Abstract
Long noncoding RNAs (lncRNAs) are proved tobe deregulated in a number of cancers and play important roles in tumor biology. Colon cancer-associated transcript-1 (CCAT1) was initially found in colon cancer and promoted cancer cell development. However, the role of CCAT1 in laryngeal squamous cell carcinoma (LSCC) is still unknown. In this study, we demonstrated that CCAT1 expression was upregulated in the LSCC tissues compared with the normal adjacent tissues. Moreover, CCAT1 expression was higher in LSCC tissues thanin adjacent normal tissues in 70% of patients. In addition, higher expression of CCAT1 was correlated with T3-4 grade and advanced clinical stage. Overexpression of CCAT1 promoted Hep-2 cell proliferation, migration and invasion. Ectopic expression of CCAT1 suppressed E-cadherin expression while enhanced the expressionof N-cadherin and Vimentin. Furthermore, CCAT1 overexpression increased Hep-2 cell colony formation and cell cycle. We also demonstrated that CCAT1 overexpression suppressed the expression of let-7 and enhanced the expression of HMGA2 and Myc, the direct target genes of let-7. To conclude, our data demonstrated that CCAT1 played an oncogenic role in the development of LSCC.
Keywords: Long noncoding RNAs, lncRNAs, CCAT1, laryngeal squamous cell carcinoma
Introduction
Laryngeal squamous cell carcinoma (LSCC) is the second most common malignancy of the head and neck worldwide [1-5]. Patients with LSCC demonstratelow cure rate and high morbidity, thereby requiring novel diagnostic and therapeutic procedures [6-9]. Despite great advances achieved in the diagnosisof LSCC, there is no ideal biomarker for diagnosis and treatment of LSCC patients [10-12]. Therefore, it is crucial to identify noveluseful molecular biomarkers for diagnosis and therapy of LSCC.
Long noncoding RNAs (lncRNAs) are a group of RNA molecules with length longer than 200 nucleotides [13-16]. Recent studies have demonstrated that lncRNAs play crucial roles in many biological and pathological processes such as development, proliferation, apoptosis, migration and invasion [17-21]. Moreover, lncRNAs can act as tumor suppressor genes or oncogenes in different tumors including lung cancer, breast cancer, bladder cancer, colorectal cancer and osteosarcoma [22-26].
In our study, we demonstrated that colon cancer associated transcript-1 (CCAT1) expression was upregulated in the LSCC tissues. Overexpression of CCAT1 promoted Hep-2 cell proliferation, migration and invasion. Ectopic expression of CCAT1 suppressed the expression of E-cadherinand enhanced the expression of N-cadherin and Vimentin.
Materials and methods
Tissue samples
Our study was approved by the clinical Ethics Committee of The First Affiliated Hospital of Harbin Medical University. Written consents were obtained from all patients. Paired tissues of LSCC and their matched noncancerous samples were obtained between 2012 and 2014 at our department. These patients did not receive chemotherapy or radiotherapy before admission. All samples were immediately frozen andstored in the liquid nitrogen following surgery.
Cell culture and tranfected
Hep-2 cell (LSCC cell line) was collected from Cell Bank of the Shanghai of the Chinese Academy of Science. Cells were seeded in the DMEM medium (Dulbecco’s modified Eagle’s medium) (GIBCO, USA) supplemented with fetal bovine serum (FBS). pCDNA-CCAT1 and control vector was obtained from the GenePharma (Shanghai, China) and cell tranfection was done using Lipofectamine 2000 (Invitrogen, USA) following to the instructions.
RNA extraction and real-time PCR
Total RNA was extracted from cell or tissues by TRIzol reagent (Invitrogen, USA) according to the manufacturer’s information. Real-time PCR (qRT-PCR) was done to measure the miRNA and mRNA expression on the 7500 Real Time PCR System (Applied Biosystems, CA). The relative expression of CCAT1 and mRNA was normalized to GAPDH. The primers were used as following: CCAT1, forward 5’-TTTATGCTTGAGCCTTGA-3’ and reverse 5’-CTTGCCTGAAATACTTGC-3’; GAPDH, forward 5’-GTCAACGGATTTGGTCTGTATT-3’ and reverse 5’-AGTCTTCTGGGTGGCAGTGAT-3’.
Cell proliferation,cell cycle and colony formationanalysis
CCK-8 (Cell counting kit-8; Dojindo, Japan) was used to determine the cell proliferation. Hep-2 cell was cultured in the 96-well plates and the cell proliferation was measured for 0 hour, 24 hours, 48 hours and 72 hours. Cell proliferation was measured through absorbance detection at the 450 nm. For cell cycle analysis, Hep-2 cells were harvested, washed and fixed using cold 70% ethanol. Then, cells were stained by PI (propidium iodide) and RNase A for about 30 minutes and measured using flow cytometry (BD, Becton Dickinson). For colony formation, cell was trypsinized into the single-cell and seeded in the 6-well plates. After cultured for 2 weeks, the colony was fixed and counted with crystal violet (Sigma, Germany).
Cell invasion and migration
Wound healing analysis was performed to measure the cell migration. Cells were plated in the 6 well plates and wound was done with the pipette tip. Then, cells were cultured for additional 48 hours and the wound was detected by photomicrographs. For cell invasion analysis, transwell with Matrigel was used. Cells were cultured in the upper chamber with serum-free media and 10% FBS was added into the lower chamber. After 48 hours, the invaded cells in the lower membrane were stained with crystalviolet and counted using a microscope (Olympus, Japan).
Western blot assay
Total protein was isolated from cell or tissues using lysis buffer. Protein was separated by 12% SDS-PAGE and then transferred to PVDF (polyvinylidenefluoride) membranes (Millipore, MA). The PVDF membrane was blocked with milk and incubated with primary antibodies against (HMGA2, Myc, GAPDH, abcam) overnight. Protein was measured using ECL (enhanced chemiluminescence reagent) (Pierce) following to manufacturer’s instructions.
Statistical analysis
Data were shown as mean ± SD (standard deviation). The significance differences between more than two groups were determinedone-way ANOVA and the difference between two groups was analyzed using the Student t-test. P value with less than 0.05 was considered statistically significant.
Result
CCAT1 expression was upregulated in LSCC tissues
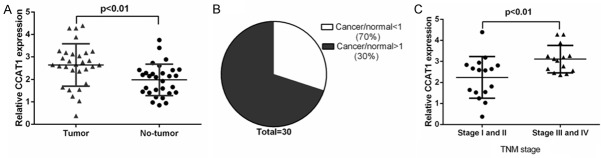
We measured the CCAT1 expression in the 30 pairs of LSCC tissues. CCAT1 expression was upregulated in LSCC tissues compared with the normal adjacent tissues (Figure 1A). Moreover, CCAT1 expression was increased in LSCC tissues compared with the adjacent normal tissues in 21 patients (70%, 21/30) (Figure 1B). Inaddition, higher expression of CCAT1 was correlated with T3-4 grade and advanced clinical stage (Figure 1C).
Figure 1.
The expression of CCAT1 was upregulated in LSCC tissues. A. The expression of CCAT1 in the LSCC tissues and normal adjacent tissues was measured using qRT-PCR. B. CCAT1 expression was increased in LSCC tissues compared with adjacent normal tissues in 21 patients. C. Higher expression of CCAT1 was related with T3-4 clinical stage.
CCAT1 increased LSCC cell proliferation andpromoted LSCC cell colony formation and cell cycle
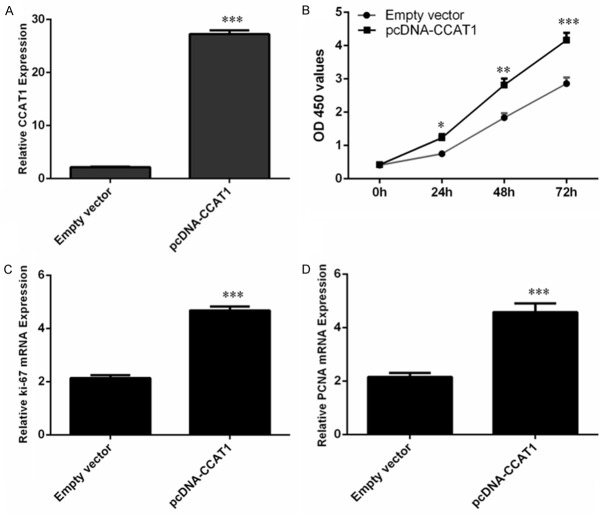
We used the pcDNA-CCAT1 to measure the function of CCAT1 in Hep-2 cell. We found that pcDNA-CCAT1 could promote the CCAT1 expression inHep-2 cell (Figure 2A). Ectopic expression of CCAT1 promoted Hep-2 cell proliferation (Figure 2B). Moreover, overexpression of CCAT1 increased the expression of ki-67 (Figure 2C) and PCNA (Figure 2D). CCAT1 overexpression increased Hep-2 cell colony formation (Figure 3A, 3B). Moreover, ectopic expression of CCAT1 also promoted Hep-2 cell cycle (Figure 3C).
Figure 2.
CCAT1 increased the LSCC cell proliferation. A. The expression of CCAT1 was detected in the Hep-2 cell using qRT-PCR. B. Ecotpic expression of CCAT1 increased the Hep-2 cell proliferation. C. The expression of ki-67 was measured using qRT-PCR. D. The expression of PCNA was measured using qRT-PCR.
Figure 3.
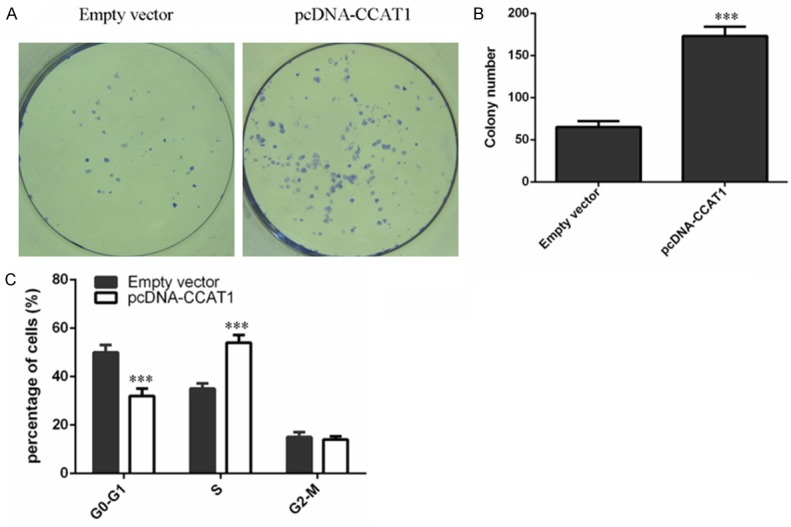
CCAT1 promoted the LSCC cell colony formation and cell cycle. A. Ecotpic expression of CCAT1 promoted the Hep-2 cell colony formation. B. The relative colony number was shown. C. Ecotpic expression of CCAT1 increased the Hep-2 cell cycle.
CCAT1 enhanced LSCC cell migration and invasion
Wound healing analysis showed that CCAT1 overexpression enhanced Hep-2 cell migration (Figure 4A and 4C). Moreover, cell invasion analysis demonstrated that CCAT1 promoted Hep-2 cell invasion (Figure 4B, 4D). In addition, overexpression of CCAT1 could suppress the expression of E-cadherinand enhance the expression of N-cadherin and Vimentin (Figure 4E).
Figure 4.
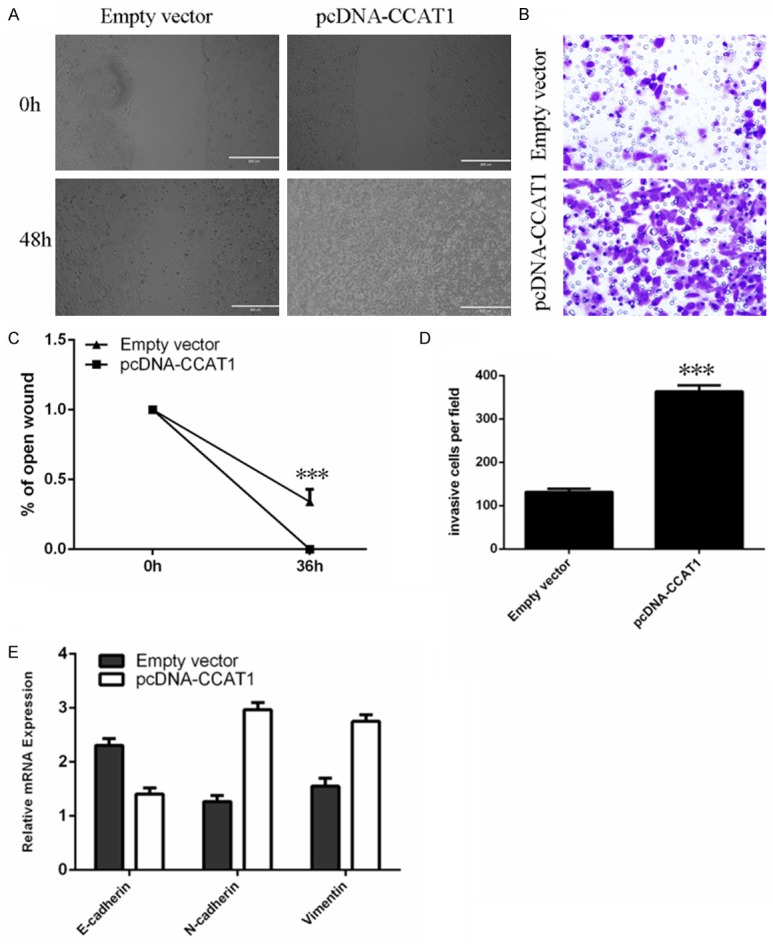
CCAT1 enhanced the LSCC cell migration and invasion. A. Ecotpic expression of CCAT1 promoted the Hep-2 cell migration. B. Overexpression of CCAT1 enhanced the Hep-2 cell invasion. C. The relative open wound was shown. D. The relative invasive cells were shown. E. The expression level of E-cadherin, N-cadherin and Vimentin was measured using qRT-PCR.
CCAT1 suppressed let-7 expression and enhanced HMGA2 and Myc expression
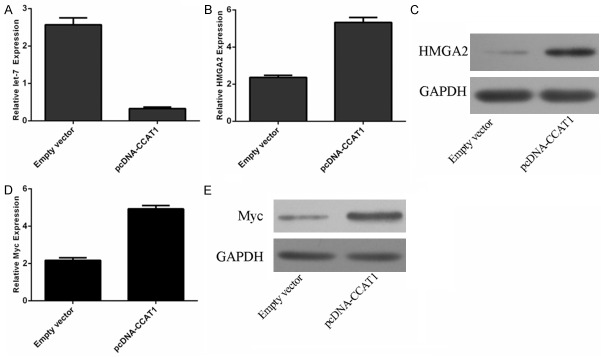
Overexpression of CCAT1 could promote the expression of let-7 expression (Figure 5A). Moreover, ectopic expression of CCAT1 enhanced the expression of HMGA2 (Figure 5B, 5C). In addition, ectopic expression of CCAT1 also promoted the expression of Myc (Figure 5D, 5E).
Figure 5.
CCAT1 suppressed the let-7 expression and enhanced the HMGA2 and Myc expression. A. The expression of let-7 was detected using qRT-PCR. B. The mRNA expression of HMGA2 was measured using qRT-PCR. C. The protein expression of HMGA2 was measured using western blot. D. The mRNA expression of Myc was measured using qRT-PCR. E. The protein expression of Myc was measured using western blot.
Discussion
In our study, CCAT1 expression was upregulated in the LSCC tissues compared with the normala djacent tissues. Moreover, CCAT1 expression was higher in LSCC tissues compared with adjacent normal tissues in 70% patients. Inaddition, higher expression of CCAT1 correlated with T3-4 grade and advanced clinical stage. Overexpression of CCAT1 promoted Hep-2 cell proliferation, migration and invasion. Ectopic expression of CCAT1 could suppress the expression of E-cadherinand enhance the expression of N-cadherin and Vimentin. Furthermore, CCAT1 overexpression increased Hep-2 cell colony formation and cell cycle. We also demonstrated that CCAT1 overexpression suppressed let-7 expression and enhanced the expression of HMGA2 and Myc, the direct target genes of let-7. These data demonstrated that CCAT1 played an oncogenic role in the development of LSCC.
Previous studies had showed that CCAT1 played an important role in several tumors such as colorectal cancer, breast cancer, hepatocellular carcinoma, gallbladder cancer and gastric cancer [27-31]. For example, Ye et al [27] demonstrated that CCAT1 expression was higher in the colorectal cancer tissues than in normal para-carcinoma tissues. The expression level of CCAT1 was correlated with tumor staging, local infiltration depth, vascular invasion and CA19-9 level. Zhang et al [28] showed that CCAT1 expression in breast tissues were higher than those in non-tumor tissues. High CCAT1 expression was correlated with advanced TNM stage, low differentiation grade and more lymph node metastases. Patients with high CCAT1 expression had a lower overall survival (OS) and progression-free survival. Yang et al [32] demonstrated that CCAT1 expression was upregulated in gastric cancer tissues compared with normal adjacent tissues. Deng et al [33] found that CCAT1 expression was upregulated in hepatocellular carcinoma tissues compared with non-cancerous hepatictissues. Higher expression of CCAT1 was associated with microvascular invasion, tumor size, poor prognosis and AFP. CCAT1 overexpression increased the hepatocellular carcinoma cell migration and proliferation. However, the role of CCAT1 in LSCC is still unknown. In our study, we found that CCAT1 expression was increased in LSCC tissues compared with the normal adjacent tissues. Inaddition, higher expression of CCAT1 was correlated with advanced clinical stage. Overexpression of CCAT1 promoted Hep-2 cell proliferation, migration and invasion. Ectopic expression of CCAT1 could suppress the E-cadherin expression and enhance the expressionof N-cadherin and Vimentin. Furthermore, CCAT1 overexpression increased the Hep-2 cell colony formation and cell cycle. These results suggested that CCAT1 acted as an oncogene in the development of LSCC.
Increasing evidences have showed that lncRNAsexert functions by targeting miRNAs. For example, Ma et al [30] found that CCAT1 expression was increased in gallbladder cancertissues. Overexpression of CCAT1 suppressed the expression of miR-218-5p and promoted the expression of miR-218-5 ptarget gene Bmi1. Moreover, another study also showed that CCAT1 inhibited the let-7 expression and enhanced c-Myc and HMGA2 expression, the target genesof let-7 [33]. In line with these data, we also found that CCAT1 overexpression suppressed the let-7 expression and enhanced the expression of HMGA2 and Myc, which were the direct target genes of let-7. Our data suggested that CCAT1 played an important role in LSCC development partly through targeting let-7 expression.
In conclusion, we demonstrated that CCAT1 expression was upregulated in LSCC tissues and CCAT1 overexpression promoted LSCC cell proliferation, colony formation, cell cycle, migration and invasion. In addition, overexpression of CCAT1 suppressed let-7 expression and enhanced HMGA2 and Myc expression, which were the target genes of let-7. These data demonstrated that CCAT1 served as an oncogene in the development of LSCC.
References
- 1.Yungang W, Xiaoyu L, Pang T, Wenming L, Pan X. miR-370 targeted FoxM1 functions as a tumor suppressor in laryngeal squamous cell carcinoma (LSCC) Biomed Pharmacother. 2014;68:149–154. doi: 10.1016/j.biopha.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Ren J, Zhu D, Liu M, Sun Y, Tian L. Downregulation of miR-21 modulates Ras expression to promote apoptosis and suppress invasion of Laryngeal squamous cell carcinoma. Eur J Cancer. 2010;46:3409–3416. doi: 10.1016/j.ejca.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 3.Yu X, Li Z. The role of microRNAs expression in laryngeal cancer. Oncotarget. 2015;6:23297–23305. doi: 10.18632/oncotarget.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito K, Inagaki K, Kamimoto T, Ito Y, Sugita T, Nakajo S, Hirasawa A, Iwamaru A, Ishikura T, Hanaoka H, Okubo K, Onozaki T, Zama T. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS One. 2013;8:e71480. doi: 10.1371/journal.pone.0071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo HN, Wang ZH, Sheng Y, Zhang Q, Yan J, Hou J, Zhu K, Cheng Y, Xu YL, Zhang XH, Xu M, Ren XY. MiR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med Oncol. 2014;31:789. doi: 10.1007/s12032-013-0789-z. [DOI] [PubMed] [Google Scholar]
- 6.Zhao XD, Zhang W, Liang HJ, Ji WY. Overexpression of miR-155 promotes proliferation and invasion of human laryngeal squamous cell carcinoma via targeting SOCS1 and STAT3. PLoS One. 2013;8:e56395. doi: 10.1371/journal.pone.0056395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M, Tian L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. 2014;31:148. doi: 10.1007/s12032-014-0148-8. [DOI] [PubMed] [Google Scholar]
- 8.Gao C, Li X, Tong B, Wu K, Liu Y, Anniko M, Duan M. Up-regulated expression of Dicer reveals poor prognosis in laryngeal squamous cell carcinoma. Acta Otolaryngol. 2014;134:959–963. doi: 10.3109/00016489.2014.920962. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Wu Y, Liu Y, Deng H, Shen Z, Xiao B, Guo J. miR-21, miR-106b and miR-375 as novel potential biomarkers for laryngeal squamous cell carcinoma. Curr Pharm Biotechnol. 2014;15:503–508. doi: 10.2174/1389201015666140519110616. [DOI] [PubMed] [Google Scholar]
- 10.Hu A, Huang JJ, Xu WH, Jin XJ, Li JP, Tang YJ, Huang XF, Cui HJ, Sun GB. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: association with patient survival. Am J Transl Res. 2014;6:604–613. [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Lei DP, Jin T, Zhao XN, Li G, Pan XL. Altered expression of miR-21 and PTEN in human laryngeal and hypopharyngeal squamous cell carcinomas. Asian Pac J Cancer Prev. 2011;12:2653–2657. [PubMed] [Google Scholar]
- 12.Nohata N, Hanazawa T, Kinoshita T, Inamine A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer. 2013;108:1648–1658. doi: 10.1038/bjc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Yu X, Shen J. Long non-coding RNAs: emerging players in osteosarcoma. Tumour Biol. 2016;37:2811–6. doi: 10.1007/s13277-015-4749-4. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Li Z. Long non-coding RNA HOTAIR: A novel oncogene (Review) Mol Med Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, Li Z. Long non-coding RNA growth arrest-specific transcript 5 in tumor biology. Oncol Lett. 2015;10:1953–1958. doi: 10.3892/ol.2015.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–585. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 20.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iranpour M, Soudyab M, Geranpayeh L, Mirfakhraie R, Azargashb E, Movafagh A, Ghafouri-Fard S. Expression analysis of four long noncoding RNAs in breast cancer. Tumour Biol. 2016;37:2933–40. doi: 10.1007/s13277-015-4135-2. [DOI] [PubMed] [Google Scholar]
- 23.Hu X, Bao J, Wang Z, Zhang Z, Gu P, Tao F, Cui D, Jiang W. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol. 2016;37:3497–504. doi: 10.1007/s13277-015-4023-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Li X, Song Y, Zhang P, Xiao Y, Xing Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem Biophys Res Commun. 2015;467:223–228. doi: 10.1016/j.bbrc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, Liu X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2016;34:932–41. doi: 10.1002/jor.23105. [DOI] [PubMed] [Google Scholar]
- 26.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Z, Zhou M, Tian B, Wu B, Li J. Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal cancer and its clinical significance. Int J Clin Exp Med. 2015;8:3707–3715. [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XF, Liu T, Li Y, Li S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2015;8:9440–9445. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 30.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizrahi I, Mazeh H, Grinbaum R, Beglaibter N, Wilschanski M, Pavlov V, Adileh M, Stojadinovic A, Avital I, Gure AO, Halle D, Nissan A. Colon Cancer Associated Transcript-1 (CCAT1) Expression in Adenocarcinoma of the Stomach. J Cancer. 2015;6:105–110. doi: 10.7150/jca.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 33.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]