Abstract
In the past decade, thousands of long noncoding RNAs (lncRNAs) have been identified, and emerging data indicate that lncRNAs can have important biological functions and roles in human diseases including cancer. Many lncRNAs appear to be expressed specifically in the brain, and the roles of lncRNAs in neural stem cells (NSCs) and brain development are now beginning to be discovered. Here we review recent advances in understanding the diversity of lncRNA structure and functions in NSCs and brain development. NSCs in the adult mouse ventricular-subventricular zone (V-SVZ) generate new neurons throughout life, and we discuss how key elements of this adult neurogenic system have facilitated the discovery and functional characterization of known and novel lncRNAs. A review of lncRNAs described in other NSC systems reveals a variety of molecular mechanisms, including binding and recruitment of transcription factors, epigenetic modifiers, and RNA-splicing factors. Finally, we review emerging evidence indicating that specific lncRNAs can be key players driving glial malignancies, and discuss next steps towards an in vivo understanding of lncRNA function in development and disease.
Keywords: long noncoding RNA, lncRNA, chromatin, epigenetics, neural stem cell, neurogenesis, brain development, brain tumor, glioma
Introduction
The human genome transcribes many thousands of lncRNAs – transcripts >200 nucleotides long with no evidence of protein coding potential, and it is now clear that lncRNAs can have critical biological functions and roles in human disease [1–4]. In particular, aberrant lncRNA expression may underlie some of the most devastating human neurological disorders including Alzheimer’s disease [5], schizophrenia [6, 7], developmental delay and autism [8, 9]. However, very few lncRNAs have been characterized in terms of function. This knowledge gap is a critical barrier to progress in this emerging field. New detailed studies of individual lncRNAs are vital to understand how this class of noncoding transcript can contribute to disease and/or serve as therapeutic targets [10].
Before high-throughput sequencing technologies, very few lncRNAs were known, and many fundamental discoveries were made by their individual study. For instance, the lncRNA Xist was discovered in the early 1990’s, and an elegant combination of in vivo and in vitro studies demonstrated its critical role in X-chromosome inactivation [4, 11] and new insights into its mechanism of action [12–14]. In the past decade, many more lncRNAs have been identified. There are over 50,000 human lncRNAs [15–17], and similar catalogs have been generated from various mouse tissues and model organisms [18–22], leading to novel insights into their genomic structure and patterns of expression. While there has been a remarkable surge in their bioinformatic characterization, there is comparatively little data regarding the developmental and molecular function of individual lncRNAs, especially in terms of neural development. For the field of lncRNA biology, the challenge now is to (1) select specific lncRNAs for further study, (2) determine their roles in vivo, and (3) understand their mechanisms of action at the molecular level.
Annotation via the GENCODE consortium suggests that 40% of differentially expressed lncRNAs are specific to the brain [17]. Even within the brain, lncRNA expression patterns are noted to be particularly region-specific [22, 23] and dynamically expressed during neural differentiation in vitro as well as NSC populations in vivo [22, 24, 25]. Here, we describe the use of the V-SVZ to characterize lncRNAs with roles in neurogenesis, including the lncRNA Pnky, which plays a unique role in adult and embryonic NSC lineages. We also review the lncRNAs that have been characterized in other NSC populations and their putative mechanisms of action. Finally, we discuss the potential roles for lncRNAs in the transformation and propagation of central nervous system tumors, and suggest therapeutic strategies informed by careful study of lncRNA biology.
The V-SVZ: a tractable system for molecular-genetic studies of NSC differentiation
In the developing brain, most NSCs are spatially and temporally ephemeral in nature, continually changing their location and developmental potential as the brain grows in term of both size and anatomic complexity. In contrast, NSCs in the adult neurogenic germinal zones are harbored in relatively stable cellular niches and generate a specific subset of neurons for the life of the animal. Furthermore, because adult neurogenesis in the mouse can be studied over longer time periods (e.g., months), it is possible that subtle phenotypes observed in the short-term studies of embryonic neurogenesis might be much more prominent in systems of adult neurogenesis. Thus, the reduced developmental complexity and increased duration of adult neurogenesis provides certain experimental advantages for the study of fundamentally new biological mechanisms.
In the adult mouse brain, several thousand NSCs are located in the V-SVZ of the lateral ventricle [26, 27]. (Fig. 1A, B). Throughout adult life, V-SVZ NSCs (B1 cells) give rise to transit-amplifying cells (C cells), which generate neuroblasts (A cells) that migrate to the olfactory bulb (OB) [28–32] where they differentiate into interneurons (Fig. 1C). Like radial glia – the NSC population of the embryonic brain – B1 cells have many glial cell characteristics [28, 33, 34], including expression of the glial-fibrillary acidic protein (GFAP), glutamate aspartate transporter (GLAST), and brain lipid binding protein (BLBP). In the embryo, radial glia have distinct regional identities, generating different populations of neurons related to the “geographic” location of NSC population in the developing brain. Similarly, B1 cells located in geographically distinct regions of the ventricle wall generate specific interneuron subtypes, and these differences in NSC regional identity appear to be cell-intrinsic [35, 36]. Throughout development, some radial glia generate astrocytes and oligodendrocytes. Indeed, B1 cells are also capable of generating astrocytes and oligodendrocytes [37–39]. Thus, while providing the advantage of generating neurons for long periods of time, B1 cells of the V-SVZ exhibit many characteristics fundamentally important to NSCs in the developing brain.
Figure 1. The V-SVZ lineage.
(A) Coronal view of V-SVZ. (B) Enlarged view of V-SVZ: Type B cells (blue) can contact the ventricle with a thin process extended between ependymal cells (white); type C cells (green) are transit-amplifying cells that give rise to migratory type A cells (red). (C) Sagittal view showing paths of migratory neuroblasts (red, type A cells) to the OB. (D) Schematic of NSC lineages. (E) V-SVZ NSC culture in self-renewal (left) and differentiation (right) conditions. NSCs are GFAP+ (stained in green). Neuroblasts stain for marker Tuj1 (red).
In vivo, B1 cells can exist in a quiescent or activated state [40, 41]. Activated B1 cells express the epidermal growth factor receptor (EGFR) and GFAP. These activated NSCs undergo asymmetric division for self-renewal and the production of C cells (EGFR+,GFAP−), also called transit-amplifying progenitors (TAPS) [42, 43]. C cells express proneural transcription factor Ascl1 and neurogenic Dlx2 and divide symmetrically approximately three times before becoming migratory A cells (Fig. 1D) [27]. A cells travel along the ventricle wall within cellular aggregates called chains that coalesce into the rostral migratory stream (RMS) [44]. DLX2+ migratory neuroblasts are distinguished from TAPs by their expression of doublecortin (DCX) and polysialylated neural cell adhesion molecule (PSA-NCAM) and CD24 [33, 45]. Upon reaching the olfactory bulb, these neuroblasts migrate out radially from the RMS and differentiate into functional interneurons.
V-SVZ NSCs can be efficiently cultured for molecular and biochemical studies, and when grown as monolayers, these cells recapitulate neurogenesis in vitro (Fig. 1E) [46, 47]. In self-renewal conditions, nearly all cells express NSC marker Sox2, and the majority (60–70%) co-stain for additional NSC markers Nestin and GFAP [48]. Upon differentiation, these cultures generate large numbers of neuroblasts (Tuj1+, DCX+, ~40% of cells), oligodendrocytes (Olig2+, O4+, ~15%), and other astrocytes (GFAP+, Nestin-, ~40%) [22, 46, 47]. Importantly, these cultured NSCs transplanted back to the V-SVZ generate neurons for the OB [36], suggesting that these cultured NSCs retain many key in vivo regulatory mechanisms despite their propagation in vitro.
Over the past two decades, by integrating in vivo and in vitro V-SVZ studies, many different laboratories have been able to elucidate key developmental principles regarding the role of signaling molecules, transcription factors, microRNAs, and chromatin modifiers [49]. Such discoveries have often provided unique insights into mechanistic themes relevant to both embryonic and postnatal brain development [35]. Below, we describe how the V-SVZ has been utilized as a model system in which to make new strides into the emerging field of lncRNA biology.
Long Noncoding RNAs in V-SVZ neurogenesis
While several studies of neural lncRNAs had been performed in ESC-derived NPCs or cultured cell lines [24, 50–54], comparatively less is known about the expression and function of lncRNAs in neural lineages in vivo. As noted above, the V-SVZ harbors NSCs with a relatively well-understood daughter cell lineage that generates new neurons for the OB. To generate a reference transcriptome of lncRNAs expressed in V-SVZ neurogenesis, RNA-seq was performed on the V-SVZ and OB from the adult mouse brain [22]. NSCs are also located in the subgranular zone of the dentate gyrus (DG) [55], and DG was also included in the RNA-seq analysis. This sequencing data of the two major adult neurogenic brain regions was combined with data from ESCs and ESC-derived NSCs to generate a catalog of 8992 lncRNAs. Interestingly, 2108 lncRNA transcripts were uniquely recovered from sequencing data of the adult neurogenic brain regions, suggesting that some lncRNAs underlie the long-term self-renewal and neurogenic capacity of the V-SVZ and DG germinal zones.
To confirm the expression and genomic structure of novel lncRNAs RNA Capture-Seq [56] was used to capture novel transcripts and re-sequence them at increased depth. These efforts both confirmed the expression and structure of novel transcripts and revealed that neural lncRNAs can have complex alternative splice variants. For instance, sequences corresponding to an entire coding gene could be identified within the excised “intron” of some lncRNAs. Conversely, some lncRNAs are found within the intron of specific mRNAs. Future studies may provide evolutionary insights into how such complex genomic structures can arise, and whether these genomic arrangements relate to lncRNA function.
After comprehensive annotation of lncRNAs expressed in the tissue or system of interest, an important next step is to identify candidates for functional studies. One method of approaching this issue is to first filter the large set of lncRNAs for those that are differentially expressed among the cells in the lineage under study. While it is certainly possible that lncRNAs expressed ubiquitously have important developmental functions, differential gene expression has generally been a useful first step in the characterization of coding genes. V-SVZ NSCs and their neurogenic daughter cells can be isolated with immunocytochemistry and fluorescent-activated cell sorting (FACS) [45]. To identify which lncRNAs are differentially expressed in the adult V-SVZ neurogenic lineage, a FACS protocol [45] was employed to prospectively isolate EGFR+GFAP+ activated stem cells, EGFR+GFAP-TAPs, and CD24+ young neuroblasts for lncRNA expression analysis. Using the same custom microarrays, the dynamic expression of lncRNAs during neurogenesis in vitro was also determined and correlated with the in vivo lineage. For instance, lncRNA Dlx1as is increased in the TAPs in vivo and exhibit increased expression after 2 days of differentiation in vitro, when the TAP population is rapidly expanding.
The methylation of specific histone lysine residues correlates with transcriptional activity [57] and several analyses of chromatin state maps and transcript expression indicate that histone modifications correlate with lncRNA expression in a manner similar to that of protein-coding genes [21, 22, 24]. Trimethylation of histone 3 lysine 4 (H3K4me3) at promoter regions correlates with active transcription, whereas trimethylation of histone 3 lysine 27 (H3K27me3) corresponds to repressed loci. Loci enriched for both H3K4me3 and H3K27me3 have been termed “bivalent” and are transcriptionally repressed but “poised” for activation [58]. In ESCs, bivalent coding genes are enriched for key developmental regulators. Given that lncRNAs appear to share transcriptional regulatory mechanisms and strategies with mRNAs, those lncRNAs that are bivalent in ESC and/or NSC populations may also be enriched for those that regulate development. Thus, chromatin-state maps of H3K4me3 and H3K27me3 were generated from V-SVZ NSCs, and these data were merged with similar datasets from mouse ESCs, ESC-derived NPCs, and non-neurogenic fibroblasts. Together, with the in vivo expression analysis of FACS-isolated V-SVZ cells, the in vitro analysis of lncRNA expression dynamics during neural differentiation, these chromatin state maps were integrated into an online resource (http://neurosurgery.ucsf.edu/danlimlab/lncRNA/) that allows for browsing and filtering based on chromatin and expression criteria to generate gene signatures for different stages of neurogenesis in the V-SVZ lineage (Figure 2). This resource was used to identify and begin characterization of several lncRNA transcripts, including Six3os, Dlx1as, and Pnky, discussed in sections below.
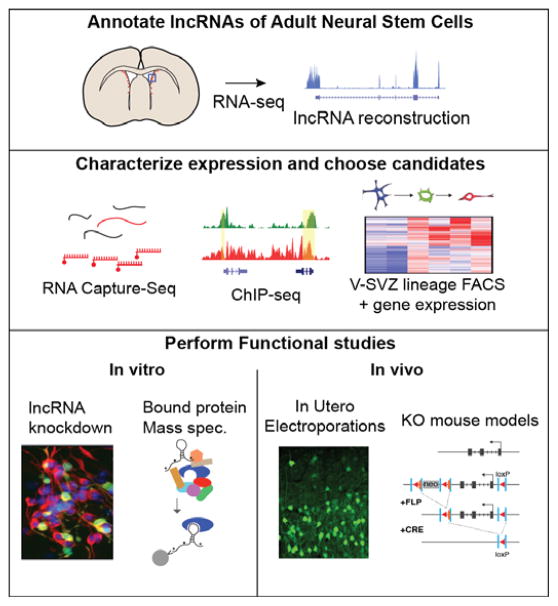
Figure 2. Workflow for identification and characterization of lncRNAs active in neural development.
First, unbiased RNA-seq is performed and long noncoding RNA transcripts are reconstructed from these reads. To better characterize gene structure and expression levels, RNA Capture-Seq is performed. LncRNAs (shown here in red) are captured and enriched for further sequencing. ChIP-seq is used to characterize chromatin marks across embryonic stem cells, neural stem cells, and non-neural cell types. Finally, expression profiling is performed using FACS-isolated cells of the V-SVZ lineage. Together these data are integrated to choose candidates for further study. Functional assays performed in vitro include shRNA-mediated knockdown in NSC cultures. To determine lncRNA protein-binding partners, RNA pulldown assays are performed in which biotinylated RNA is mixed with cell extract, followed by pulldown and characterization of bound proteins by mass spectrometry. In vivo models for probing lncRNA function include in utero electroporation of embryonic NSCs, and generation of knockout mouse models.
Integration of genome-wide datasets led to the discovery of Pnky, a 825 bp lncRNA divergently transcribed 2.5 kb upstream of the Pou3f2 locus [48]. Pnky is bivalent in ESCs, and this locus becomes monovalent for H3K4me3 in both ESC-NPCs and V-SVZ NSCs, suggesting its importance as a regulator of neural lineage activation. Pnky is only detected in neural tissues, and this transcript is enriched in the nucleus. Analysis of FACS-purified cells indicates that Pnky is most highly expressed in V-SVZ NSCs and becomes downregulated upon differentiation into neuroblasts.
Short-hairpin RNA (shRNA) mediated knockdown of Pnky in postnatal V-NSC cultures increases neurogenesis by 3–4 fold [48]. Time-lapse imaging of individual Pnky-depleted V-SVZ NSCs demonstrated that the potentiation of neurogenesis relates to: (1) an increase in neuronal lineage commitment, (2) an increase in the number of transit-amplifying divisions, and (3) a decrease in death of neuronal progenitors (Figure 3). This phenotype suggests a developmental function for Pnky that is distinct from that of other lncRNAs with known neurodevelopmental function. For instance, as discussed in sections below, knockdown of lncRNAs TUNA, RMST, Six3OS, and Dlx1as all strongly inhibits neurogenesis [22, 50, 54, 59], suggesting that these lncRNAs are required to positively regulate neuronal differentiation. Similarly, genetic deletion of linc-Brn1b results in decreased proliferation of embryonic cortical intermediate progenitors and decreased neurogenesis [60]. lncRNA Evf2 is required for proper expression of Dlx5/6, which encodes key neurogenic transcription factors, and loss of Evf2 results in defective interneuron production [61–63]. Thus, unlike the aforementioned lncRNAs that appear to potentiate neurogenesis, Pnky appears to “restrain” neurogenesis from NSCs, perhaps serving to control their long-term self-renewal and rate of neuronal production.
Figure 3. Knockdown of the lncRNA Pnky enhanced neurogenesis from V-SVZ NSCs.
Left: Normal lineage progression of neuronal production from wild-type V-SVZ NSCs (blue) to transit amplifying cells (TA, green) to neuroblasts (NB, red). Pnky is expressed highest in NSCs and becomes down-regulated during lineage progression. Right: Pnky knockdown promotes neuronal production through two mechanisms: 1) a greater proportion of NSCs commit to the neurogenic lineage, and 2) TA cells undergo more cell divisions, resulting in a greater total number of cell divisions and an increased number of “generations” per initial progenitor.
Although the mechanisms of action for most lncRNAs is not known, an emerging theme is that lncRNAs interact with cellular proteins to form functional ribonucleoprotein complexes. Mass spectrometry analysis of proteins enriched by biotinylated Pnky transcripts led to the identification of Polypyrimidine tract-binding protein 1 (PTBP1) as a protein that associates with Pnky transcripts in the nucleus of V-SVZ NSCs cells. PTBP1 regulates mRNA transcript levels and pre-mRNA splicing during neuronal differentiation [51, 64, 65]. In V-SVZ NSCs, Pnky and PTBP1 regulate an overlapping set of transcripts as well as alternative splicing isoforms, and epistasis experiments with Pnky/PTBP1 double knockdown cells suggest they work in the same genetic pathway to regulate neurogenesis from V-SVZ-NSCs. These studies highlight the interaction between a lncRNA and RNA-binding protein can regulate key aspect of NSC biology. Below, we discuss lncRNAs that have been identified in other NSC populations, including neuroblastoma cell lines, ESC-derived NSCs, and mouse developing cortex (Table 1).
Table 1.
Summary of lncRNAs active in neural development
| lncRNA | Tissue/Cell Line | Effect of knockdown/deletion | Protein binding partner | Refs |
|---|---|---|---|---|
| Cyrano | Zebrafish embryonic brain and notocord | Microcephaly, tail curling, neural tube defects, loss of NeuroD-positive neurons in zebrafish embryos treated with morpholino | [20] | |
| Dali | N2A mouse neuroblastoma cell line | Decreased neurite growth and size during differentiation | POU3F3 DNMT1 |
[53] |
| Dlx1as | V-SVZ transit-amplifying cells and neuroblasts Embryonic interneuron progenitors |
Decreased neurogenesis from VSVZ-NSCs. Increased DLX1 expression in adult mouse Dlx1as-depleted brains. |
[22, 71] | |
| Evf2 | Embryonic interneuron progenitors | Decreased number of GABAergic interneurons in early postnatal hippocampus of Evf-2 depleted brains | DLX1 DLX2 BRG1 MECP2 |
[61–63, 72] |
| Linc-Brn1b | Cortical progenitors | Reduction of intermediate progenitors in the developing cortex. Reduced cortical layer II-IV thickness and barrel cortex disruption in knockout mice. |
[60, 78] | |
| Megamind/TUNA | mESCs and neural progenitors H9 human ESCs Mouse and zebrafish embryonic brain |
Reduction of neural commitment of ESCs and neuronal differentiation Hydrocephalus and loss of NeuroD-positive neurons in zebrafish embryos treated with morpholino |
PTBP1 HNRNP-K NCL |
[20, 54] |
| Paupar | N2A mouse neuroblastoma cell line | Cell cycle arrest and increased neurite outgrowth upon differentiation. | PAX6 | [52] |
| Pinky | V-SVZ NSCs Cortical progenitors |
Increased production of neurons from V-SVZ NSCs and expansion of TA population. Increased production of neuroblasts and depletion of embryonic stem cell population. |
PTBP1 | [48] |
| RMST | H9-derived human neural stem cells | Decreased neuronal differentiation | SOX2 HNRNPA2/B1 |
[50] |
| Six3os | V-SVZ NSCs Retinal progenitor cells |
Decreased generation of neurons and oligodendrocytes from V-SVZ-NSCS. Decreased production of rod bipolar cells and increase in production of Muller glia from retinal progenitors. |
EZH2 EYA1 EYA3 EYA4 |
[22, 59] |
LncRNAs associated with homeobox transcription factors
Many high-throughput sequencing efforts and ISH analyses have identified a set of homeodomain associated opposite strand transcripts (HOSTs) [66]. These transcripts can show either reciprocal or coordinated expression with their protein-coding neighbor in vivo. Some homeobox transcription factors such as Six3, Pax6, the Dlx gene family, and the Pou family of transcription factors are critical in neural development. One such lncRNA, Six3os, can modulate SIX3 function in the developing retina, and physically interacts with chromatin-modifier EZH2 as well as the EYA family of transcriptional coactivators [59]. In V-SVZ NSCs, Six3os is specifically enriched in V-SVZ stem cells and is down-regulated upon differentiation; depletion of Six3os results in a decrease in production of both neuronal and oligodendroglial lineages [22].
DLX2 is a homeobox transcription factor that is required for interneuron development in both the OB [67] and forebrain [68], and its proper expression is critical for postnatal neurogenesis in the V-SVZ [46]. Dlx1 and Dlx2 are oriented in an inverted configuration separated by a 8.3 kb intergenic region with several ultraconserved elements that contain known binding sites for pro-neurogenic transcription factors [69]. The lncRNA Dlx1as is also transcribed from this intergenic region. The transcriptional start site overlaps an ultraconserved enhancer region, while its 3′ end partially overlaps Dlx1 on the opposite strand. This transcript has been described in embryonic development [70], where it demonstrates increased expression in more mature cell types relative to the expression of Dlx1/2. In adult neurogenesis, ISH analysis of brain sections and custom microarray analysis of FACS-isolated V-SVZ cell populations demonstrates that Dlx1as is preferentially expressed in the TAPs and migratory neuroblasts of the RMS and throughout the OB of adult mice. In contrast, Dlx1 and Dlx2 are strongly expressed throughout the V-SVZ, RMS, and OB [22, 23]. Dlx1as knockdown in V-SVZ neural progenitors results in decreased neurogenesis without an apparent loss of oligodendroglial differentiation, in contrast to the results observed with Six3os, whose knockdown affected both neuronal and glial lineages. These data demonstrate that lncRNA depletion from a stem cell population can selectively disrupt the neuronal lineage [22].
A Dlx1as-null mouse has been generated by inserting a poly-A cassette in the first intron of Dlx1as [71]. Importantly, this mouse does not efficiently produce full-length Dlx1as, however transcription can still initiate at the TSS. Dlx1as mutant mice are born at expected ratios, survive to adulthood, and are grossly normal. Dlx1as null brains have a slight increase in Dlx1 as measured by ISH, however there is no obvious increase in interneuron generation. The mild phenotype found in the Dlx1as could be caused by several factors: (1) the premature-polyA strategy did not produce a strong enough depletion of the lncRNA, (2) the act of transcription at the Dlx1as promoter is sufficient for its partial function, (3) this straight knockout of Dlx1as could be developmentally compensated by redundant mechanisms. It is, of course, entirely possible that lncRNA Dlx1as is not required for proper neurogenesis or development in vivo, but additional and/or alternative experimental manipulations of this locus may be required to reveal its potential function.
Similar to the Dlx1/2 locus, the Dlx5/6 locus encodes a lncRNA, Evf2, from an ultraconserved intergenic enhancer region. Initial studies in cell lines suggested that Evf2 forms a complex with Dlx2 and enhances activation of the Dlx5/6 intergenic enhancer [63], consistent with models proposed for Paupar and Six3os. An Evf-2 knockout mouse was generated using a premature poly-A signal strategy [62]. Evf-2-null mice have reduced numbers of interneurons in the embryonic hippocampus, however numbers return to normal in the adult. Nevertheless, electrophysiological recordings of the adult hippocampus revealed loss of synaptic inhibition, demonstrating that an early lack of interneurons cause aberrant circuitry in the adult brain.
In contrast to what would be expected based on in vitro results, in vivo deletion of Evf-2 led to an increase of Dlx5 and Dlx6 levels, similar to what was seen at the Dlx1 locus with Dlx1as knockout. Consistent with in vitro findings, Evf-2 null mice had a loss of DLX2 recruitment to the intergenic enhancer, however they also lacked the recruitment of MECP2, methyl-CpG-binding protein and transcriptional repressor of the Dlx5/6 locus. Further studies revealed that Evf-2 exists in a complex with DLX1 and the BRG1 chromatin-remodeling factor. Evf-2 inhibits BRG1 activity, thereby providing a mechanism through which a lncRNA can exert a repressive effect while forming a complex with transcriptional activators [72]. These data demonstrate that the lncRNA Evf-2 can recruit both activators and repressors to the Dlx5/6 locus, and highlights the importance of using genetic knockout strategies to complement in vitro knockdown experiments in cell culture.
Regulation of DNA Methylation
To further characterize the interaction between MECP2, DLX1/2, and Evf-2, the Khotz laboratory conducted genetic epistasis experiments by crossing the Evf-2-null mouse with Mecp2 −/− and Dlx1/2+/− mutant strains [61]. Mecp2-null mice display a 2-fold increase in Dlx5 transcript levels. This increase in Dlx5 expression is abrogated on a Dlx1/2 +/− background, suggesting that DLX1/2 and MECP2 are antagonistic at the Dlx5/6 locus. Analysis of methylation of the DLX5/6 enhancer revealed that Evf-2 inhibits the methylation of the DLX5/6 enhancer in trans. Taken together, these data suggested a model in which Evf-2 can regulate the methylation state of the Dlx5/6 enhancer, which modulates the recruitment of the methyl-CpG repressor MECP2 and DLX1/2 transcriptional activators. Interestingly, a recent study identified hundreds of transcripts that can bind DNA methyltransferase DNMT1 and negatively regulate DNA methylation [73], and the lncRNA Dali has been shown to bind both its neighbor POU3F3 and DNMT1. These studies suggest a similar mechanism may be utilized by Evf-2 at the Dlx5/6 enhancer.
Transcriptional Activation by lncRNAs in Neurogenesis
Genome-wide expression studies and siRNA screens in embryonic stem cells identified lncRNAs required for the maintenance of pluripotency and repression of the neural lineage [24]. Ng, et al. used custom lncRNA microarrays to identify 35 lncRNAs that are upregulated during neuronal differentiation from human embryonic stem cells (ESCs) [74]. Four of these candidates were chosen for shRNA-mediated knockdown, and depletion of any of the four lncRNAs decreases in neurogenesis. One of the identified lncRNAs, RMST, localizes to chromatin at the promoters of key neurogenic regulators, including Sp8 and Dlx2 [50]. Intriguingly, RMST binds directly to SOX2 as well as RNA-binding protein HnRNPA2/B1. RMST co-occupies loci bound by SOX2, and SOX2 occupancy is lost upon RMST knockdown, suggesting that a lncRNA, perhaps through an hnRNP adapter, can modulate the recruitment of transcription factors to downstream targets.
Paupar is a lncRNA transcribed upstream from Pax6, a key homeobox transcription factor expressed in neural progenitor populations [52]. Paupar associates with chromatin, and depletion of Paupar causes cell-cycle arrest and drives the neuronal differentiation of the N2A neuroblastoma cell line. Genome-wide studies indicate that Paupar and Pax6 regulate a common set of transcriptional targets, and genome-wide analysis of Paupar localization on chromatin demonstrates that it binds hundreds of gene promoters, including key neural differentiation genes. Interestingly, Paupar transcript is enriched at regions containing the DNA-binding motif of Pax6, and further studies demonstrate that Paupar directly binds Pax6, and the lncRNA and transcription factor co-occupy several gene promoters. Unlike the role of lncRNAs in targeting epigenetic complexes, knockdown of Paupar does not affect Pax6 localization; rather, Paupar can enhance transcriptional activation or repression by Pax6. These data suggest that PAX6 recruits Paupar to loci genome-wide, where it then acts to modify Pax6 function through as of yet undefined mechanisms.
Similar to RMST and Paupar, the lncRNA Dali binds to a key neurogenic transcription factor, POU3F3 [53]. Depletion of this lncRNA in the N2A neuroblastoma cell line inhibits proper differentiation into neurons. Genome-wide analysis of Dali occupancy on chromatin indicate that Dali binds more than one thousand genes, many involved in cell cycle regulation and neuronal function. Pulldown of biotinylated Dali revealed a direct interaction with DNA methyltransferase DNMT1 as well as POU3F3, and depletion of Dali results in an increase of DNA methylation at target genes. Taken together, these data suggest a model whereby a lncRNA can form a complex with both a neural-specific transcription factor and chromatin-modifying enzyme to affect gene expression and DNA methylation at distant genomic targets.
Conservation across species of lncRNA function in neurogenesis
Sequencing and annotation of lncRNAs in the zebrafish genome led to the identification of two brain-enriched lncRNAs, Megamind and Cyrano [20]. Depletion of Cyrano during development in vivo results in morphological defects in the head and eyes, and neural tube defects. Embyronic depletion of Megamind leads to small heads with hydrocephalus and loss of neuronal populations throughout the brain. Treatment with lower doses allows survival to adulthood, however these fish exhibit locomotor defects [54]. Remarkably, the developmental phenotypes caused by depletion of megamind or Cyrano can be rescued with the mouse or human homologues of these transcripts, suggesting lncRNAs play a conserved role in development of the nervous system despite their relative lack of primary sequence conservation.
Further study of the mammalian homologue of Megamind, called TUNA, revealed that this lncRNA is required for neuronal differentiation from ESCs [54]. TUNA binds RNA-binding proteins PTBP1, hnRNP K, and NCL. These three proteins exist in a complex that requires RNA to form, suggesting that TUNA can serve as a scaffold for complex assembly. TUNA localizes to chromatin at the promoters of several key neurogenic genes, including SOX2. TUNA depletion causes a failure to recruit HNRNP-K to the SOX2 promoter, and subsequent down-regulation of SOX2 and a loss of neurogenesis. TUNA therefore represents another example of neurogenesis being controlled through the cooperation of a lncRNA and RNA-binding proteins.
LncRNAs in cortical development
Neurogenesis in the cortex follows a stereotyped series of cell divisions and transitions similar to those found in the V-SVZ lineage. Radial glial cells are ventricle-contacting stem cells that reside in the ventricular zone (VZ) and can either self renew or divide to give rise to TBR2+ transit-amplifying progenitors that differentiate and migrate through the SVZ and intermediate zone (SVZ/IZ) [75]. These cells divide once or more to give rise to neurons that will then populate the six cortical layers. The layers are born in an “inside out” fashion, with the inner most layers born at the onset of neurogenesis around E11.5, and the outer layers of neurons being born towards the end of the neurogenic period [76]. Aprea and colleagues used a transgenic fluorescent reporter system to separate and purify proliferating progenitors (PPs), differentiating progenitors (DPs), and newborn neurons (N) from the embryonic cortex [25]. These cells were subject to whole-transcriptome sequencing, and lncRNAs from each of these cell types were identified, including 9 ‘switch genes,’ defined as genes either enriched (on-switch) or depleted (off-switch) in DPs compared to other cell types. Among these 9 switch genes were RMST and Gomafu (Miat), which had previously been shown to play key roles in neural development in other experimental systems.
Gomafu overexpression or knockdown via in utero electroporation of developing cortex causes an expansion of TBR2+ progenitors in the VZ, a decrease in the number of differentiating progenitors, and a decrease of neurons reaching the cortical plate [25]. This loss of neurons is chiefly related to an increase in cell death of neurons in the IZ, suggesting that Gomafu regulates progenitor production and neuronal survival in vivo. Mechanistically, Gomafu can affect the splicing of Wnt7b, a known regulator of progenitor proliferation and differentiation in the embryonic cortex. Further studies are required to fully characterize the differential splicing induced by Gomafu manipulation, but differential splicing of Wnt raises the intriguing possibility that a lncRNA can control cell fate decisions through the splicing of the ligands and possibly receptors of key signaling pathways. Interestingly, further studies have implicated Gomafu in aberrant alternative splicing in neurons seen in schizophrenia [6].
Neurog1 is a neurogenic transcription factor expressed in the developing cortex. Like Dlx1as and Evf-2, utNgn1 is a long noncoding RNA transcribed from a conserved enhancer element near Neurog1 [77]. This transcript’s expression is tightly correlated with Ngn1 expression in vivo, and depletion of this lncRNA in acutely dissected neural progenitor cultures results in a failure to upregulate Ngn1 upon differentiation.
The lncRNA Pnky was also studied in the context of cortical development; Pnky transcript is expressed in the embryonic brain of both mouse and human in the VZ [48]. Electroporation of Pnky knockdown constructs at E13.5 and analysis two days later causes a decrease in the proportion of GFP+ cells in the VZ and a corresponding increase of GFP+ cells in the SVZ/IZ. Further analyses demonstrated that Pnky-knockdown decreases SOX2+ stem cells in the VZ and increases the proportion of SATB2+ young neurons in the SVZ/IZ. These data indicate that the lncRNA Pnky can regulate the differentiation of cortical NSCs in vivo, and further demonstrates a lncRNA can play a key role in both embryonic and adult NSC populations.
Loss-of-function studies in mice
In addition to the Dlx1as and Evf-2 mutant mice discussed above, a recent set of studies reported the generation and characterization of 18 lncRNA knockout mice, including 13 with evidence for expression in the brain [60, 78]. In contrast to Dlx1as and Evf-2 mice, these knockouts were generated by complete or near-complete deletion of the targeted lncRNA, and knock-in of a Lac-Z reporter. Of the 20, three lncRNA knockout mice, Fendrr −/−, Mdgt −/−, and Peril −/− demonstrated neonatal or perinatal lethality with variable penetrance. Peril is located 110 kb downstream of key neural transcription factor Sox2 and is expressed in germinal zones of E14.5 and E18.5 brain and spinal cord. RNA-seq of Peril −/− brains vs. controls revealed a down-regulation of pathways involved in cell cycle, energy processing, and protein translation processes, suggesting that Peril may be involved in the regulation of the cell cycle or metabolism of neurogenic progenitors.
Like Pou3f2 (Brn2), the Pou3f3 (Brn1) locus is located near putative lncRNAs. Two lncRNAs are transcribed near the Brn1 locus: linc-Brn1a is bi-directionally transcribed from the Brn1 promoter, while linc-Brn1b is ~10 kb downstream of Brn1. Linc-Brn1b is expressed in germinal zones of the developing brain starting at E13.5 [60]. By E18.5 expression is absent in the VZ and SVZ and is restricted to neurons of the cortical plate, and in adult mice expression is maintained in upper cortical layers of the somatosensory and visual cortex.
Linc-Brn1b-knockout mice exhibit a significant decrease in Brn1 mRNA and protein expression. RNA-seq of null-cortices revealed downregulated genes were enriched in gene ontology terms associated with cellular proliferation and uprgulated genes were enriched for terms related to neuronal differentiation. Consistent with these results, there was a decrease in proliferating intermediate progenitors in the embryonic brain and a selective reduction of upper layer neurons. Interestingly, this was accompanied by an expansion of deep layer neurons, suggesting a mis-specification of cortical progenitor fate.
Long Non Coding RNAs in Gliomagenesis
A deeper understanding of lncRNAs in normal developmental processes can provide insight into the aberrant hijacking of these pathways leading to uncontrolled growth and tumor formation. Glioblastoma multiforme (GBM) is a highly malignant primary brain tumor (WHO Grade IV tumor). The most common of the primary brain tumors, GBM occurs in 7.2 per 100,000 adults per year, and this diagnosis carries a median prognosis of just over one year when treated with surgery, chemotherapy and radiation [79, 80]. Recent analysis [81–83] using microarray gene expression assays has demonstrated lncRNA expression signatures can be used to predict clinical phenotypes and prognosis in glioma. While it is clear that lncRNA expression can be correlated with tumor molecular subtype and clinical behavior, the functional significance of lncRNA in glioma is now just beginning to be explored: Several lncRNAs have been analyzed in preclinical models of GBM, including putative lncRNA tumor suppressors linc-RoR [84] and ADAMTS9-AS2[85], and lncRNA oncogenes CRNDE [86], and H19 [87], however the most well-studied and illustrative example of a lncRNA functioning in tumorigenesis is lncRNA HOTAIR.
HOTAIR, Hox transcript antisense intergenic RNA, is transcribed in the antisense direction of the HOXC gene. HOTAIR is well described for its ability of the 5′ domain to recruit EZH2 to target genes. It further has the ability to function as a molecular scaffold with binding surfaces to assemble histone modification enzymes [88]. HOTAIR overexpression is correlated with multiple cancer lines including breast [89], hepatocellular [90], and colorectal cancer [91], with increased expression correlating with poor prognosis, tumor invasiveness, and metastatic disease. HOTAIR expression is similarly upregulated in GBM samples, with its expression levels serving as an independent prognostic factor in patients with GBM [92]. HOTAIR controls cell cycle progression in glioma via interaction with EZH2, and HOTAIR knockdown or EZH2 inhibition block cell cycle progression in cultured cells [93]. Furthermore, knockdown of HOTAIR is sufficient to block tumor formation in an orthotopic GBM model [94]. Bromodomain and extraterminal (BET) domain proteins represent a class of therapeutic targets in cancer and GBM. Treatment of GBM samples with a BET inhibitor decreases GBM growth and also causes a decrease in HOTAIR expression; furthermore, BET protein family member BRD4 was found to directly bind the HOTAIR promoter [95].
Knockdown of the lncRNA Pnky causes a strong differentiation phenotype and stem cell depletion. Recent study of glioblastoma tumor-propagating cells demonstrated that these “cancer stem cells” rely on a core set of neurodevelopmental transcription factors for their propagation. Included in this set is the gene neighbor of Pnky, BRN2, and chromatin-state maps of the Pnky/Brn2 locus in tumor-propagating cells revealed widespread active chromatin marks at the promoters [96]. Further, the protein-binding partner of Pnky, PTBP1 [97], is upregulated in GBM and functions as a driver of tumor growth. The Pnky/PTBP1 complex is therefore an attractive therapeutic target in GBM that was first discovered through careful study of the lncRNAs of the V-SVZ lineage. Careful characterization of both lncRNAs and their protein-interacting partners in normal developmental lineages and glioma has the potential to broaden therapeutic targets to that may simultaneously target upstream factors, the lncRNA itself, and its protein-interacting partner.
It is clear that lncRNAs can play key roles in both normal neural development and in cancer progression. As we and others [98, 99] have reviewed, studies of lncRNA function in vivo can yield different and sometimes contradictory phenotypes compared to cell culture models. Studies in the V-SVZ and embryonic cortex represent an important first step towards describing the function of lncRNAs in vivo, while the next wave of functional lncRNA discoveries will be made with knockout and conditional knockout mouse models. Here, the challenge will be in the specific design of the lncRNA knockout strategy and careful dissection of RNA function from the potential function of underlying DNA regulatory elements. Finally, transgenic rescue experiments, long considered the ‘gold standard’ for establishing the function of protein-coding genes in vivo, should be performed for any lncRNA expected to exert trans-acting effects [99, 100]. Further understanding of the pathways governed by lncRNAs, and especially their function in vivo, has the potential to unlock new strategies for both regenerative medicine and the treatment of CNS malignancies.
Acknowledgments
This work was supported by VA 5I01 BX000252-06, American Brain Tumor Association, Sontag Foundation, Shurl and Kay Curci Foundation, NCI-SPORE DRP, and the Hana Jabsheh Initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature structural & molecular biology. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 4.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 5.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, Nayler SP, Nones K, Hu J, Bredy TW, Nakagawa S, Rigo F, Taft RJ, Cairns MJ, Blackshaw S, Wolvetang EJ, Mattick JS. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular psychiatry. 2013 doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 7.Millar JK, James R, Brandon NJ, Thomson PA. DISC1 and DISC2: discovering and dissecting molecular mechanisms underlying psychiatric illness. Annals of medicine. 2004;36:367–378. doi: 10.1080/07853890410033603. [DOI] [PubMed] [Google Scholar]
- 8.Talkowski ME, Maussion G, Crapper L, Rosenfeld JA, Blumenthal I, Hanscom C, Chiang C, Lindgren A, Pereira S, Ruderfer D, Diallo AB, Lopez JP, Turecki G, Chen ES, Gigek C, Harris DJ, Lip V, An Y, Biagioli M, Macdonald ME, Lin M, Haggarty SJ, Sklar P, Purcell S, Kellis M, Schwartz S, Shaffer LG, Natowicz MR, Shen Y, Morton CC, Gusella JF, Ernst C. Disruption of a large intergenic noncoding RNA in subjects with neurodevelopmental disabilities. American journal of human genetics. 2012;91:1128–1134. doi: 10.1016/j.ajhg.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. Journal of molecular neuroscience : MN. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Lessing D, Anguera MC, Lee JT. X Chromosome Inactivation and Epigenetic Responses to Cellular Reprogramming. Annual review of genomics and human genetics. 2013 doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- 12.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, Sweredoski MJ, Shishkin AA, Su J, Lander ES, Hess S, Plath K, Guttman M. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC neuroscience. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71:605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos AD, Diaz A, Nellore A, Delgado RN, Park KY, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of Genome-wide Approaches Identifies lncRNAs of Adult Neural Stem Cells and Their Progeny In Vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D, Lesche M, Dahl A, Groszer M, Hiller M, Calegari F. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32:3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponti G, Obernier K, Alvarez-Buylla A. Lineage progression from stem cells to new neurons in the adult brain ventricular-subventricular zone. Cell Cycle. 2013;12 doi: 10.4161/cc.24984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 29.Luskin MB. Neuroblasts of the postnatal mammalian forebrain: their phenotype and fate. J Neurobiol. 1998;36:221–233. [PubMed] [Google Scholar]
- 30.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 31.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 32.Peretto P, Merighi A, Fasolo A, Bonfanti L. Glial tubes in the rostral migratory stream of the adult rat. Brain Res Bull. 1997;42:9–21. doi: 10.1016/s0361-9230(96)00116-5. [DOI] [PubMed] [Google Scholar]
- 33.Doetsch F, Garcia-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Verdugo JM, Doetsch F, Wichterle H, Lim DA, Alvarez-Buylla A. Architecture and cell types of the adult subventricular zone: in search of the stem cells. J Neurobiol. 1998;36:234–248. doi: 10.1002/(sici)1097-4695(199808)36:2<234::aid-neu10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 35.Lim DA, Alvarez-Buylla A. Adult neural stem cells stake their ground. Trends Neurosci. 2014 doi: 10.1016/j.tins.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- 37.Marshall CA, Suzuki SO, Goldman JE. Gliogenic and neurogenic progenitors of the subventricular zone: who are they, where did they come from, and where are they going? Glia. 2003;43:52–61. doi: 10.1002/glia.10213. [DOI] [PubMed] [Google Scholar]
- 38.Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nait-Oumesmar B, Decker L, Lachapelle F, Avellana-Adalid V, Bachelin C, Van Evercooren AB. Progenitor cells of the adult mouse subventricular zone proliferate, migrate and differentiate into oligodendrocytes after demyelination. Eur J Neurosci. 1999;11:4357–4366. doi: 10.1046/j.1460-9568.1999.00873.x. [DOI] [PubMed] [Google Scholar]
- 40.Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, Doetsch F. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron. 2014;82:545–559. doi: 10.1016/j.neuron.2014.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mich JK, Signer RA, Nakada D, Pineda A, Burgess RJ, Vue TY, Johnson JE, Morrison SJ. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife. 2014;3:e02669. doi: 10.7554/eLife.02669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortega F, Berninger B, Costa MR. Primary culture and live imaging of adult neural stem cells and their progeny. Methods in molecular biology (Clifton NJ. 2013;1052:1–11. doi: 10.1007/7651_2013_22. [DOI] [PubMed] [Google Scholar]
- 43.Ortega F, Gascon S, Masserdotti G, Deshpande A, Simon C, Fischer J, Dimou L, Chichung Lie D, Schroeder T, Berninger B. Oligodendrogliogenic and neurogenic adult subependymal zone neural stem cells constitute distinct lineages and exhibit differential responsiveness to Wnt signalling. Nature cell biology. 2013;15:602–613. doi: 10.1038/ncb2736. [DOI] [PubMed] [Google Scholar]
- 44.Doetsch F, Alvarez-Buylla A. Network of tangential pathways for neuronal migration in adult mammalian brain. Proc Natl Acad Sci U S A. 1996;93:14895–14900. doi: 10.1073/pnas.93.25.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci U S A. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheffler B, Walton NM, Lin DD, Goetz AK, Enikolopov G, Roper SN, Steindler DA. Phenotypic and functional characterization of adult brain neuropoiesis. Proc Natl Acad Sci U S A. 2005;102:9353–9358. doi: 10.1073/pnas.0503965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz CC, Salinas RD, Zarabi H, Kriegstein AR, Lim DA. The long noncoding RNA pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. doi: 10.1016/j.stem.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 51.Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci. 2012;15:381–388. S381. doi: 10.1038/nn.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014;33:296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, Ponting CP. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin N, Chang KY, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang CS, Cunningham TJ, Head SR, Duester G, Dong PD, Rana TM. An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell. 2014;53:1005–1019. doi: 10.1016/j.molcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 58.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 59.Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural development. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keppetipola N, Sharma S, Li Q, Black DL. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Critical reviews in biochemistry and molecular biology. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yap K, Lim ZQ, Khandelia P, Friedman B, Makeyev EV. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26:1209–1223. doi: 10.1101/gad.188037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rapicavoli NA, Blackshaw S. New meaning in the message: noncoding RNAs and their role in retinal development. Dev Dyn. 2009;238:2103–2114. doi: 10.1002/dvdy.21844. [DOI] [PubMed] [Google Scholar]
- 67.Long JE, Garel S, Alvarez-Dolado M, Yoshikawa K, Osumi N, Alvarez-Buylla A, Rubenstein JL. Dlx-dependent and -independent regulation of olfactory bulb interneuron differentiation. J Neurosci. 2007;27:3230–3243. doi: 10.1523/JNEUROSCI.5265-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 69.Poitras L, Ghanem N, Hatch G, Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development. 2007;134:1755–1765. doi: 10.1242/dev.02845. [DOI] [PubMed] [Google Scholar]
- 70.Liu JK, Ghattas I, Liu S, Chen S, Rubenstein JL. Dlx genes encode DNA-binding proteins that are expressed in an overlapping and sequential pattern during basal ganglia differentiation. Dev Dyn. 1997;210:498–512. doi: 10.1002/(SICI)1097-0177(199712)210:4<498::AID-AJA12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 71.Kraus P, Sivakamasundari V, Lim SL, Xing X, Lipovich L, Lufkin T. Making sense of Dlx1 antisense RNA. Dev Biol. 2013;376:224–235. doi: 10.1016/j.ydbio.2013.01.035. [DOI] [PubMed] [Google Scholar]
- 72.Cajigas I, Leib DE, Cochrane J, Luo H, Swyter KR, Chen S, Clark BS, Thompson J, Yates JR, 3rd, Kingston RE, Kohtz JD. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015;142:2641–2652. doi: 10.1242/dev.126318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 77.Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci U S A. 2012;109:16939–16944. doi: 10.1073/pnas.1202956109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goff LA, Groff AF, Sauvageau M, Trayes-Gibson Z, Sanchez-Gomez DB, Morse M, Martin RD, Elcavage LE, Liapis SC, Gonzalez-Celeiro M, Plana O, Li E, Gerhardinger C, Tomassy GS, Arlotta P, Rinn JL. Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2015;112:6855–6862. doi: 10.1073/pnas.1411263112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO R. European Organisation for, T. Treatment of Cancer Brain, G. Radiotherapy, G. National Cancer Institute of Canada Clinical Trials. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 80.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-oncology. 2012;14(Suppl 5):v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang XQ, Kiang KM, Wang YC, Pu JK, Ho A, Cheng SY, Lee D, Zhang PD, Chen JJ, Lui WM, Fung CF, Leung GK. IDH1 mutation-associated long non-coding RNA expression profile changes in glioma. Journal of neuro-oncology. 2015;125:253–263. doi: 10.1007/s11060-015-1916-9. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 83.Chen Y, Wu JJ, Lin XB, Bao Y, Chen ZH, Zhang CR, Cai Z, Zhou JY, Ding MH, Wu XJ, Sun W, Qian J, Zhang L, Jiang L, Hu GH. Differential lncRNA expression profiles in recurrent gliomas compared with primary gliomas identified by microarray analysis. Int J Clin Exp Med. 2015;8:5033–5043. [PMC free article] [PubMed] [Google Scholar]
- 84.Feng S, Yao J, Chen Y, Geng P, Zhang H, Ma X, Zhao J, Yu X. Expression and Functional Role of Reprogramming-Related Long Noncoding RNA (lincRNA-ROR) in Glioma. Journal of molecular neuroscience : MN. 2015;56:623–630. doi: 10.1007/s12031-014-0488-z. [DOI] [PubMed] [Google Scholar]
- 85.Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu Y, Zhu W. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 87.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS ONE. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, Sugimachi K, Mimori K, Wakabayashi G, Mori M. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 91.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 92.Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, Yu SZ, Pu PY, Liu N, You YP, Jiang T, Kang CS G. Chinese Glioma Cooperative. HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-oncology. 2013;15:1595–1603. doi: 10.1093/neuonc/not131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang K, Sun X, Zhou X, Han L, Chen L, Shi Z, Zhang A, Ye M, Wang Q, Liu C, Wei J, Ren Y, Yang J, Zhang J, Pu P, Li M, Kang C. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6:537–546. doi: 10.18632/oncotarget.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L, Kong L, Wei J, Chen L, Yang J, Wang Q, Zhang J, Yang Y, Jiang T, Li M, Kang C. HOTAIR is a therapeutic target in glioblastoma. Oncotarget. 2015;6:8353–8365. doi: 10.18632/oncotarget.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad NG, Wahlestedt C. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A. 2015;112:8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ferrarese R, Harsh GRt, Yadav AK, Bug E, Maticzka D, Reichardt W, Dombrowski SM, Miller TE, Masilamani AP, Dai F, Kim H, Hadler M, Scholtens DM, Yu IL, Beck J, Srinivasasainagendra V, Costa F, Baxan N, Pfeifer D, von Elverfeldt D, Backofen R, Weyerbrock A, Duarte CW, He X, Prinz M, Chandler JP, Vogel H, Chakravarti A, Rich JN, Carro MS, Bredel M. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124:2861–2876. doi: 10.1172/JCI68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kohtz JD. Long non-coding RNAs learn the importance of being in vivo. Frontiers in genetics. 2014;5:45. doi: 10.3389/fgene.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, Higgs DR, Miska EA, Ponting CP. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]