus, where it associates with chromatin and binds transcription factors and their regulators to control gene expression, including that of the chemokine Ccl5, leading to regulatory T cell (Treg) recruitment and immune evasion [57••].
Other canonical adhesion proteins have been observed in the nucleus, e.g. zyxin, α-actinin and paxillin [60, 61, 62], and several have been shown to regulate hormone receptor signalling to influence gene transcription [63]. Indeed, the presence of a LIM domain appears to characterise a number of proteins that shuttle between integrin adhesion sites and the nucleus [64]. In addition, catenins, components of cell-cell adhesions, can enter the nucleus and control gene expression [65]. These findings raise a number of important questions about the generality, or specificity, of adhesion protein roles at sites far from adhesions. We do not know whether the role of nuclear FAK — or other nuclear adhesion proteins — is related to their better-understood adhesion roles (linking, for example, transcription to microenvironmental sensing) and, if so, how this is regulated or linked to disease phenotypes. It may be that some adhesion proteins have evolved scaffolding functions in other subcellular sites that are not related to their adhesion functions. The answers will undoubtedly come from further, ever more sophisticated, proteomic and network analyses, coupled to molecular intervention via genome editing and in physiologically relevant disease models. Nevertheless, these emerging data demonstrate a need to be cautious when addressing subcellular localisation solely by immunofluorescence microscopy, and they challenge our current view of the architecture and function of adhesion protein complexes. Moreover, they imply that long-range signalling by adhesion proteins, at sites distal to adhesions, will contribute to diverse cellular processes.
Conclusions
For the coordination of a cellular process such as migration, networks of scaffolds and signals must combine to manifest a cohesive biological response. It is now clear that such molecular systems are extremely complex. Knowledge of the components that form the machinery of cell adhesion has become more complete over the past decade, and a consensus adhesome has now been defined. A universal adhesion ‘module’ appears to exist, comprising a set of ubiquitous, standard components, to which other modules of adhesion proteins associate and dissociate during adhesion site maturation and turnover. Such a modular system of protein complex assembly/disassembly and signalling could permit modulation of adhesion strength and rapid response to the ECM with fidelity. Of course, major questions remain about the precise molecular mechanisms that operate at the extracellular-intracellular interface, the ‘modular’ dynamics needed to respond rapidly to changing physical and chemical stimuli and transduce resulting signals, and the features that define context-dependent adhesion regulation and mis-regulation — such as in wound healing and disease. Immediate challenges have to include defining the roles of unexpected classes of molecules at adhesion sites and the roles of adhesion proteins at non-canonical, distal subcellular locations, and these represent active areas of investigation. Network analyses will provide systems-level answers.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
A.B. and M.C.F. are funded by Cancer Research UK (grant C157/A15703).
References
- 1.Byron A., Humphries J.D., Humphries M.J. Defining the extracellular matrix using proteomics. Int J Exp Pathol. 2013;94:75–92. doi: 10.1111/iep.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hynes R.O. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charras G., Sahai E. Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- 4.Engler A.J., Humbert P.O., Wehrle-Haller B., Weaver V.M. Multiscale modeling of form and function. Science. 2009;324:208–212. doi: 10.1126/science.1170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphries J.D., Byron A., Humphries M.J. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byron A., Morgan M.R., Humphries M.J. Adhesion signalling complexes. Curr Biol. 2010;20:R1063–R1067. doi: 10.1016/j.cub.2010.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger B., Yamada K.M. Molecular architecture and function of matrix adhesions. Cold Spring Harb Perspect Biol. 2011;3:a005033. doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaidel-Bar R., Itzkovitz S., Ma’ayan A., Iyengar R., Geiger B. Functional atlas of the integrin adhesome. Nat Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Winograd-Katz S.E., Fässler R., Geiger B., Legate K.R. The integrin adhesome: from genes and proteins to human disease. Nat Rev Mol Cell Biol. 2014;15:273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]; This analysis of the literature and online databases synthesised current understanding of the roles of adhesion molecules, and their mutations, in disease. It also provided an updated list of literature-curated adhesome components.
- 10.Byron A., Humphries J.D., Bass M.D., Knight D., Humphries M.J. Proteomic analysis of integrin adhesion complexes. Sci Signal. 2011;4:pt2. doi: 10.1126/scisignal.2001827. [DOI] [PubMed] [Google Scholar]
- 11.Kuo J.C., Han X., Yates J.R., 3rd, Waterman C.M. Isolation of focal adhesion proteins for biochemical and proteomic analysis. Methods Mol Biol. 2012;757:297–323. doi: 10.1007/978-1-61779-166-6_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gieger T., Zaidel-Bar R. Opening the floodgates: proteomics and the integrin adhesome. Curr Opin Cell Biol. 2012;24:562–568. doi: 10.1016/j.ceb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Jones M.C., Humphries J.D., Byron A., Millon-Frémillon A., Robertson J., Paul N.R., Ng D.H., Askari J.A., Humphries M.J. Isolation of integrin-based adhesion complexes. Curr Protoc Cell Biol. 2015;66:9.8.1–9.8.15. doi: 10.1002/0471143030.cb0908s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson J., Humphries J.D., Paul N.R., Warwood S., Knight D., Byron A., Humphries M.J. Characterisation of the phospho-adhesome by mass spectrometry-based proteomics. Methods Mol Biol. 2016 doi: 10.1007/978-1-4939-7154-1_15. (in press) [DOI] [PubMed] [Google Scholar]
- 15••.Horton E.R., Byron A., Askari J.A., Ng D.H., Millon-Frémillon A., Robertson J., Koper E.J., Paul N.R., Warwood S., Knight D. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol. 2015;17:1577–1587. doi: 10.1038/ncb3257. [DOI] [PMC free article] [PubMed] [Google Scholar]; Computational integration of multiple adhesion proteomes generated a meta-adhesome that provided insights into the functional diversity of adhesome components and established a consensus adhesome of core adhesion machinery. Further, the study characterised temporal adhesome dynamics during adhesion assembly and disassembly.
- 16.Humphries J.D., Byron A., Bass M.D., Craig S.E., Pinney J.W., Knight D., Humphries M.J. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal. 2009;2:ra51. doi: 10.1126/scisignal.2000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiller H.B., Friedel C.C., Boulegue C., Fässler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuo J.C., Han X., Hsiao C.T., Yates J.R., 3rd, Waterman C.M. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byron A., Humphries J.D., Craig S.E., Knight D., Humphries M.J. Proteomic analysis of αrot integrin adhesion complexes reveals α-subunit-dependent protein recruitment. Proteomics. 2012;12:2107–2114. doi: 10.1002/pmic.201100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiller H.B., Hermann M.R., Polleux J., Vignaud T., Zanivan S., Friedel C.C., Sun Z., Raducanu A., Gottschalk K.E., Théry M. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat Cell Biol. 2013;15:625–636. doi: 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- 21•.Bachir A.I., Zareno J., Moissoglu K., Plow E.F., Gratton E., Horwitz A.R. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr Biol. 2014;24:1845–1853. doi: 10.1016/j.cub.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used fluorescence imaging to determine distinct stoichiometries and dynamics of several adhesion proteins during early adhesion assembly and maintenance.
- 22•.Hoffmann J.E., Fermin Y., Stricker R.L., Ickstadt K., Zamir E. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. Elife. 2014;3:e02257. doi: 10.7554/eLife.02257. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using fluorescence imaging techniques, the authors showed that a number of adhesion proteins form complexes in the cytosol. These pre-formed ‘building blocks’ could enable rapid, accurate and modular assembly of adhesion sites.
- 23.Yao M., Goult B.T., Chen H., Cong P., Sheetz M.P., Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagarrigue F., Vikas Anekal P., Lee H.S., Bachir A.I., Ablack J.N., Horwitz A.F., Ginsberg M.H. A RIAM/lamellipodin-talin-integrin complex forms the tip of sticky fingers that guide cell migration. Nat Commun. 2015;6:8492. doi: 10.1038/ncomms9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y.L., Lu S., Szeto K.W., Sun J., Wang Y., Lasheras J.C., Chien S. FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci Rep. 2014;4:6024. doi: 10.1038/srep06024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidel-Bar R., Ballestrem C., Kam Z., Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 27.Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., Horwitz A.R. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Changede R., Xu X., Margadant F., Sheetz M.P. Nascent integrin adhesions form on all matrix rigidities after integrin activation. Dev Cell. 2015;35:614–621. doi: 10.1016/j.devcel.2015.11.001. [DOI] [PubMed] [Google Scholar]; This work used super-resolution microscopy to reveal the consistency of size and density of very early adhesions under a range of conditions, including integrin ligand density and mobility.
- 29.Iskratsch T., Yu C.H., Mathur A., Liu S., Stévenin V., Dwyer J., Hone J., Ehler E., Sheetz M. FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev Cell. 2013;27:545–559. doi: 10.1016/j.devcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolat L., Hunyara J.L., Bowen J.R., Karasmanis E.P., Elgawly M., Galkin V.E., Spiliotis E.T. Septins promote stress fiber-mediated maturation of focal adhesions and renal epithelial motility. J Cell Biol. 2014;207:225–235. doi: 10.1083/jcb.201405050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kai F., Fawcett J.P., Duncan R. Synaptopodin-2 induces assembly of peripheral actin bundles and immature focal adhesions to promote lamellipodia formation and prostate cancer cell migration. Oncotarget. 2015;6:11162–11174. doi: 10.18632/oncotarget.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Stehbens S.J., Paszek M., Pemble H., Ettinger A., Gierke S., Wittmann T. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat Cell Biol. 2014;16:561–573. doi: 10.1038/ncb2975. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using live-cell imaging, this investigation showed that microtubule-associated CLASP proteins tether microtubules to adhesion sites. This promotes the disassembly of mature adhesions, a process that is proposed to be linked to local exocytosis, ECM degradation and release of cell-matrix connections.
- 33.Yue J., Xie M., Gou X., Lee P., Schneider M.D., Wu X. Microtubules regulate focal adhesion dynamics through MAP4K4. Dev Cell. 2014;31:572–585. doi: 10.1016/j.devcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng D.H., Humphries J.D., Byron A., Millon-Frémillon A., Humphries M.J. Microtubule-dependent modulation of adhesion complex composition. PLOS ONE. 2014;9:e115213. doi: 10.1371/journal.pone.0115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrail D.J., Khambhati N.N., Qi M.X., Patel K.S., Ravikumar N., Brandenburg C.P., Dawson M.R. Alterations in ovarian cancer cell adhesion drive taxol resistance by increasing microtubule dynamics in a FAK-dependent manner. Sci Rep. 2015;5:9529. doi: 10.1038/srep09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villari G., Jayo A., Zanet J., Fitch B., Serrels B., Frame M., Stramer B.M., Goult B.T., Parsons M. A direct interaction between fascin and microtubules contributes to adhesion dynamics and cell migration. J Cell Sci. 2015;128:4601–4614. doi: 10.1242/jcs.175760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Byron A., Askari J.A., Humphries J.D., Jacquemet G., Koper E.J., Warwood S., Choi C.K., Stroud M.J., Chen C.S., Knight D., Humphries M.J. A proteomic approach reveals integrin activation state-dependent control of microtubule cortical targeting. Nat Commun. 2015;6:6135. doi: 10.1038/ncomms7135. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mass spectrometric analyses of adhesion complexes induced by conformation-specific anti-integrin monoclonal antibodies identified preferential recruitment of microtubule-associated proteins to active integrin. Cell biological analyses showed that active integrin stabilises microtubules at the cell periphery. The dataset also provides a resource of activation state-dependent integrin-associated proteins and reveals their striking functional diversity.
- 38.Carisey A., Tsang R., Greiner A.M., Nijenhuis N., Heath N., Nazgiewicz A., Kemkemer R., Derby B., Spatz J., Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Hernández-Varas P., Berge U., Lock J.G., Strömblad S. A plastic relationship between vinculin-mediated tension and adhesion complex area defines adhesion size and lifetime. Nat Commun. 2015;6:7524. doi: 10.1038/ncomms8524. [DOI] [PMC free article] [PubMed] [Google Scholar]; A systems microscopy approach was used here to characterise adhesion site size and stability in the context of vinculin-based tension. Probabilistic mapping and modelling predicted the balance between microtubule-mediated and mechanotransduction-mediated mechanisms of adhesion complex regulation.
- 40.Austen K., Ringer P., Mehlich A., Chrostek-Grashoff A., Kluger C., Klingner C., Sabass B., Zent R., Rief M., Grashoff C. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat Cell Biol. 2015;17:1597–1606. doi: 10.1038/ncb3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atherton P., Stutchbury B., Wang D.Y., Jethwa D., Tsang R., Meiler-Rodriguez E., Wang P., Bate N., Zent R., Barsukov I.L. Vinculin controls talin engagement with the actomyosin machinery. Nat Commun. 2015;6:10038. doi: 10.1038/ncomms10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong V.W., Rustad K.C., Akaishi S., Sorkin M., Glotzbach J.P., Januszyk M., Nelson E.R., Levi K., Paterno J., Vial I.N. Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat Med. 2011;18:148–152. doi: 10.1038/nm.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae Y.H., Mui K.L., Hsu B.Y., Liu S.L., Cretu A., Razinia Z., Xu T., Puré E., Assoian R.K. A FAK-Cas-Rac-lamellipodin signaling module transduces extracellular matrix stiffness into mechanosensitive cell cycling. Sci Signal. 2014;7:ra57. doi: 10.1126/scisignal.2004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J., Aponte-Santamaría C., Sturm S., Bullerjahn J.T., Bronowska A., Gräter F. Mechanism of focal adhesion kinase mechanosensing. PLoS Comput Biol. 2015;11:e1004593. doi: 10.1371/journal.pcbi.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton E.R., Astudillo P., Humphries M.J., Humphries J.D. Mechanosensitivity of integrin adhesion complexes: role of the consensus adhesome. Exp Cell Res. 2016 doi: 10.1016/j.yexcr.2015.10.025. (in press) [DOI] [PubMed] [Google Scholar]
- 46.Hytönen V.P., Wehrle-Haller B. Mechanosensing in cell-matrix adhesions – converting tension into chemical signals. Exp Cell Res. 2016 doi: 10.1016/j.yexcr.2015.10.027. (in press) [DOI] [PubMed] [Google Scholar]
- 47.Chicurel M.E., Singer R.H., Meyer C.J., Ingber D.E. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- 48•.Robertson J., Jacquemet G., Byron A., Jones M.C., Warwood S., Selley J.N., Knight D., Humphries J.D., Humphries M.J. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat Commun. 2015;6:6265. doi: 10.1038/ncomms7265. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported the first phosphoproteomic characterisation of adhesion complexes, identifying recruitment of both constitutively phosphorylated adhesion proteins and those that undergo adhesion-induced phosphorylation.
- 49.LaFlamme S.E., Nieves B., Colello D., Reverte C.G. Integrins as regulators of the mitotic machinery. Curr Opin Cell Biol. 2008;20:576–582. doi: 10.1016/j.ceb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byron A., Humphries J.D., Humphries M.J. Alternative cellular roles for proteins identified using proteomics. J Proteomics. 2012;75:4184–4185. doi: 10.1016/j.jprot.2012.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel H., Zich J., Serrels B., Rickman C., Hardwick K.G., Frame M.C., Brunton V.G. Kindlin-1 regulates mitotic spindle formation by interacting with integrins and Plk-1. Nat Commun. 2013;4:2056. doi: 10.1038/ncomms3056. [DOI] [PubMed] [Google Scholar]
- 52•.Petridou N.I., Skourides P.A. FAK transduces extracellular forces that orient the mitotic spindle and control tissue morphogenesis. Nat Commun. 2014;5:5240. doi: 10.1038/ncomms6240. [DOI] [PubMed] [Google Scholar]; This work showed that FAK, but not its kinase activity, is required for spindle orientation, which is linked to the role of FAK in mechanotransduction.
- 53.Golubovskaya V.M., Finch R., Cance W.G. Direct interaction of the N-terminal domain of focal adhesion kinase with the N-terminal transactivation domain of p53. J Biol Chem. 2005;280:25008–25021. doi: 10.1074/jbc.M414172200. [DOI] [PubMed] [Google Scholar]
- 54.Lim S.T., Chen X.L., Lim Y., Hanson D.A., Vo T.T., Howerton K., Larocque N., Fisher S.J., Schlaepfer D.D., Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim S.T., Miller N.L., Chen X.L., Tancioni I., Walsh C.T., Lawson C., Uryu S., Weis S.M., Cheresh D.A., Schlaepfer D.D. Nuclear-localized focal adhesion kinase regulates inflammatory VCAM-1 expression. J Cell Biol. 2012;197:907–919. doi: 10.1083/jcb.201109067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo S.W., Zhang C., Zhang B., Kim C.H., Qiu Y.Z., Du Q.S., Mei L., Xiong W.C. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. EMBO J. 2009;28:2568–2582. doi: 10.1038/emboj.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Serrels A., Lund T., Serrels B., Byron A., McPherson R.C., von Kriegsheim A., Gómez-Cuadrado L., Canel M., Muir M., Ring J.E. Nuclear FAK controls chemokine transcription Tregs, and evasion of anti-tumor immunity. Cell. 2015;163:160–173. doi: 10.1016/j.cell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study found that, in SCC, nuclear FAK stimulates the expression of chemokine and cytokine genes via its association with a network of transcriptional regulators. This leads to recruitment of regulatory T cells and tumour immune evasion, which can be counteracted by a small-molecule inhibitor of FAK, resulting in tumour regression.
- 58.Lobo M., Zachary I. Nuclear localization and apoptotic regulation of an amino-terminal domain focal adhesion kinase fragment in endothelial cells. Biochem Biophys Res Commun. 2000;276:1068–1074. doi: 10.1006/bbrc.2000.3547. [DOI] [PubMed] [Google Scholar]
- 59•.Brami-Cherrier K., Gervasi N., Arsenieva D., Walkiewicz K., Boutterin M.C., Ortega A., Leonard P.G., Seantier B., Gasmi L., Bouceba T. FAK dimerization controls its kinase-dependent functions at focal adhesions. EMBO J. 2014;33:356–370. doi: 10.1002/embj.201386399. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ultrastructural and biochemical analyses of full-length FAK detailed the role of ligand-induced FAK dimerisation, through interactions of its FAT, FERM and kinase domains, in kinase-dependent functions of FAK.
- 60.Nix D.A., Beckerle M.C. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Honda K., Yamada T., Endo R., Ino Y., Gotoh M., Tsuda H., Yamada Y., Chiba H., Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol. 1998;140:1383–1393. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woods A.J., Roberts M.S., Choudhary J., Barry S.T., Mazaki Y., Sabe H., Morley S.J., Critchley D.R., Norman J.C. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J Biol Chem. 2002;277:6428–6437. doi: 10.1074/jbc.M109446200. [DOI] [PubMed] [Google Scholar]
- 63.Hervy M., Hoffman L., Beckerle M.C. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 64.Kadrmas J.L., Beckerle M.C. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 65.McCrea P.D., Gottardi C.J. Beyond β-catenin: prospects for a larger catenin network in the nucleus. Nat Rev Mol Cell Biol. 2016;17:55–64. doi: 10.1038/nrm.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]