Abstract
Cardiac ryanodine receptor (Ryr2) Ca2+ release channels and cellular metabolism are both disrupted in heart disease. Recently, we demonstrated that total loss of Ryr2 leads to cardiomyocyte contractile dysfunction, arrhythmia, and reduced heart rate. Acute total Ryr2 ablation also impaired metabolism, but it was not clear whether this was a cause or consequence of heart failure. Previous in vitro studies revealed that Ca2+ flux into the mitochondria helps pace oxidative metabolism, but there is limited in vivo evidence supporting this concept. Here, we studied heart-specific, inducible Ryr2 haploinsufficient (cRyr2Δ50) mice with a stable 50% reduction in Ryr2 protein. This manipulation decreased the amplitude and frequency of cytosolic and mitochondrial Ca2+ signals in isolated cardiomyocytes, without changes in cardiomyocyte contraction. Remarkably, in the context of well preserved contractile function in perfused hearts, we observed decreased glucose oxidation, but not fat oxidation, with increased glycolysis. cRyr2Δ50 hearts exhibited hyperphosphorylation and inhibition of pyruvate dehydrogenase, the key Ca2+-sensitive gatekeeper to glucose oxidation. Metabolomic, proteomic, and transcriptomic analyses revealed additional functional networks associated with altered metabolism in this model. These results demonstrate that Ryr2 controls mitochondrial Ca2+ dynamics and plays a specific, critical role in promoting glucose oxidation in cardiomyocytes. Our findings indicate that partial RYR2 loss is sufficient to cause metabolic abnormalities seen in heart disease.
Keywords: calcium intracellular release, cardiac metabolism, cardiomyocyte, heart failure, metabolomics, mitochondrial metabolism, proteomics, ryanodine receptor, transcriptomics, tricarboxylic acid cycle (TCA cycle) (Krebs cycle), metabolism, calcium, mitochondria, intracellular calcium release
Introduction
The type 2 ryanodine receptor (Ryr2)2 sarcoplasmic reticulum (SR) Ca2+ release channel plays a central role in cardiac excitation-contraction coupling (1). Ca2+ signals generated by RYR2 have also been implicated in heart rate (2–5), hypertrophic gene regulation (6, 7), and cardiomyocyte superstructure (8, 9). Partially reduced Ryr2 expression, channel density, and/or signaling have been identified in models of aging (10) and heart disease (9, 11–13), which are conditions associated with metabolic dysfunction and a lack of energy substrate flexibility (14, 15). However, it remains to be determined whether a partial reduction in Ryr2 signaling can drive adult cardiac metabolic dysfunction.
SR Ca2+ release channels are known to transmit privileged Ca2+ signals into adjacent mitochondria (16). Based on in vitro studies, it has been proposed that these Ca2+ signals stimulate mitochondrial oxidative energy metabolism via pyruvate dehydrogenase (Pdh), key tricarboxylic acid (TCA) cycle enzymes, the electron transport chain, and the mitochondrial ATPase (17–19). These concepts have been extended to cardiac biology, where studies have shown a link between mitochondrial Ca2+ and energy production in the heart (20–27). However, direct in vivo evidence that Ca2+ flux from SR specifically paces mitochondrial oxidative glucose metabolism in normally functioning cardiomyocytes has not been published. We recently reported that complete Ryr2 gene knock-out in adult mouse cardiomyocytes results in broad defects in energy metabolism (4, 20), but our previous model rapidly progressed to heart failure and sudden cardiac death (4), making it difficult to know whether mitochondrial metabolism is modulated by Ryr2 in normally functioning cardiomyocytes. Given this previous work and the association between Ryr2 dysfunction (9, 11–13) and oxidative metabolism in heart disease (14, 15), it was of interest to further explore links between Ryr2 and energy production using improved models.
To determine the potential links between Ryr2 and metabolism in the context of normal cardiac function, we devised a model where cardiomyocyte-specific, inducible Cre recombinase was used to delete only one Ryr2 allele (cRyr2Δ50 mice). Here, we report that a stable ∼50% loss of Ryr2 protein in adult cardiomyocytes causes striking changes in mitochondrial Ca2+ cycling and is sufficient to specifically inhibit oxidative glucose metabolism but not fatty acid oxidation, lactate oxidation, or glycolysis. These metabolic effects occurred without significantly affecting cellular contraction or in vivo cardiac output, although we did observe a modest decrease in cRyr2Δ50 heart rate. Our results suggest that pathophysiologically relevant loss of Ryr2 can account for the metabolic phenotype of failing hearts and that RYR2 plays a critical role in stimulating glucose oxidation in vivo.
Results
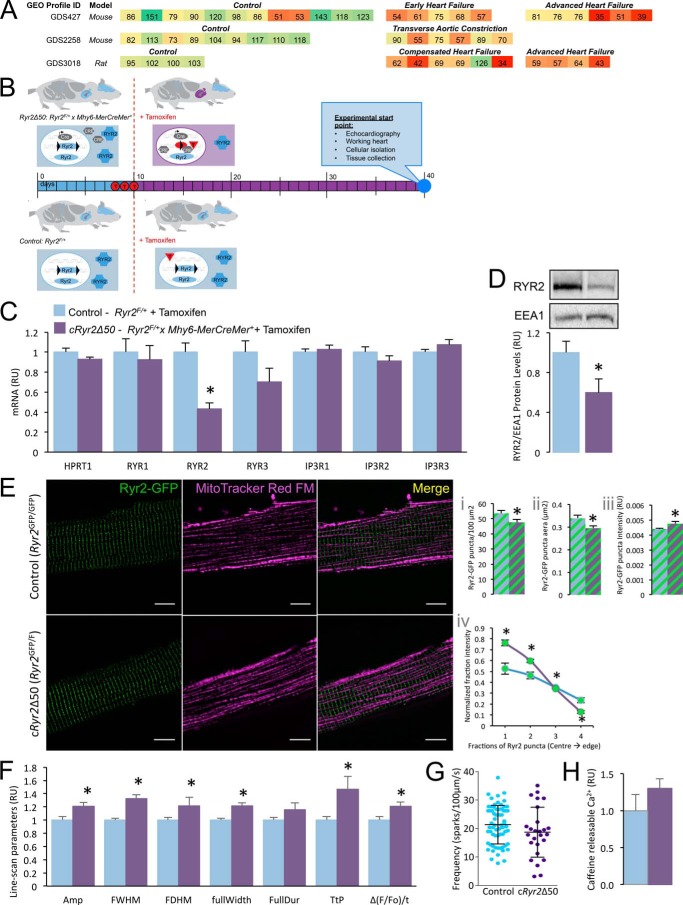
Previous studies have implicated the loss of RYR2 function or expression in disease states (9, 11–13). Similarly, analysis of publically accessible transcriptomic data on the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO) identified mouse and rat data sets with qualitatively reduced Ryr2 mRNA (Fig. 1A).
FIGURE 1.
Generation of an inducible, heart-specific, partial Ryr2 ablation model (Ryr2Δ50). A, selected Ryr2 transcriptomics data from NCBI GEO were re-normalized to the average of their control values and presented as a heat map. B, breeding scheme, experimental design, and analysis timeline. C, cRyr2Δ50 cardiomyocytes have a specific reduction in Ryr2 mRNA, and no compensation from other known endoplasmic reticulum (ER)/SR Ca2+ channels (n = 4–8, *, p ≤ 0.05). Blue bars = control (Ryr2flox/wildtype + tamoxifen), purple bars = cRyr2kΔ50 (Ryr2flox/wildtype × Mhy6-MerCreMer+ + tamoxifen) throughout. D, cardiac RYR2 protein levels in cRyr2Δ50 mice 3 weeks after tamoxifen (n = 7, *, p ≤ 0.05). E, images of Ryr2-GFP fusion proteins in isolated cardiomyocytes from control (Ryr2GFP/GFP) and cRyr2Δ50 (Ryr2GFP/flox × Mhy6-MerCreMer + tamoxifen) mice (n = 9 cells/group; scale bar, 10 μm). Images were quantified for Ryr2-GFP puncta number (panel i), area (panel ii), and intensity (panel iii). A measure of puncta intensity distribution (mean fractional brightness from the center to the edge of the puncta) was also quantified (panel iv). *, p ≤ 0.05. F, line-scanning spark measurement parameters of cRyr2Δ50 cardiomyocytes (control n = 70, cRyr2Δ50 n = 26; data from three cell isolations, *, p ≤ 0.05) Amp, amplitude; FWHM, full width at half maximum; FDHM, full duration at half maximum; fullWidth, full width; FullDur, full duration; TtP, time to peak. G, spark frequency in isolated cRyr2Δ50 cardiomyocytes (control n = 70, cRyr2Δ50 n = 26; data from three cell isolations). H, peak Fluo-4 intensity following 3 mm caffeine treatment of control (n = 5) and cRyr2Δ50 cardiomyocytes (n = 6). All data were plotted as mean ± S.E.
To model partial Ryr2 reduction, we deleted one Ryr2 allele in adult mouse hearts. We confirmed ∼50% ablation of Ryr2 mRNA and protein in the cRyr2Δ50 mice 3 weeks following the induction of haploinsufficiency with sequential tamoxifen injections (Fig. 1, B–D). Importantly, there was no compensation by other SR-Ca2+ release channel genes (Fig. 1C).
We examined the localization and organization of Ryr2 signaling structures using a Ryr2-GFP fusion protein knock-in allele (28). In cRyr2Δ50 cardiomyocytes, Ryr2 puncta exhibited a grossly normal distribution, but were fewer, smaller, and less bright, with a more compact shape (Fig. 1E). Resolution past the diffraction limit of light will be required to further characterize the (dys)organization of these structures.
Confocal line-scanning microscopy and Ca2+ spark analysis revealed that cRyr2Δ50 cardiomyocytes produced sparks with greater amplitude and slower kinetics than control cells (Fig. 1F). We did not observe a significant difference in spark frequency between the two groups (Fig. 1G), although we note that cRyr2Δ50 cardiomyocytes may not follow a normal distribution and that the majority of cRyr2Δ50 cells displayed lower spark frequency than the control average. These results suggest that there may be altered gating kinetics in cRyr2Δ50 cardiomyocytes, but do not indicate gross alterations of channel open probabilities in a basal state.
To determine whether partial RYR2 ablation had an effect on SR Ca2+ load, we measured the size of caffeine-evoked Ca2+ responses in cRyr2Δ50 cardiomyocytes (Fig. 1H). We did not observe a significant difference in caffeine-releasable Ca2+ between cRyr2Δ50 and control cardiomyocytes.
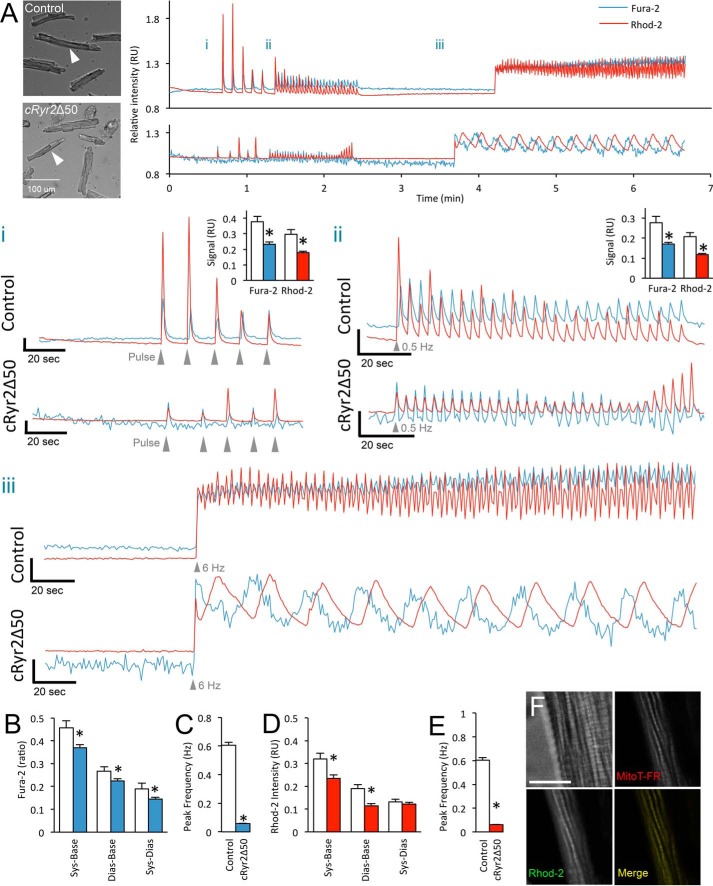
We simultaneously measured cytosolic and mitochondrial Ca2+ in isolated cells stimulated with, in order, five single pulses of field stimulation, a period of continuous 0.5-Hz stimulation, and a period of 6-Hz pulses (Fig. 2). Both control and cRyr2Δ50 cardiomyocytes responded to field stimulation and displayed similar Ca2+ dynamics to pulse or 0.5-Hz stimulation, revealing that the cells had similar viability and basal function (Fig. 2A (i and ii)). However, cRyr2Δ50 cardiomyocytes had modestly decreased cytosolic and mitochondrial systolic Ca2+ amplitudes at these frequencies (Fig. 2A (i and ii)). At 6 Hz, chosen to approximate the rapid murine heart rate, cRyr2Δ50 cardiomyocytes had significantly different cytosolic and mitochondrial Ca2+ dynamics (Fig. 2, A (iii), C, and D). Specifically, cRyr2Δ50 cytosolic and mitochondrial Ca2+ transients had a significantly lower frequency and were temporally disassociated by ∼10 s (Fig. 2, A (iii), C, and E). Co-staining with MitoTracker confirmed correct Rhod-2 loading (Fig. 2F). These data collectively indicate that ∼50% Ryr2 ablation disrupts SR-to-mitochondria communication.
FIGURE 2.
Cytosolic and mitochondrial Ca2+ in cRyr2Δ50 cardiomyocytes. A, representative simultaneous Fura-2 and Rhod-2 fluorescence traces from healthy control and cRyr2Δ50 cardiomyocytes. Bright field images with arrows indicate individual cardiomyocytes from which traces are derived. Panel i, enlargement of pulse stimulation region. Scale axis begins at 0.9 RU. Inset graphs show average peak systolic Fura-2 ratio and Rhod-2 intensity relative to baseline. Panel ii, enlargement of 0.5-Hz stimulation region. Scale axis begins at 0.9 RU. Inset graphs show average peak systolic Fura-2 ratio and Rhod-2 intensity relative to baseline. Panel iii, enlargement of 6-Hz stimulation region. Scale axis begins at 8.5 RU. *, p ≤ 0.05. B, average Fura-2 ratio in control and cRyr2Δ50 cardiomyocytes during 6-Hz stimulation. White bars = control; solid color bars = cRyr2Δ50 throughout (mean ± S.E.). Sys-Base denotes the difference between average peak systolic and baseline measurements, Dias-Base denotes the difference between average minimal diastolic and average baseline measurements, and Sys-Dias denotes the difference between peak systolic and minimum diastolic measurements. *, p ≤ 0.05. C, average frequency of cytosolic Ca2+ transients elicited during 6-Hz stimulation. *, p ≤ 0.05. D, average Rhod-2 intensity in control and cRyr2Δ50 cardiomyocytes during 6-Hz stimulation. *, p ≤ 0.05. E, average frequency of elicited mitochondrial Ca2+ transients observed during 6-Hz stimulation. For all average values: control n = 63 cells, cRyr2Δ50 n = 141 cells, three independent cell preparations from three mice per treatment; *, p ≤ 0.05. F, high resolution microscopy of isolated mouse cardiomyocyte showing colocalization of Rhod-2-AM and MitoTracker Deep Red dye (MitoT-FR). Fluorescent channel images are from a single optical plane from a deconvolved wide field z-stack. Scale bar is 10 μm. All data were plotted as mean ± S.E. Control = Ryr2flox/wildtype + tamoxifen; cRyr2Δ50 = Ryr2flox/wildtype × Mhy6-MerCreMer+ + tamoxifen.
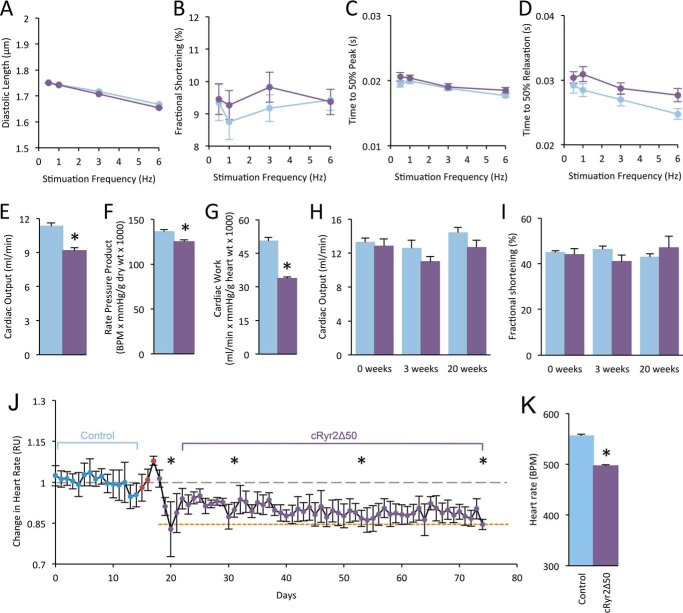
We next examined the function of cRyr2Δ50 cardiomyocytes, in vitro and in vivo. We did not observe significant changes in diastolic length, fractional shortening, the time to 50% peak contraction, or the time to 50% relaxation (Fig. 3, A–D), indicating that contractile function in individual cells is normal. Cardiac function in isolated perfused working hearts revealed small decreases in cardiac output, rate pressure product, and cardiac work in cRyr2Δ50 hearts (Fig. 3, E–G). However, these changes were modest when compared with the dramatic mechanical dysfunction observed in total Ryr2 knock-out hearts (4, 20). Echocardiography failed to show any significant differences in heart function in cRyr2Δ50 mice 3 or 20 weeks following tamoxifen (Fig. 3, H–I). We have previously demonstrated that complete Ryr2 gene knock-out results in reduced heart rate and fatal arrhythmia (4, 20). In the present study, we found an intermediate level of bradycardia in cRyr2Δ50 mice using subcutaneously implanted ECG telemetry in freely moving, unanesthetized mice (Fig. 3, J and K). These experiments suggest that a stable, ∼50% reduction in RYR2 is compatible with well preserved function in vivo and in vitro, although a mild impairment can be identified in the ex vivo working heart system.
FIGURE 3.
Cardiac function and heart rate in cRyr2Δ50 mice. A and B, isolated cardiomyocytes were assessed for average diastolic length (A) and fractional shortening (B) upon stimulation (n = 3; *, p ≤ 0.05.). C and D, isolated cardiomyocyte contraction rate reported as time to 50% peak contraction (C) and time to 50% peak relaxation (D). White circles = control; black circles = cRyr2Δ50. E–G, cardiac output (E), rate pressure product (F), and cardiac work (G) measured during working heart perfusions (control n = 10, cRyr2Δ50 n = 13; *, p ≤ 0.05.). White bars = control; black bars = cRyr2Δ50; throughout. Echocardiograms of control and cRyr2Δ50 mice 3 weeks following tamoxifen treatment are shown. BPM, beats per minute. H and I, average cardiac output (H) and fractional shortening (I) of control and cRyr2Δ50 mice 3 and 20 weeks following tamoxifen treatment (n = 5). J, average heart rate from implantable ECG radio telemetry. Blue points denote days prior to tamoxifen injections (red points), but following surgical recovery and the removal of analgesics. Heart rate is normalized to the average heart rate measured during baseline (gray dashed line). The red dashed line denotes average heart rate of the total cRyr2KO mice reported in our previous publication (4) K, average heart rate before and after tamoxifen treatment (mean ± S.E.; n = 5; *, p ≤ 0.05). All data were plotted as mean ± S.E. Control = Ryr2flox/wildtype + tamoxifen; cRyr2Δ50: Ryr2flox/wildtype × Mhy6-MerCreMer+ + tamoxifen.
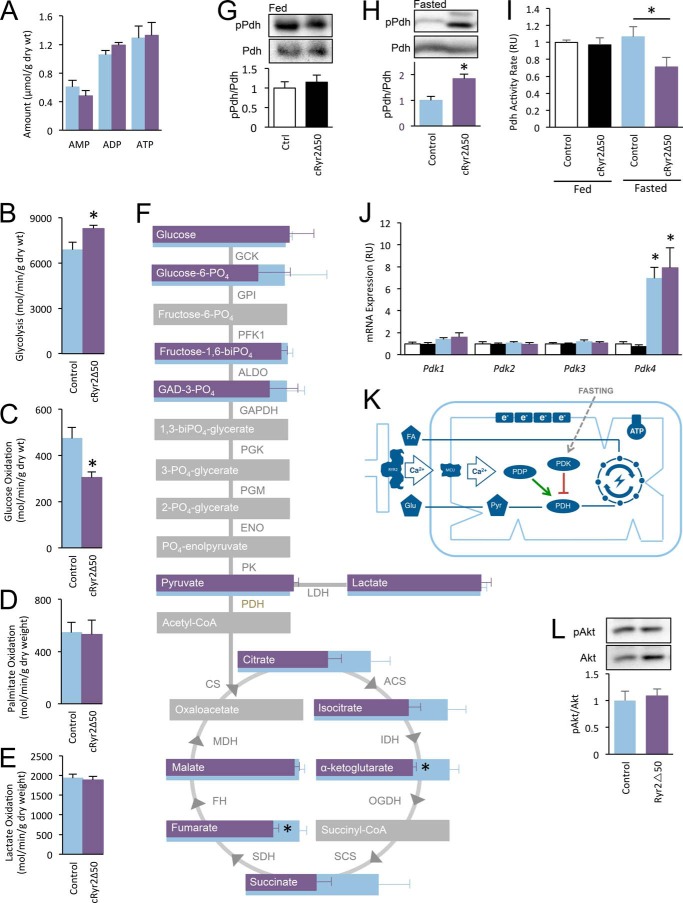
Having shown that cRyr2Δ50 cardiomyocytes have preserved function, we assessed cardiac energy metabolism. Unlike the total RyR2KO hearts (4), there was no change in total ATP levels in cRyr2Δ50 hearts (Fig. 4A), suggesting that they are not significantly energy-deprived. Perfused working cRyr2Δ50 hearts were employed to assess utilization and oxidation of multiple energy substrates. Simultaneous measurements revealed a significant decrease in glucose oxidation and an increase in glycolysis (Fig. 4, B and C). Fatty acid oxidation and lactate oxidation were not altered in these cRyr2Δ50 hearts (Fig. 4, D and E). A metabolomic survey of tricarboxylic acid cycle intermediates in cRyr2Δ50 heart tissue displayed significant decreases in fumarate and α-ketoglutarate, as well as a general trend toward decreased levels of other tricarboxylic acid cycle intermediates (Fig. 4F). Glycolytic metabolites upstream of entry into the tricarboxylic acid cycle, including pyruvate, remained unchanged in cRyr2Δ50 hearts (Fig. 4F). These data demonstrate that glucose utilization is altered in cRyr2Δ50 hearts in the absence of major cardiac dysfunction and suggest that Ryr2-mediated Ca2+ flux has a specific role in promoting the full oxidation of glucose in cardiac tissues in vivo.
FIGURE 4.
cRyr2Δ50 mice have a specific defect in glucose oxidation, associated with hyperphosphorylation and reduced activity of Pdh. A, average heart ATP, ADP, and AMP as measured by HPLC-chromatography on whole homogenized hearts (n = 3). B and C, non-oxidative glycolysis (B) and complete glucose oxidation (C) were simultaneously measured in working hearts (control n = 5, cRyr2Δ50 n = 6; *, p ≤ 0.05). Light blue bars = control, purple bars = cRyr2Δ50 throughout. D and E, at the same time, palmitate oxidation (D) and lactate oxidation (E) were measured (control n = 5, cRyr2Δ50 n = 7). F, diagram showing the results of targeted mass spectroscopy-based metabolomic analysis of TCA cycle acid intermediaries and glycolysis pathway sugar intermediates (n = 8; *, p ≤ 0.05; gray = unmeasured). GCK, germinal center kinase; GPI, glycosylphosphatidylinositol; PFK1, phosphofructokinase 1; ALDO, aldo-keto reductase; PGK, phosphoglycerate kinase; PGM, phosphoglycerate mutase; ENO, eno/pyruvate carboxylase; PK, protein kinase. G and H, pyruvate dehydrogenase kinase E1-α subunit serine 293 phosphorylation levels in mice fed ad libitum (white, control n = 3; black, cRyr2Δ50 n = 3; *, p ≤ 0.05) and fasted overnight (blue, control n = 5; purple, cRyr2Δ50 = 6; *, p ≤ 0.05). pPdh, phospho-Pdh; Ctrl, control. I, pyruvate dehydrogenase activity in ad libitum fed and overnight fasted whole homogenized hearts (fed control n = 7, fed cRyr2Δ50 n = 8, fasted control n = 8, fasted cRyr2Δ50 n = 8; *, p ≤ 0.05). J, pyruvate dehydrogenase kinase isoform mRNA levels in ad libitum fed and fasted cRyr2Δ50 hearts (n = 6, *, p ≤ 0.05). K, working model of context-dependent regulation of PDH activity by mitochondrial Ca2+. L, assessment of Akt phosphorylation levels in overnight fasted cRyr2Δ50 hearts (n = 6; *, p ≤ 0.05). All data were plotted as mean ± S.E.
To examine the molecular mechanism involved in this metabolic shift, we first focused on Pdh, the enzyme that catalyzes the conversion of pyruvate to acetyl-CoA. Pdh is a critical gatekeeper between glycolysis and complete glucose oxidation (29), and its activity is regulated by complex mechanisms involving inhibitory phosphorylation at three sites, including serine 293. Pdh phosphorylation is mediated by a family of Pdh kinases (Pdk1–4), whereas its dephosphorylation is catalyzed by a Ca2+-sensitive phosphatase (Pdp1) (17). Initially, we found that Pdh phosphorylation was unchanged in the fed state (Fig. 4G). However, when we fasted mice overnight, cRyr2Δ50 hearts displayed significantly increased Pdh phosphorylation levels (Fig. 4H). Consistent with this, fasted cRyr2Δ50 hearts had significantly lower Pdh enzyme activity (Fig. 4I). There were no significant differences in Pdk expression between control and cRyr2Δ50 hearts, although Pdk4 was profoundly up-regulated following fasting in both groups (Fig. 4J). Together, our data demonstrate that, in the context of increased inhibitory Pdk tone during fasting, cRyr2Δ50 mice are less efficient at dephosphorylating and reactivating Pdh activity than control mice. This is most likely explained by a decrease in the activity of Ca2+-sensitive Pdp1 phosphatase caused by the reduced mitochondrial Ca2+ uptake downstream of Ryr2 ablation (Figs. 2 and 3K). We did not observe a difference in Akt phosphorylation between fasted cRyr2Δ50 and control hearts (Fig. 4L), suggesting that insulin signaling through this pathway was normal under the conditions of our study.
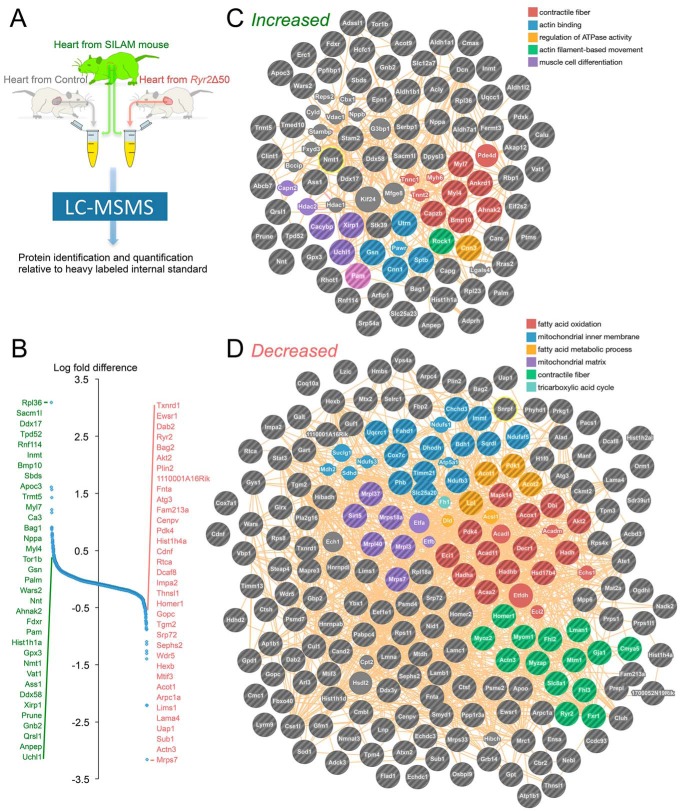
To provide unbiased insight into the functional and metabolic effects of partial Ryr2 reduction, we performed proteomic analysis of extracts from fed-state control and cRyr2Δ50 hearts, spiked with known amounts of SILAM wild type heart tissue (heavy labeled) to allow quantitative comparisons (Fig. 5A) (30). We identified 1702 unique proteins with high confidence after excluding blood-borne contaminants, and then compared their relative enrichment levels using the reference SILAM sample (Fig. 5B; supplemental Transcriptomics Data File). Most quantified proteins were similarly enriched with a few exceptions. The 35 most highly increased proteins included: Sacm1l, a regulator of inositol 1,4,5-trisphosphate (IP3) signaling; Bmp10, a key regulator of heart development; atrial natriuretic factor, a cardio-protective hormone and biomarker of cardiac dysfunction (31); and Ahnak2, a large t-tubule protein that interacts with Ryr2 (32) (Fig. 5B). On the other hand, the 35 most highly decreased proteins included: Homer, which interacts with Ryr2 to decrease its open probability (33); and Ryr2 itself. Network analysis identified clusters of interacting proteins involved in heart muscle contraction, actin binding/motility, and muscle cell differentiation that were increased (Fig. 5C), as well as clusters of proteins involved in fatty acid oxidation/metabolism, mitochondrial inner membranes/matrix, contractile fibers, and TCA cycle that were enriched in the cRyr2Δ50 heart proteome (Fig. 5D). This unbiased survey illustrates that hearts undergo metabolic and contractile remodeling as a result of partial Ryr2 deletion.
FIGURE 5.
Proteomic analysis of cRyr2Δ50 hearts. A, schematic depicting the experimental design of the proteomics experiment, including spike-in with stable isotope-labeled heart tissue (SILAM hearts) to enable quantitative normalization. Four mice per treatment group were pooled and analyzed simultaneously by mass spectroscopy. B, distribution of relative protein abundance measurements, with the 25 most enriched proteins (green) and 25 most depleted proteins (red) between treatment groups listed. C and D, analysis of up-regulated (C) and down-regulated (D) proteins identified significantly enriched networks (false discovery rate < 0.05).
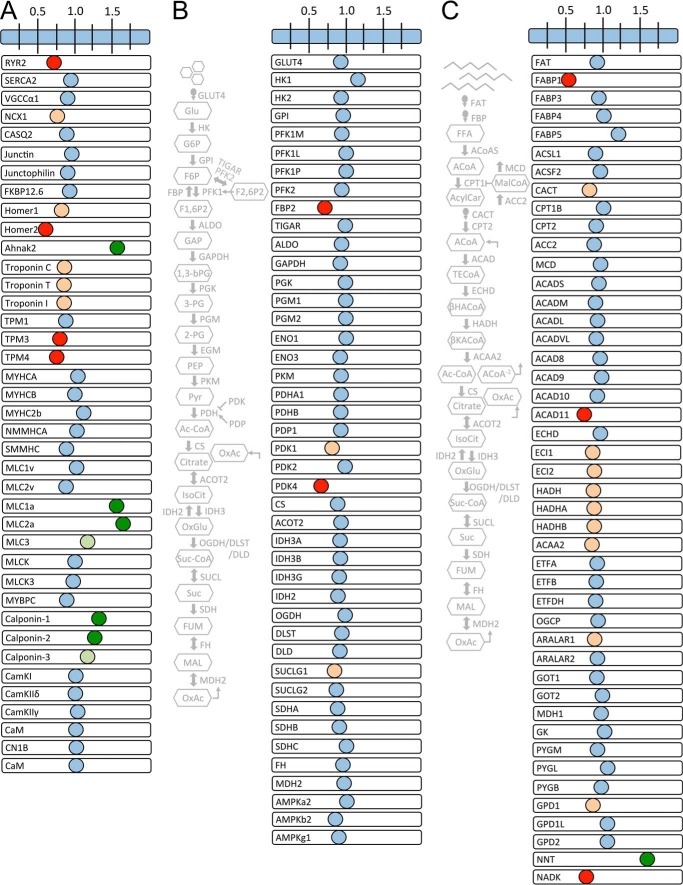
We mined our proteomic data to assess the status of cRyr2Δ50 cardiomyocyte sarcomeres and mitochondria (Figs. 6 and 7). This targeted survey of core excitation-contraction proteins identified a modest decrease in the Na/Ca2+ exchanger Ncx1 (Fig. 6A). We also observed modest decreases in the troponin/tropomyosin complex and a large increase in atrial isoforms of regulatory and essential myosin light chains, which have been associated with heart disease (34), as well as increased levels of calponins, proteins that interact with actin and tropomyosin, which, when overexpressed, can partially rescue hearts from dilated cardiomyopathy (35). A survey of the glycolytic and glucose oxidation pathways showed no changes in core pathway enzymes (Fig. 6B). Instead, we observed reductions only in fructose-1,6-bisphosphatase 2 (Fbp2), an anabolic protein that counteracts the activity of phosphofructokinase 1 (Pfk1), the key gatekeeper of glycolysis (36), as well as Pdk1 and Pdk4, inhibitory kinases of the Pdh glucose oxidation gatekeeper (29) (Fig. 6B). These data suggest a reduction of inhibitory signals to both glycolysis and glucose oxidation in cRyr2Δ50 hearts despite the reduced glucose oxidation and context-dependent Pdh hyperphosphorylation. A similar survey of the fat oxidation pathway shows modest decreases in the carnitine acyl-carnitine transporter and some β-oxidation proteins (Fig. 6C). However, because these changes failed to result in reduced palmitate oxidation rates, these changes must be within the range of dynamic regulation. We also observed a large decrease in NAD kinase and a large increase in nicotinamide N-methyltransferase (NNT), changes that are consistent with the cell trying to preserve reducing equivalents for the TCA cycle but that may also compromise the antioxidant capacity of the cell (37) (Fig. 6C). We did not observe robust changes in the electron transport chain or ATP synthase machinery sub-proteomes (Fig. 7). Collectively, this proteomic analysis revealed changes initiated by the stable 50% reduction in Ryr2 that are likely to contribute to effects on metabolism.
FIGURE 6.
Parallel analysis of functional protein categories in cRyr2Δ50 hearts. A–C, visualization of the changes of key cardiac proteins arranged into functional clusters of excitation-contraction coupling proteins (A), glycolysis and glucose oxidation proteins (B), or fat oxidation and other key metabolic proteins (C). Data are plotted as the -fold difference between cRyr2Δ50 and control hearts (one sample per group made with four pooled hearts per sample to reduce variation). Red, ≤0.75-fold change; orange, ≤0.85 change; pale green, ≥1.15 enrichment; dark green, ≥1.25 enrichment. Gray schematics show location of presented glucose and fat oxidation proteins in their respective pathways.
FIGURE 7.
Electron transport chain, ATP synthase complex, and other metabolic effectors in cRyr2Δ50 hearts. Visualization of the changes of additional cardiac proteins arranged into functional clusters of electron transport chain, metabolic, calcium-sensitive, and cardiac pathology associated proteins is shown. Data are plotted as the -fold difference between cRyr2Δ50 and control hearts (one sample per group made with four pooled hearts per sample to reduce variation). Red represents a ≤0.75-fold change, orange represents a ≤0.85 change, pale green represents a ≥1.15-fold enrichment, and dark green represents a ≥1.25-fold enrichment.
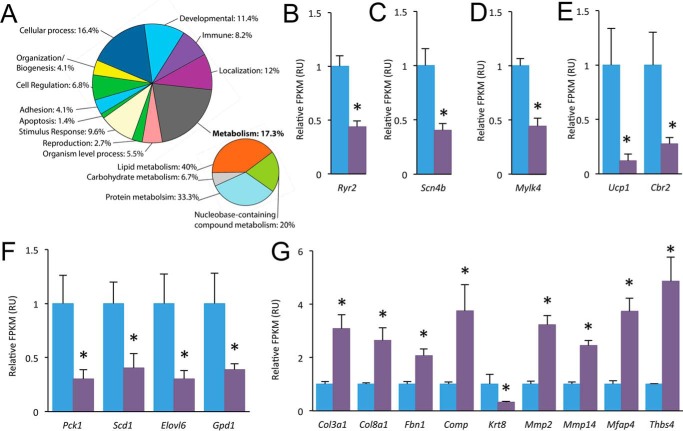
Our mass spectrometry-based proteomic analysis was not able to assess every gene product. Thus, RNA sequencing was also employed to provide genome-wide analysis of changes in fed-state cRyr2Δ50 hearts. There were relatively few changes in the transcriptome of cRyr2Δ50 hearts that reached genome-wide significance (Fig. 8A; supplemental Proteomics Data File). However, notable differences included decreased expression of Ryr2 (Fig. 8B), voltage-gated Na+ channel Scn4b (Fig. 8C), and Mylk4 (Fig. 8D), a novel isoform of myosin light chain kinase that is down-regulated in heart failure (38). We also found decreased expression of Ucp1 and Cbr2 genes that utilize TCA reducing equivalents for non-energy-producing processes but potentially at the expense of protection from reactive oxygen species (39) (Fig. 8E). Biosynthetic genes were also significantly decreased in cRyr2Δ50 hearts, including key gluconeogenesis gene Pck1 and Gpd1, a protein that scavenges glycolysis products for sugar biosynthesis (Fig. 8F). Cbr2 and Gpd1 were also observed to be decreased in the heart proteome (Figs. 6C and 7). We also observed a general up-regulation of extracellular structural and remodeling genes consistent with cardiac pathology (40) (Fig. 8G). In summary, the cRyr2Δ50 heart transcriptome also revealed contractile and metabolic modulation/compensation.
FIGURE 8.
Transcriptomics analysis of cRyr2Δ50 hearts. A, biological Gene Ontology (PANTHER) groups of significantly changed genes in the cRyr2Δ50 heart transcriptome. B–G, changes in the mRNA levels of Ryr2 (B), as well as other genes involved in excitation-contraction (C), contractility (D), energetics (E), biosynthetic pathways (F), and extracellular matrix (G) (n = 4, *, p ≤ 0.05). All data were plotted as mean ± S.E. Control = Ryr2flox/wildtype + tamoxifen; cRyr2Δ50: Ryr2flox/wildtype × Mhy6-MerCreMer+ + tamoxifen.
Discussion
The goal of the present study was to examine the effects of ∼50% reduction in Ryr2 on heart metabolism and function. We found that a stable 50% loss of Ryr2 protein impaired mitochondrial Ca2+ signaling, reduced Ca2+-dependent Pdh activation, and specifically reduced oxidative glucose metabolism. Importantly, these changes occurred in the absence of robust mechanical dysfunction, meaning that they are primary effects downstream of Ryr2. Unbiased proteomic and transcriptomic surveys of cRyr2Δ50 hearts revealed alterations in contractile and metabolic gene networks predicted to increase glucose oxidation and Ryr2 Ca2+ release, suggesting mechanisms for compensation in this model.
Our principle finding is that Ryr2 specifically promotes glucose oxidation in cardiomyocytes. A wealth of in vitro research has shown that mitochondrial Ca2+ signaling is important for the oxidation of energy fuel substrates by stimulating TCA cycle enzymes, the electron transport chain, and ATP synthase (17–19). It had been previously proposed that glucose oxidation is controlled by Ca2+ signaling via the key gatekeeper for pyruvate entry into the TCA cycle, Pdh, which is activated by a Ca2+-sensitive phosphatase (Pdp) (17, 29). To the best of our knowledge, our work is the first in vivo demonstration of an ionic mechanism that specifically controls glucose oxidation, but not fat oxidation. The finding that Ryr2-to-mitochondria Ca2+ signals play a specific and sensitive role in promoting glucose oxidation may represent a key mechanism by which the metabolic demands of excitation-contraction are directly coupled to the rate of ATP production in cardiomyocytes.
Glucose oxidation contributes to cardiomyocyte energy metabolism and is preferentially up-regulated in periods of oxidative stress or increased metabolic demand (14). It is notable that we only observed the effects of Ryr2 haploinsufficiency on Pdh phosphorylation/activity in fasted cRyr2Δ50 mice. Fasting increased Pdk4 expression, which would be expected to increase the inhibitory phosphorylation tone on the Pdh system (41). In this metabolic stress situation, the phosphatase activity of Pdp may be more important for reactivating Pdh to allow for effective glucose oxidation (42). Our findings suggest that, in this context, Ryr2 becomes necessary to fully drive Pdp activity and prevent hyperphosphorylation of Pdh. The effects of Ryr2 on Pdh activation may have been partially obscured by basal reductions in Pdk1 and Pdk4 in the fed-state cRyr2Δ50 heart proteome, suggesting the possibility that Pdh activation may normally be even more sensitive to Ryr2 than our experiments revealed. Nevertheless, our data clearly demonstrate, for the first time, that Ryr2 is critically involved in preferentially increasing glucose oxidation during metabolic stress. The fasting dependence of this system is reminiscent of the conditions required to uncover changes in Pdh regulation in mice lacking the mitochondrial Ca2+ uniporter, which required either a metabolic or β-adrenergic stress to display altered Pdh regulation (23, 25).
Our previous research measured the effects of total Ryr2 deletion on oxidative metabolism and reported reduced heart ATP levels and a general reduction in total oxidative energy metabolism (20). The present study demonstrates that a 50% Ryr2 reduction specifically disrupts glucose oxidation, which suggests that there may be a hierarchy where oxidative metabolism of multiple substrates is generally Ca2+-sensitive, but glucose oxidation is attuned to more subtle changes in mitochondrial Ca2+. Because glucose oxidation and glycolysis are frequently uncoupled in models of heart disease before the general impairment of oxidative metabolism seen in heart failure (14, 15), these data also suggest that progressive Ryr2 dysfunction in disease may contribute to mounting metabolic dysfunction in a “dose-dependent” manner.
Our results indicate that a 50% reduction in Ryr2 changes the frequency and amplitude of cytosolic and mitochondrial Ca2+ signals. Because groups of Ryr2 normally function in concert, due to proximity and physical interaction (8), it is possible that decreasing Ryr2 abundance reduced the functional coupling within the Ryr2 signaling apparatus, leading to more gradual activation and deactivation of Ca2+ transients. Our data also suggest disrupted privileged communication between Ryr2 and mitochondria. Mitochondria rely on microdomain signaling where the mitochondrial Ca2+ uniporter is paired with Ca2+ release channels to drive mitochondrial Ca2+ uptake (16). Our data suggest that when Ryr2 is reduced, Ca2+ cannot flow as quickly between these two organelles via microdomain signaling. It is possible that deleting 50% of Ryr2 proteins disrupts SR-mitochondria tethering (43, 44). Ultra-structural/superresolution analysis of Ryr2 in this mouse model could form the basis of a future study.
Another important finding of this study is that a 50% decrease in Ryr2 is sufficient to significantly reduce heart rate. In our previous work on total cRyr2 knock-out mice, we noted a substantial reduction in heart rate, as well as tachycardic arrhythmias (4). Our results here show that even in the absence of acute mechanical dysfunction, stable Ryr2 haploinsufficiency is sufficient to reduce heart rate, which provides evidence that this is a bona fide effect of Ryr2 signaling and not a consequence of heart dysfunction (4). This provides further strong evidence that Ryr2 is critical for cardiac pace-making and supports a model where heart rate is regulated by an ensemble of SR and plasma membrane ion channels (2).
The cRyr2Δ50 mouse line is a major improvement on previous loss-of-function Ryr2 models as it is not lethal and does not show the dramatic changes in cardiac mechanical function found in total cRyr2KO mice (heart-specific, inducible Ryr2 gene knockout mice). Our cRyr2Δ50 model also greatly reduces the potential for confounding long-term compensatory effects of global, life-long mutations (7), as well as the putative period of tamoxifen drug effects observed in some studies (45). Additionally, induced Ryr2 haploinsufficiency models a situation analogous to disease conditions with a decrease in either Ryr2 expression or function (9–13) (Fig. 1A). Indeed, numerous studies have reported reduced Ryr2 levels or function in diabetic cardiomyopathy (11), heart failure (12, 13), and aging (10), implicating Ryr2 dysfunction as an element of heart disease and its predisposing conditions. Heart disease is also associated with reductions in the oxidative capacity of the heart and reductions in the metabolic flexibility including reduced glucose oxidation and increased glycolysis (4). Our study shows that a chronic 50% reduction of Ryr2 in adult animals, similar to what is seen in multiple disease states, is sufficient to disrupt normal glucose oxidation in cardiomyocytes. Thus, Ryr2 dysfunction may contribute to the impaired metabolic flexibility seen in heart disease.
Materials and Methods
Experimental Animals
All animal protocols were approved by the University of British Columbia Animal Care Committee in accordance with international guidelines. Mice with “floxed” Ryr2 alleles were generated and characterized previously (4, 5, 20). Tamoxifen-inducible, cardiomyocyte-specific Ryr2 haploinsufficiency mice (cRyr2Δ50 mice) were generated by crossing C57Bl6 Ryr2flox/wildtype mice with C57Bl6 mer-cre-mer; strain: 005657, The Jackson Laboratory, Bar Harbor, ME). Tamoxifen was injected i.p. into 8–16-week-old Ryr2flox/wildtype:mer-Cre-mer mice and littermate control mice (Ryr2flox/wildtype mice injected with tamoxifen) for 3 consecutive days at 3 mg/40 g of body weight. All mice were given >3 weeks recovery after tamoxifen to reach stable Ryr2 ablation and to circumvent any possible effects of tamoxifen, which is cleared from mice within 21 days (Fig. 1B) (45).
Ryr2 puncta quantification employed a Ryr2-GFP fusion protein knock-in allele (28). Cre-expressing, tamoxifen injected Ryr2flox/GFP mice were compared with tamoxifen-injected Ryr2GFP/GFP littermate controls.
C56Bl6 SILAM mice (30) were generated by feeding mice MouseExpress l-LYSINE (13C6, 99%) at 8 g/kg of diet (Cambridge Isotope Laboratories, Andover, MA) for two generations. Amino acid incorporation within long-lived neuronal tissue was confirmed to be >97%.
To assess differences in Pdh activity, some mice were fasted for 16 h. All other data were collected from ad libitum fed mice with a rodent chow diet.
Imaging and Functional Measurements
All live cell analysis was conducted on cardiomyocytes isolated from cRyr2Δ50 and control mice using Langendorff reverse perfusion to introduce collagenase to the cardiac vasculature (46). All live cardiomyocyte analysis occurred within 8 h of cell isolation, and only rod-shaped cardiomyocytes with typical cell morphology were considered in analysis.
Ryr2-GFP puncta were imaged in cardiomyocytes counterstained with 5 μm MitoTracker Red FM (Thermo Fisher) for 15 min, followed by a 5-min wash, using a 100×/1.45 oil objective on a spinning disk confocal system based on Zeiss Axiovert 200M microscope. 3D image stacks, acquired with identical settings, were deblurred by the Nearest Neighbor Deconvolution module of the SlideBook 6 software (Intelligent Imaging Innovations, Denver, CO), and five representative planes evenly distributed within each cell were selected for further quantifications using the CellProfiler software (Broad Institute). Unprocessed images were used for the analysis related to the fluorescence intensity of the identified Ryr2-GFP puncta.
Cardiomyocyte Ca2+ sparks of isolated cells loaded with 5 μm Fluo-4-AM were imaged through the 63× objective of a Zeiss LSM 700 confocal microscope in line scan mode (1053 lines per second, 0.142-μm pixel size) in minimal media. Sparks were quantified using SparkMaster, an ImageJ plugin (47).
To measure whole-cell Ca2+ signals, we employed a ratiometric Ca2+-sensitive fluorescent dye, Fura-2-acetoxymethyester (AM) (Invitrogen). Mitochondrial Rhod-2-AM (Invitrogen) localization was confirmed by co-loading cardiomyocytes with 5 μm MitoTracker Deep Red (644/655 nm) (Invitrogen) and imaging at high resolution using a 100 × 1.45 NA objective on a Zeiss Axiovert-200M microscope with a CoolSnapHQ2 CCD Camera (Intelligent Imaging Innovations). Simultaneous cytosolic Ca2+ and mitochondrial Ca2+ measurements were performed on isolated cardiomyocytes, which were incubated with 5 μm Fura-2-AM (340 and 380 nm excitation; >510 nm emission) and 5 μm Rhod-2-AM (550 nm excitation/580 nm emission) for 15 min and washed for 5 min before imaging using a 10 × 0.5 NA objective Zeiss Axiovert-200M microscope with a CoolSnapHQ2 CCD Camera (Intelligent Imaging Innovations). Additionally, imaged cells were provided a field stimulation of 80 mA, 5-ms pulses with a hybrid system using a stimulus isolator (World Precision Instruments, Sarasota, CA) and a custom-made programmable stimulator (McAfee Scientific) with periods of: individual analog pulses (to test for responding cells), 0.5-Hz continuous stimulation (to measure Ca2+ transients in the context of full cell relaxation), or 6-Hz continuous stimulation (to approximate the rapid stimulation of an in vivo heartbeat). This range was also chosen to correlate with the cardiomyocyte function and contractility experiment. Fura-2 ratios to measure relative cytosolic Ca2+ and Rhod-2 intensities to measure mitochondrial Ca2+ were quantified using the SlideBook software (Intelligent Imaging Innovations). Along with the prior conditions, only cardiomyocytes that displayed stable non-stimulated Fura-2 and Rhod-2 baselines as well as excitability to an initial five field stimulation pulses were considered in this analysis. Cardiomyocytes from at least three independent isolations per treatment group were studied.
Cardiomyocyte function and contractility were assessed as described elsewhere (48). Briefly, a suspension of the isolated ventricular cardiomyocytes was transferred to a chamber with stimulating electrodes (Cell MicroControls, Norfolk, VA) that was precoated each day with laminin (1 mg/ml) to help immobilize the cells, and then fixed to the heated stage of an Olympus IX70 inverted microscope with 400× quartz optics. The cells were continuously superfused with Tyrode's solution consisting of 1.5 mmol/l CaCl2 in 95% O2, 5% CO2 at 0.5–1 ml/min at 34–36 °C. An IonOptix (Milton, MA) video system imaged the cells at 240 Hz, allowing measurement of the degree and rate of myocyte shortening during field stimulation at various frequencies using a programmable stimulator.
In Vivo and ex Vivo Functional Analysis
Cardiac function was examined by echocardiography as described previously (4). Briefly, we utilized a Vevo 770 high-resolution image system (FUJIFILM VisualSonics, Inc., Toronto, Ontario, Canada) (49). Left ventricular mass and systolic function, left ventricular end-systolic and end-diastolic dimensions, interventricular septum thickness, and posterior wall were measured from M-mode traces. Shortening and ejection fraction were calculated as described (49).
Heart rate was assessed by echocardiography and concurrent ECG as described (4). In vivo heart rate was also assessed using implantable ETA-F10 ECG radiotelemetry as described (4). Heart rate and rhythmicity were analyzed following recovery from surgery and after the discontinuation of all analgesic drugs.
Ex vivo analyses of heart function and metabolism were carried out using the working heart perfusion model (4, 20, 50). Myocardial substrate utilization was measured in working hearts as detailed elsewhere (20, 51). Glycolysis, as well as myocardial rates of oxidation of palmitate, glucose, and lactate were determined by the quantitative collection of 3H2O or 14CO2 produced by hearts perfused with Krebs-Henseleit solution-containing either [9,10-3H]palmitate and [U-14C]lactate or [U-14C]glucose and [5-3H]glucose.
Metabolomics
Mouse hearts were weighed, and then homogenized in 375 μl of 50% MeOH/100 mg of tissue weight with the aid of two 5-mm metal balls shaken for 1 min at 30 Hz. Sample temperature was kept below −10 °C during homogenization. 375 μl of methanol/100 mg of homogenate weight were then added, and the sample was homogenized again for 2 × 1 min. The tube was placed on ice for 30 min and centrifuged at 4 °C and 13,000 rpm for 20 min. The supernatant was collected and stored at −20 °C for UPLC-FTMS and UPLC-MRM/MS. Quantification of carboxylic acids was performed using chemical derivatization with 3-nitrophenylhydrazine followed by subsequent UPLC-MRM/MS determination according to a protocol as described elsewhere (52). Quantitation of glucose, other datable aldoses, and reducing sugar phosphates in the mouse heart tissues was performed using chemical derivatization with anion exchange chromatography followed by UPLC-MRM/MS determination using a previously described protocol (53). Selective quantitation of fructose 1,6-bisphosphate was performed using UPLC-MRM/MS with chemical derivatization. AMP, ADP, and ATP levels were assessed using high-performance liquid chromatography in freeze-clamped hearts.
Pdh Activity Assay
Pdh activity was measured in proteins isolated from clamp-frozen, homogenized heart. Buffers included 2 mm EDTA, 2 mm NaF, 1× Complete mini EDTA-free protease inhibitor cocktail tablet per 10 ml of buffer (Roche Applied Science, Basel, Switzerland), and 100 μl of sodium orthovanadate gel to preserve Pdh phosphorylation. Pdh activity and quantity were measured using a Pdh Microplate Assay KIT (Abcam, Cambridge, UK). Pdh activity was normalized to total Pdh in each sample, as well as an internal normalization factor to account for run-to-run variations.
Transcriptomics
RNA was isolated from heart tissue using TRIzol, followed by cleanup (RNeasy; Qiagen, Venlo, The Netherlands). After reverse transcription (SuperScript III; Invitrogen), TaqMan quantitative RT-PCR (qPCR) was conducted using probes from Applied Biosystems (Carlsbad, CA) and PerfeCTa qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD) on a StepOnePlus thermocycler (Applied Biosystems). SYBR Green quantitative RT-PCR was conducted using PerfeCTa SYBR Green qPCR SuperMix (Quanta Biosciences, Gaithersburg MD). Relative gene expression changes were analyzed by the 2−ΔCt method and plotted in normalized relative units (RU). Hypoxanthine-guanine phosphoribosyltransferase (HPRT) and cyclophilin were used as internal controls, after ensuring that they were not altered in cRyr2KO cardiomyocytes. TaqMan cyclophilin primers were 5′-GTCTGCAAACAGCTCGAA-3′, 5′-ACGCCACTGTCGCTTT-3′, and 5′-/56-FAM/TGCAGCCATGGTCAAC-3′ (produced by Integrated DNA Technologies, Coralville, IA), where FAM is 6-carboxyfluorescein. Additional primer details are available in supplemental Tables S1–S4.
RNA sequencing employed Ion Torrent® technology (Life Technologies). Specifically, total RNA was quantified using the QubitTM RNA Assay Kit and the Qubit® 2.0 Fluorometer, and its RNA integrity number was measured with the Agilent RNA 6000 Nano Kit on the 2100 Bioanalyzer instrument with the 2100 Expert software. Isolation of mRNA was then performed using the Dynabeads® mRNA DIRECT Micro Kit. Whole transcriptome libraries were constructed using the Ion Total RNA-Seq Kit on the AB Library BuilderTM System. Yield and size distribution of the transcriptome libraries were assessed using the Agilent High Sensitivity DNA Kit on the 2100 Bioanalyzer. Templating was constructed using the Ion OneTouchTM 2 System using the Ion PITM Template OT2 200 Kit V3 before sequencing using the Ion PITM Sequencing 200 Kit V3 on the Ion ProtonTM System. Initial analysis and gene counts were performed on the Torrent SuiteTM software (version 4.2.1). Downstream analysis was conducted using the Tuxedo pipeline as well as the CummeRbund software suite in R software (54).
Proteomics
Targeted protein measurement was conducted using Western blots on lysates from mechanically disrupted hearts, which were homogenized and sonicated in ice-cold lysis buffer. Samples were quantified and boiled with loading dye, and 15–50 μg of protein were used in SDS-PAGE electrophoresis. Proteins were then transferred to PVDF membranes using standard semi-dry (≤120 kDa) or wet transfer (>120 kDa) approaches and subsequently treated with targeted primary and horseradish peroxidase-conjugated secondary antibodies. Bands were visualized using an enhanced chemiluminescence detection kit, and then quantified by densitometry. The following commercial antibodies were used: anti-Eea1 (Ab2900, Abcam, Cambridge, UK), anti-PdhS293 (NB110-93429, Novus Biologicals, Littleton, CO), anti-Pdh (Ab110330, Abcam), anti-pAktS473 (9271, Cell Signaling, Danvers, MA), and anti-Akt (2966, Cell Signaling). Rabbit polyclonal anti-RYR2 antibodies were provided by Dr. Anthony Lai. For Ryr2 Western blots, we utilized anti-Eea1 as a large protein loading control, which we previously validated (as described in Ref. 4).
Hearts were briefly perfused to reduce the number of red blood cells and blood-borne proteins, and then rapidly frozen, prior to proteomic analysis. Frozen hearts were mechanically homogenized, and then proteins were isolated and treated as described previously (55). Isolated protein samples from four individual mice per treatment group were then pooled, quantified, and mixed 1:1 by weight with SILAM heart tissue (30) that was prepared in the same manner, before final sample processing as described (55). Sample digestion and offline fractionation were also conducted as described (55) with the exception that endoproteinase LysC was used in place of trypsin for the full digestion. High-pH reverse phase fractionation and mass spectroscopy were conducted as described previously (55). Briefly, each sample was fractionated by high-pH reversed phase on an Agilent Zorbax Extend-C18 analytical column (5 μm, 4.6 × 50 mm) into eight fractions per sample, which were then measured using a Thermo Q-Exactive Hybrid Quadrupole Mass Spectrometer coupled to a Thermo Easy nLC-1000 Liquid Chromatograph. Each fraction was resolved on a 180-min gradient and analyzed on the mass spectrometer in positive ion mode, at 70,000 resolution, from 300 to 20,000 m/z. All mass spectrometry data were captured in profile mode. The 10 most intense peaks were selected (underfill ratio of 10%, 2.2 m/z isolation window, and a normalized collision energy of 28 with 20% stepping) for fragmentation by higher-energy collisional dissociation, and were excluded thereafter for 30 s. Peptides with unassigned charge states, and those with charge 1, were not selected. Peptide identification was carried out using the Andromeda algorithm using the MaxQuant software version 1.5.0.0 from the mouse UniProt database (July 2014), largely using the following settings: endoproteinase LysC cleavage specificity, maximum two missed cleavages, including carbamidomethyl cysteine as a fixed modification and protein N-terminal acetylation, methionine oxidation, and asparagine and glutamine deamination as variable modifications. Match between runs and dependent peptide search were both used in addition to the default 1% false discovery rate settings. Razor peptides were used for quantification, and MaxQuant re-quantification was enabled. Obvious blood-borne, non-cardiac proteins (e.g. albumin) were ignored.
Statistical Analysis
Data are expressed as mean ± S.E., unless otherwise indicated. Results were considered statistically significant when p ≤ 0.05 using Student's t test or, where appropriate, two-factor mixed design analysis of variance with repeated measures and Bonferroni's post test. All experiments were repeated on at least three cRyr2Δ50 mice and at least three of their tamoxifen-injected littermate controls (Ryr2flox/wildtype) unless otherwise specified.
Author Contributions
M. J. B. conceived studies and wrote the manuscript. M. J. B., R. W., H. C., P. A., R. F. A., J. H., D. M., and M. P. designed and performed experiments and analyzed data. R. W., H. C., P. A., R. F. A., J. H., D. M., M. P., N. E. S., L. B., J. E. K., S. R. W. C., D. F., R. W. B., C. H. B., L. J. F., T. M., and M. F. A. reviewed the manuscript. N. E. S. designed and performed experiments. L. B. and J. E. K. performed experiments and analyzed data. S. R. W. C. and D. F. provided access to essential resources. R. W. B., C. H. B., L. J. F., T. M., and E. D. W. M. designed studies and provided access to essential resources. M. F. A. conceived studies, designed studies, and provided access to essential resources. J. D. J. conceived studies, analyzed data, co-wrote/edited the manuscript, and is the ultimate guarantor of this work.
Supplementary Material
Acknowledgments
We thank Nikolay Stoynov for assistance with mass spectrometry. We thank Dr. A. Lai for the Ryr2 antibody. We thank Dr. Carles Vilariño-Güell and Daniel Bong for access to RNA-seq resources. Mass spectrometry infrastructure used here was supported by the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund.
This work was supported by University of British Columbia Start-Up Funds (to J. D. J.), funding from the Canadian Institutes of Health Research (CIHR) (MOP-77688) (to L. J. F.) and (MOP 115158) (to E. D. W. M.), a scholarship from CIHR (to M. J. B.), a scholarship from the Michael Smith Foundation for Health Research (to N. E. S.), and Natural Sciences and Engineering Research Council of Canada (NSERC) and the Heart and Stroke Foundation/Libin Cardiovascular Institute Professorship in Cardiovascular Research (to S. R. W. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- Ryr2
- ryanodine receptor type 2
- Pdh
- pyruvate dehydrogenase
- Pdk
- pyruvate dehydrogenase kinase
- Pdp
- pyruvate dehydrogenase phosphatase
- SILAM
- stable isotope labeling of amino acids in mice
- SR
- sarcoplasmic reticulum
- TCA
- tricarboxylic acid
- AM
- acetoxymethyester
- UPLC-FTMS
- ultra performance-Fourier transform mass spectrometry
- UPLC-MRM/MS
- ultra performance-liquid chromatography-multiple reaction monitoring-mass spectrometry
- qPCR
- quantitative RT-PCR
- RU
- relative units.
References
- 1. Lanner J. T., Georgiou D. K., Joshi A. D., and Hamilton S. L. (2010) Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb. Perspect. Biol. 2, a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Monfredi O., Maltsev V. A., and Lakatta E. G. (2013) Modern concepts concerning the origin of the heartbeat. Physiology 28, 74–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. George C. H., Jundi H., Thomas N. L., Fry D. L., and Lai F. A. (2007) Ryanodine receptors and ventricular arrhythmias: emerging trends in mutations, mechanisms and therapies. J. Mol. Cell. Cardiol. 42, 34–50 [DOI] [PubMed] [Google Scholar]
- 4. Bround M. J., Asghari P., Wambolt R. B., Bohunek L., Smits C., Philit M., Kieffer T. J., Lakatta E. G., Boheler K. R., Moore E. D. W., Allard M. F., and Johnson J. D. (2012) Cardiac ryanodine receptors control heart rate and rhythmicity in adult mice. Cardiovasc. Res. 96, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang H. T., Tweedie D., Wang S., Guia A., Vinogradova T., Bogdanov K., Allen P. D., Stern M. D., Lakatta E. G., and Boheler K. R. (2002) The ryanodine receptor modulates the spontaneous beating rate of cardiomyocytes during development. Proc. Natl. Acad. Sci. U.S.A. 99, 9225–9230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkins B. J., and Molkentin J. D. (2004) Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem. Biophys. Res. Commun. 322, 1178–1191 [DOI] [PubMed] [Google Scholar]
- 7. Zou Y., Liang Y., Gong H., Zhou N., Ma H., Guan A., Sun A., Wang P., Niu Y., Jiang H., Takano H., Toko H., Yao A., Takeshima H., Akazawa H., et al. (2011) Ryanodine receptor type 2 is required for the development of pressure overload-induced cardiac hypertrophy. Hypertension 58, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 8. Scriven D. R. L., Asghari P., and Moore E. D. W. (2013) Microarchitecture of the dyad. Cardiovasc. Res. 98, 169–176 [DOI] [PubMed] [Google Scholar]
- 9. Crossman D. J., Ruygrok P. N., Ruygrok P. R., Soeller C., and Cannell M. B. (2011) Changes in the organization of excitation-contraction coupling structures in failing human heart. PLoS ONE 6, e17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kandilci H. B., Tuncay E., Zeydanli E. N., Sozmen N. N., and Turan B. (2011) Age-related regulation of excitation-contraction coupling in rat heart. J. Physiol. Biochem. 67, 317–330 [DOI] [PubMed] [Google Scholar]
- 11. Bidasee K. R., Dinçer Ü. D, and Besch H. R. Jr. (2001) Ryanodine receptor dysfunction in hearts of streptozotocin-induced diabetic rats. Mol. Pharmacol. 60, 1356–1364 [DOI] [PubMed] [Google Scholar]
- 12. Matsui H., MacLennan D. H., Alpert N. R., and Periasamy M. (1995) Sarcoplasmic reticulum gene expression in pressure overload-induced cardiac hypertrophy in rabbit. Am. J. Physiol. Cell Physiol. 268, C252–C258 [DOI] [PubMed] [Google Scholar]
- 13. Milnes J. T., and MacLeod K. T. (2001) Reduced ryanodine receptor to dihydropyridine receptor ratio may underlie slowed contraction in a rabbit model of left ventricular cardiac hypertrophy. J. Mol. Cell. Cardiol. 33, 473–485 [DOI] [PubMed] [Google Scholar]
- 14. Stanley W. C., Recchia F. A., and Lopaschuk G. D. (2005) Myocardial substrate metabolism in the normal and failing heart. Physiol. Rev. 85, 1093–1129 [DOI] [PubMed] [Google Scholar]
- 15. Doenst T., Nguyen T. D., and Abel E. D. (2013) Cardiac metabolism in heart failure: implications beyond ATP production. Circ. Res. 113, 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rizzuto R., and Pozzan T. (2006) Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol. Rev. 86, 369–408 [DOI] [PubMed] [Google Scholar]
- 17. Denton R. M. (2009) Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta. 1787, 1309–1316 [DOI] [PubMed] [Google Scholar]
- 18. Glancy B., Willis W. T., Chess D. J., and Balaban R. S. (2013) Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry 52, 2793–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glancy B., and Balaban R. S. (2012) Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry 51, 2959–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bround M. J., Wambolt R., Luciani D. S., Kulpa J. E., Rodrigues B., Brownsey R. W., Allard M. F., and Johnson J. D. (2013) Cardiomyocyte ATP production, metabolic flexibility, and survival require calcium flux through cardiac ryanodine receptors in vivo. J. Biol. Chem. 288, 18975–18986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., Csordás G., Jowdy C., Schneider T. G., Csordás N., Wang W., Liu Y., Kohlhaas M., Meiser M., Bergem S., Nerbonne J. M., Dorn G. W. 2nd, and Maack C. (2012) Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca2+ crosstalk. Circ. Res. 111, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu T., and O'Rourke B. (2008) Enhancing mitochondrial Ca2+ uptake in myocytes from failing hearts restores energy supply and demand matching. Circ. Res. 103, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pan X., Liu J., Nguyen T., Liu C., Sun J., Teng Y., Fergusson M. M., Rovira I. I., Allen M., Springer D. A., Aponte A. M., Gucek M., Balaban R. S., Murphy E., and Finkel T. (2013) The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 15, 1464–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gong G., Liu X., and Wang W. (2014) Regulation of metabolism in individual mitochondria during excitation-contraction coupling. J. Mol. Cell. Cardiol. 76, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luongo T. S., Lambert J. P., Yuan A., Zhang X., Gross P., Song J., Shanmughapriya S., Gao E., Jain M., Houser S. R., Koch W. J., Cheung J. Y., Madesh M., and Elrod J. W. (2015) The mitochondrial calcium uniporter matches energetic supply with cardiac workload during stress and modulates permeability transition. Cell Rep. 12, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwong J. Q., Lu X., Correll R. N., Schwanekamp J. A., Vagnozzi R. J., Sargent M. A., York A. J., Zhang J., Bers D. M., and Molkentin J. D. (2015) The mitochondrial calcium uniporter selectively matches metabolic output to acute contractile stress in the heart. Cell Rep. 12, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu Y., Rasmussen T. P., Koval O. M., Joiner M.-L. A., Hall D. D., Chen B., Luczak E. D., Wang Q., Rokita A. G., Wehrens X. H. T., Song L.-S., and Anderson M. E. (2015) The mitochondrial uniporter controls fight or flight heart rate increases. Nat. Commun. 6, 6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hiess F., Vallmitjana A., Wang R., Cheng H., ter Keurs H. E. D. J., Chen J, Hove-Madsen L., Benitez R., and Chen S. R. W. (2015) Distribution and function of cardiac ryanodine receptor clusters in live ventricular myocytes. J. Biol. Chem. 290, 20477–20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patel M. S., Nemeria N. S., Furey W., and Jordan F. (2014) The pyruvate dehydrogenase complexes: structure-based function and regulation. J. Biol. Chem. 289, 16615–16623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zanivan S., Krueger M., and Mann M. (2012) In vivo quantitative proteomics: the SILAC mouse. Methods Mol. Biol. 757, 435–450 [DOI] [PubMed] [Google Scholar]
- 31. Song W., Wang H., and Wu Q. (2015) Atrial natriuretic peptide in cardiovascular biology and disease (NPPA). Gene 569, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komuro A., Masuda Y., Kobayashi K., Babbitt R., Gunel M., Flavell R. A., and Marchesi V. T. (2004) The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc. Natl. Acad. Sci. U.S.A. 101, 4053–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pouliquin P., Pace S. M., and Dulhunty A. F. (2009) In vitro modulation of the cardiac ryanodine receptor activity by Homer1. Pflugers Arch. 458, 723–732 [DOI] [PubMed] [Google Scholar]
- 34. Hernandez O. M., Jones M., Guzman G., and Szczesna-Cordary D. (2007) Myosin essential light chain in health and disease. Am. J. Physiol. Heart Circ. Physiol. 292, H1643–H1654 [DOI] [PubMed] [Google Scholar]
- 35. Lu D., Zhang L., Bao D., Lu Y., Zhang X., Liu N., Ge W., Gao X., Li H., and Zhang L. (2014) Calponin1 inhibits dilated cardiomyopathy development in mice through the ϵPKC pathway. Int. J. Cardiol. 173, 146–153 [DOI] [PubMed] [Google Scholar]
- 36. Mor I., Cheung E. C., and Vousden K. H. (2011) Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb. Symp. Quant. Biol. 76, 211–216 [DOI] [PubMed] [Google Scholar]
- 37. Sheeran F. L., Rydström J., Shakhparonov M. I., Pestov N. B., and Pepe S. (2010) Diminished NADPH transhydrogenase activity and mitochondrial redox regulation in human failing myocardium. Biochim. Biophys. Acta 1797, 1138–1148 [DOI] [PubMed] [Google Scholar]
- 38. Herrer I., Roselló-Lletí E., Rivera M., Molina-Navarro M. M., Tarazón E., Ortega A., Martínez-Dolz L., Triviño J. C., Lago F., González-Juanatey J. R., Bertomeu V., Montero J. A., and Portolés M. (2014) RNA-sequencing analysis reveals new alterations in cardiomyocyte cytoskeletal genes in patients with heart failure. Lab. Invest. 94, 645–653 [DOI] [PubMed] [Google Scholar]
- 39. Hoerter J., Gonzalez-Barroso M.-D.-M., Couplan E., Mateo P., Gelly C., Cassard-Doulcier A.-M., Diolez P., and Bouillaud F. (2004) Mitochondrial uncoupling protein 1 expressed in the heart of transgenic mice protects against ischemic-reperfusion damage. Circulation 110, 528–533 [DOI] [PubMed] [Google Scholar]
- 40. de Haas H. J., Arbustini E., Fuster V., Kramer C. M., and Narula J. (2014) Molecular imaging of the cardiac extracellular matrix. Circ. Res. 114, 903–915 [DOI] [PubMed] [Google Scholar]
- 41. Jeong J. Y., Jeoung N. H., Park K.-G., and Lee I.-K. (2012) Transcriptional regulation of pyruvate dehydrogenase kinase. Diabetes Metab J. 36, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun W., Liu Q., Leng J., Zheng Y., and Li J. (2015) The role of Pyruvate Dehydrogenase Complex in cardiovascular diseases. Life Sci. 121, 97–103 [DOI] [PubMed] [Google Scholar]
- 43. García-Pérez C., Schneider T. G., Hajnóczky G., and Csordás G. (2011) Alignment of sarcoplasmic reticulum-mitochondrial junctions with mitochondrial contact points. Am. J. Physiol. Heart Circ. Physiol. 301, H1907–H1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Min C. K., Yeom D. R., Lee K. E., Kwon H. K., Kang M., Kim Y. S., Park Z. Y., Jeon H., and Kim D. H. (2012) Coupling of ryanodine receptor 2 and voltage-dependent anion channel 2 is essential for Ca2+ transfer from the sarcoplasmic reticulum to the mitochondria in the heart. Biochem. J. 447, 371–379 [DOI] [PubMed] [Google Scholar]
- 45. Koitabashi N., Bedja D., Zaiman A. L., Pinto Y. M., Zhang M., Gabrielson K. L., Takimoto E., and Kass D. A. (2009) Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ. Res. 105, 12–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fernando V. (2006) Cardiovascular Proteomics: Methods and Protocols, Humana Press, NJ, 10.1385/1597452149 [DOI] [Google Scholar]
- 47. Picht E., Zima A. V., Blatter L. A., and Bers D. M. (2007) SparkMaster: automated calcium spark analysis with ImageJ. Am. J. Physiol. Cell Physiol. 293, C1073–C1081 [DOI] [PubMed] [Google Scholar]
- 48. Williams S., Pourrier M., McAfee D., Lin S., and Fedida D. (2014) Ranolazine improves diastolic function in spontaneously hypertensive rats. Am. J. Physiol. Heart Circ. Physiol. 306, H867–H881 [DOI] [PubMed] [Google Scholar]
- 49. Gomez A. M., and Richard S. (2004) Mutant cardiac ryanodine receptors and ventricular arrhythmias: is “gain-of-function” obligatory? Cardiovasc. Res. 64, 3–5 [DOI] [PubMed] [Google Scholar]
- 50. Allard M. F., Parsons H. L., Saeedi R., Wambolt R. B., and Brownsey R. (2007) AMPK and metabolic adaptation by the heart to pressure overload. Am. J. Physiol. Heart Circ. Physiol. 292, H140–H148 [DOI] [PubMed] [Google Scholar]
- 51. Allard M. F., Schönekess B. O., Henning S. L., English D. R., and Lopaschuk G. D. (1994) Contribution of oxidative metabolism and glycolysis to ATP production in hypertrophied hearts. Am. J. Physiol. 267, H742–H750 [DOI] [PubMed] [Google Scholar]
- 52. Han J., Gagnon S., Eckle T., and Borchers C. H. (2013) Metabolomic analysis of key central carbon metabolism carboxylic acids as their 3-nitrophenylhydrazones by UPLC/ESI-MS. Electrophoresis 34, 2891–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han J., Tschernutter V., Yang J., Eckle T., and Borchers C. H. (2013) Analysis of selected sugars and sugar phosphates in mouse heart tissue by reductive amination and liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 85, 5965–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., and Pachter L. (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Albu R. F., Chan G. T., Zhu M., Wong E. T. C., Taghizadeh F., Hu X., Mehran A. E., Johnson J. D., Gsponer J., and Mayor T. (2015) A feature analysis of lower solubility proteins in three eukaryotic systems. J. Proteomics 118, 21–38 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.