Abstract
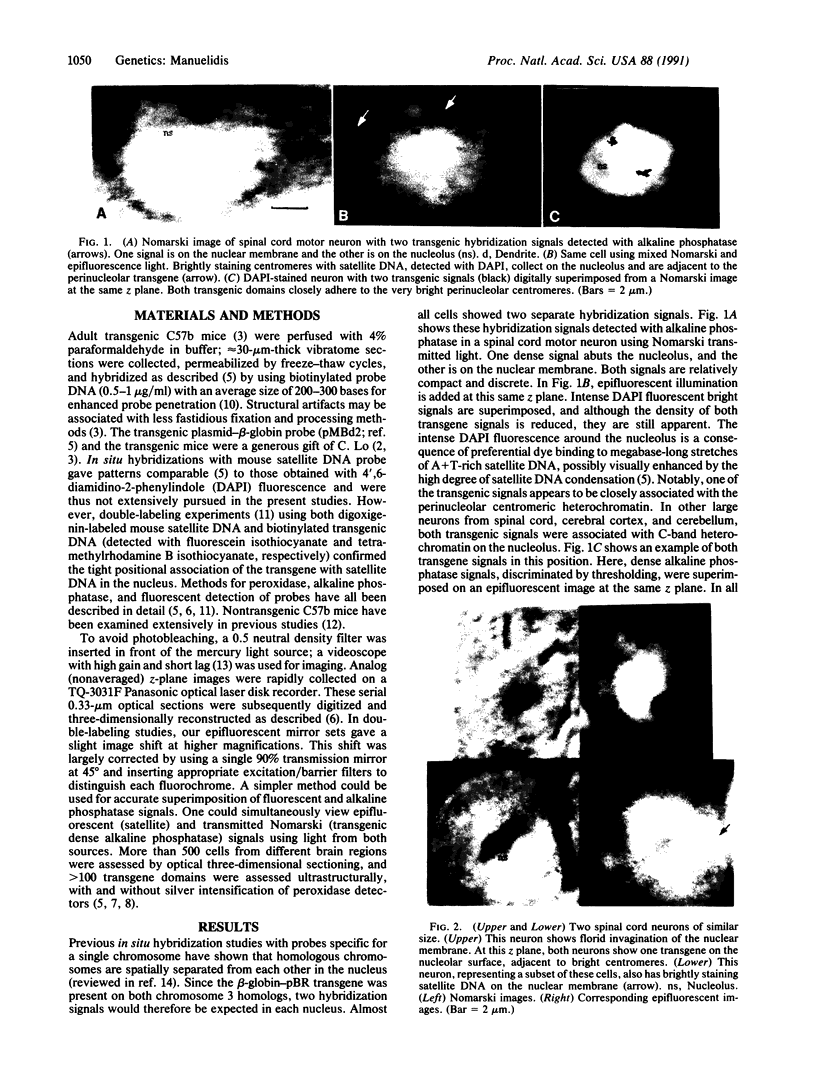
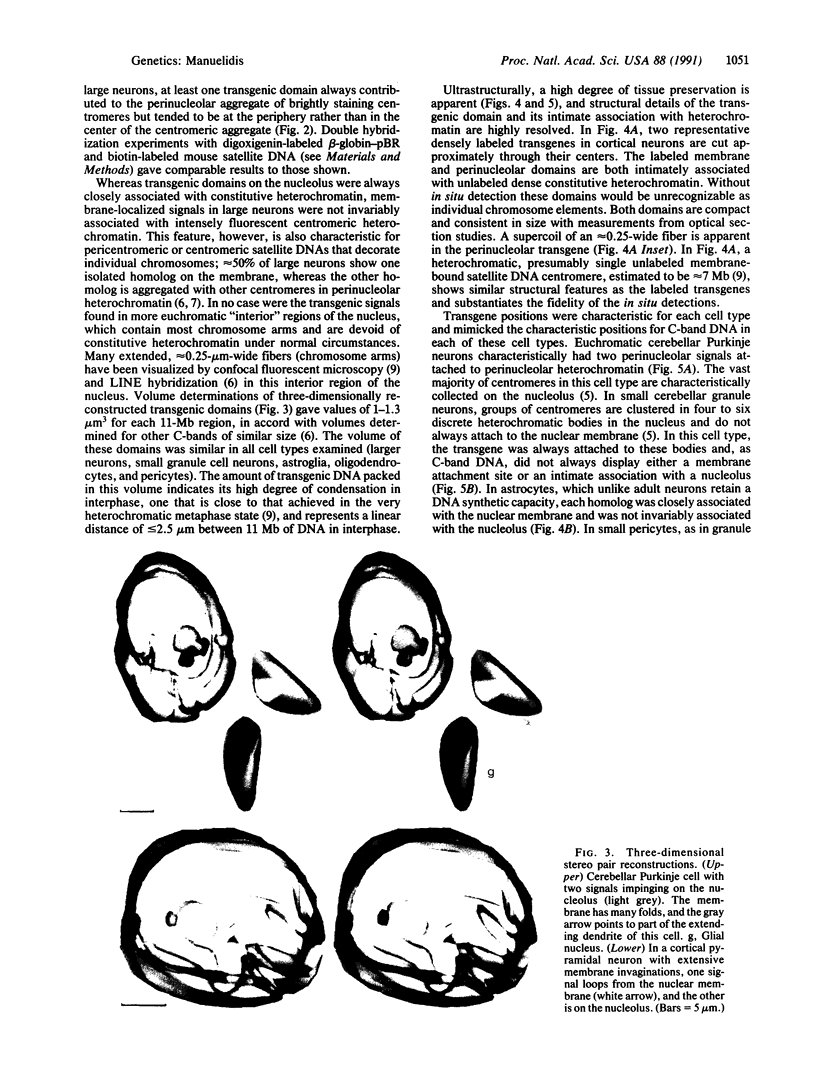
Transgenic mice provide a remarkable experimental setting for the study of nuclear architecture. The three-dimensional localization and fine structure of a foreign DNA within the mouse genome can be conveniently followed by using high-resolution in situ hybridization. Foreign DNAs designed with specific characteristics, such as base bias, sequence motif(s), and size can stably integrate into finite positions on host chromosomes. Thus the relative importance of each of these characteristics in determining the three-dimensional nuclear position and the detailed morphology of the transgene can be evaluated in different cell types. The aim of this study was to evaluate a transgene with sequence characteristics that might contribute to the de novo formation of heterochromatin in interphase nuclei. The structure of a phenotypically silent 11-megabase transgene, containing tandem repeats of beta-globin-pBR sequences integrated into the peritelomeric region of both mouse chromosome 3 homologs, was determined in adult brain cells. Neurons that are largely euchromatic were especially informative in three-dimensional studies of transgene position. The two transgenic loci behaved much like centromeric or paracentromeric A + T-rich satellite DNAs of comparable length from a single chromosome; one or both transgene domains localized together with centromeric satellite DNA on the nucleolus. This is an unusual nuclear position for a telomeric or chromosome arm region that does not contain a substantial amount of constitutively heterochromatic satellite DNA. G + C richness did not prevent these regions from assembling as dense heterochromatic bodies of approximately 1 micron3 in volume. Ultrastructurally, transgenic domains were often intimately connected with constitutive heterochromatin and were highly condensed. Labeled supercoils, formed by a discrete approximately 250-nm-wide fiber, were observed in oblique thin sections through the center of the domain. The structural data were consistent with negligible transcriptional activity detected for this locus, as well as the predicted conformation of constitutive heterochromatin. Interestingly, in transgenic but not control mice, a substantial number of large neurons, including approximately 30% of cerebellar Purkinje cells, showed excessive invaginations of the nuclear membrane.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ankenbauer T., Kleinschmidt J. A., Walsh M. J., Weiner O. H., Franke W. W. Identification of a widespread nuclear actin binding protein. Nature. 1989 Dec 14;342(6251):822–825. doi: 10.1038/342822a0. [DOI] [PubMed] [Google Scholar]
- Borden J., Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988 Dec 23;242(4886):1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- Chen T. L., Manuelidis L. SINEs and LINEs cluster in distinct DNA fragments of Giemsa band size. Chromosoma. 1989 Nov;98(5):309–316. doi: 10.1007/BF00292382. [DOI] [PubMed] [Google Scholar]
- Cremer T., Tesin D., Hopman A. H., Manuelidis L. Rapid interphase and metaphase assessment of specific chromosomal changes in neuroectodermal tumor cells by in situ hybridization with chemically modified DNA probes. Exp Cell Res. 1988 Jun;176(2):199–220. doi: 10.1016/0014-4827(88)90325-4. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- He D. C., Nickerson J. A., Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990 Mar;110(3):569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M. M., Hittelman W. N., Earnshaw W. C. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata M., Lo C. W. Organization of chromosomes in the mouse nucleus: analysis by in situ hybridization. J Cell Sci. 1988 Jun;90(Pt 2):193–199. doi: 10.1242/jcs.90.2.193. [DOI] [PubMed] [Google Scholar]
- Lo C. W., Coulling M., Kirby C. Tracking of mouse cell lineage using microinjected DNA sequences: analyses using genomic Southern blotting and tissue-section in situ hybridizations. Differentiation. 1987;35(1):37–44. doi: 10.1111/j.1432-0436.1987.tb00149.x. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science. 1990 Dec 14;250(4987):1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma. 1988;96(6):397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Chen T. L. A unified model of eukaryotic chromosomes. Cytometry. 1990;11(1):8–25. doi: 10.1002/cyto.990110104. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Different central nervous system cell types display distinct and nonrandom arrangements of satellite DNA sequences. Proc Natl Acad Sci U S A. 1984 May;81(10):3123–3127. doi: 10.1073/pnas.81.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Indications of centromere movement during interphase and differentiation. Ann N Y Acad Sci. 1985;450:205–221. doi: 10.1111/j.1749-6632.1985.tb21494.x. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Ward D. C. Chromosomal and nuclear distribution of the HindIII 1.9-kb human DNA repeat segment. Chromosoma. 1984;91(1):28–38. doi: 10.1007/BF00286482. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984 Nov 16;226(4676):792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Moroi Y., Hartman A. L., Nakane P. K., Tan E. M. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J Cell Biol. 1981 Jul;90(1):254–259. doi: 10.1083/jcb.90.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. G., Pienta K. J., Barrack E. R., Coffey D. S. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- Reuter G., Giarre M., Farah J., Gausz J., Spierer A., Spierer P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990 Mar 15;344(6263):219–223. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- Spring K. R. The study of epithelial function by quantitative light microscopy. Pflugers Arch. 1985;405 (Suppl 1):S23–S27. doi: 10.1007/BF00581775. [DOI] [PubMed] [Google Scholar]