Abstract
DNA-protein cross-links (DPCs) are bulky DNA lesions that form both endogenously and following exposure to bis-electrophiles such as common antitumor agents. The structural and biological consequences of DPCs have not been fully elucidated due to the complexity of these adducts. The most common site of DPC formation in DNA following treatment with bis-electrophiles such as nitrogen mustards and cisplatin is the N7 position of guanine, but the resulting conjugates are hydrolytically labile and thus are not suitable for structural and biological studies. In this report, hydrolytically stable structural mimics of N7-guanine-conjugated DPCs were generated by reductive amination reactions between the Lys and Arg side chains of proteins/peptides and aldehyde groups linked to 7-deazaguanine residues in DNA. These model DPCs were subjected to in vitro replication in the presence of human translesion synthesis DNA polymerases. DPCs containing full-length proteins (11–28 kDa) or a 23-mer peptide blocked human polymerases η and κ. DPC conjugates to a 10-mer peptide were bypassed with nucleotide insertion efficiency 50–100-fold lower than for native G. Both human polymerase (hPol) κ and hPol η inserted the correct base (C) opposite the 10-mer peptide cross-link, although small amounts of T were added by hPol η. Molecular dynamics simulation of an hPol κ ternary complex containing a template-primer DNA with dCTP opposite the 10-mer peptide DPC revealed that this bulky lesion can be accommodated in the polymerase active site by aligning with the major groove of the adducted DNA within the ternary complex of polymerase and dCTP.
Keywords: DNA damage, DNA polymerase, gel electrophoresis, mass spectrometry (MS), molecular dynamics, DNA-protein conjugates, reductive amination, steady-state kinetics, translesion synthesis
Introduction
Covalent entrapment of cellular proteins on genomic DNA is a common process that occurs upon exposure to a variety of endogenous and exogenous bis-electrophiles, heavy metals, and free radicals (1–7). Specifically, common chemotherapeutics such as nitrogen mustards, platinum compounds, and alkylnitrosoureas (3, 4, 8); environmental carcinogens such as formaldehyde and 1,3-butadiene (5, 9, 10); toxic metals (11–13); nitric oxide (14); free radicals (15, 16); UV light (17); and ionizing radiation (6) mediate the formation of covalent DNA-protein cross-links (DPCs).3 Mass spectrometry-based proteomic studies have identified a large number of cellular proteins that participate in DPC formation in cells treated with cross-linking agents, including DNA repair proteins, DNA polymerases, transcription factors, and structural proteins such as histones, heat shock proteins, and tubulins (3–6, 8, 10).
Because of their enormous size as compared with other nucleobase lesions and their ability to disrupt DNA duplex structure and DNA-protein interactions, DPCs are hypothesized to interfere with DNA replication and transcription (1, 2). We reported previously that protein monoepoxide reagents that selectively induce DPC lesions cause toxicity and mutations in human cells (18). In a more recent study, we have shown that human cells deficient in nucleotide excision repair were sensitized toward nitrogen mustard toxicity and contained increased numbers of N7-guanine-conjugated DPC lesions (19). Covalent DPCs have been detected in peripheral blood lymphocytes from breast cancer patients undergoing treatment with cyclophosphamide.4 Recent studies by Jentsch and co-workers (20, 21) and Walter and co-workers (22) have provided evidence that DPCs are subject to proteolytic cleavage to the corresponding DNA-peptide lesions, which are subsequently removed from DNA in a replication-dependent repair process. Thus, it is critically important to investigate the activity and accuracy of DNA polymerases on DNA-protein and DNA-peptide lesions.
The most common site on DNA that participates in DNA-protein cross-linking mediated by bis-electrophiles is the N7 atom of guanine (3–5, 10). However, to our knowledge, no reports are available in the literature in regard to the replication bypass of N7-guanine adducts, probably because of their labile nature. Alkylation at the N7 atom of 2′-deoxyguanosine creates a positive charge at the affected nucleobase, destabilizing the N-glycosidic bond and leading to spontaneous depurination and abasic site formation (23, 24).
In the present work, we generated a range of hydrolytically stable model DPCs structurally resembling N7-guanine cross-links. Template-primer complexes containing model DPCs were subjected to in vitro polymerization in the presence of human lesion bypass DNA polymerases under steady-state kinetic conditions, and the resulting primer extension products were characterized by gel electrophoresis and HPLC-ESI-MS/MS. Finally, molecular dynamics (MD) simulations were conducted to examine how DNA-peptide lesions can be accommodated within the active site of human lesion bypass polymerase κ.
Results
Synthesis and Characterization of DNA-Protein and DNA-Peptide Cross-links
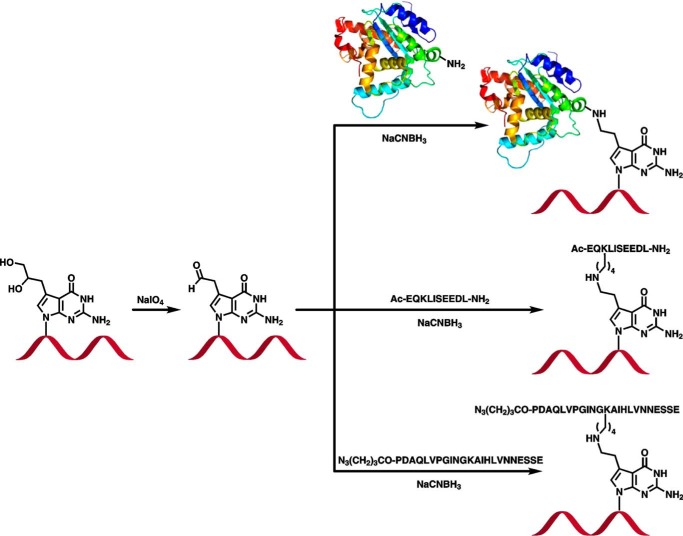
Model DNA-protein and DNA-peptide conjugates were prepared by reductive amination reactions between the Lys/Arg side chains of proteins/peptides and 7-deaza-7-(2-oxoethyl)-2′-deoxyguanosine of DNA (Fig. 1). The latter were formed in situ from 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine-containing DNA as described in our earlier publication (25). In the resulting model cross-links, Lys or Arg side chains of proteins and peptides are conjugated to the 7-deazaguanine residues of DNA via a two-carbon linker, creating a structure that is analogous to DPC lesions induced in cells by antitumor nitrogen mustards (3) and ethylene dibromide (26).
FIGURE 1.
Synthesis of DNA-protein and DNA-peptide cross-links by postsynthetic reductive amination.
DNA sequences (5′-G TCA CTG GTA XCA AGC ATT G-3′ and 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ where X is the modified base; Table 1) were selected to minimize self-annealing and were used previously in studies of DNA-DNA cross-links (27, 28). Two peptides of increasing size, a 10-mer peptide derived from c-Myc protein (EQKLISEEDL) and a 23-mer derived from tetanus toxoid (N3(CH2)3CO-PDAQLVPGINGKAIHLVNNESSE) were used. The same two peptides were used in our earlier publication investigating replication bypass of DPCs conjugated to the C5 position of thymidine using Cu(I)-catalyzed azide-alkyne cycloaddition (29). In addition, histone H2A (14 kDa), histone H4 (11.5 kDa), and AlkB (22.9 kDa) were chosen for these experiments based on the documented ability of histones and DNA repair proteins to engage in the formation of DNA-protein cross-links (3, 4, 8, 10). We also included enhanced green fluorescent protein (eGFP; 28 kDa), which was used in our earlier study of pyrimidine-conjugated DPCs (29). DNA-peptide and DNA-protein conjugates were isolated by gel electrophoresis, and their purity was confirmed by PAGE (see representative gel image in Fig. 2). Synthetic DNA-protein and DNA-peptide cross-links were structurally characterized by nano-HPLC-nanoelectrospray ionization-MS/MS (25).
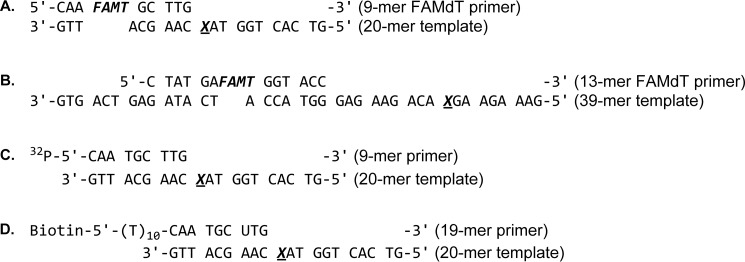
TABLE 1.
Synthetic DNA sequences used in the present study
X is dG, 7-DHP-7-deaza-dG, or DPC as indicated.
| Abbreviation | Sequence |
|---|---|
| P1 | 5′-CAA FAMTGC TTG-3′ |
| P2 | 5′-CTA TGA FAMTGG TAC C-3′ |
| P3 | 32P-5′-CAA TGC TTG-3′ |
| P4 | Biotin-5′-T10-CAA TGC UTG-3′ |
| T1 | 5′-G TCA CTG GTA GCA AGC ATT G-3′ |
| T2 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = deaza-DHP-dG) |
| T3 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = N3 (CH2)3CO-EQKLISEEDL-NH2) |
| T4 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = Ac-EQKLISEEDL-NH2) |
| T5 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = azido- (CH2)3CO-PDAQLVPGINGKAIHLVNNESSE) |
| T6 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = AlkB protein) |
| T7 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = histone H4) |
| T8 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = histone H2A) |
| T9 | 5′-G TCA CTG GTA XCA AGC ATT G-3′ (X = eGFP) |
| T10 | 5′-GAA AGA AGG ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ |
| T11 | 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ (X = deaza-DHP-dG) |
| T12 | 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ (X = N3 (CH2)3CO-EQKLISEEDL-NH2) |
| T13 | 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ (X = N3 (CH2)3CO-PDAQLVPGINGKAIHLVNNESSE) |
| T14 | 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ (X = AlkB protein) |
| T15 | 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ (X = histone H4) |
| T16 | 5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ (X = eGFP) |
FIGURE 2.
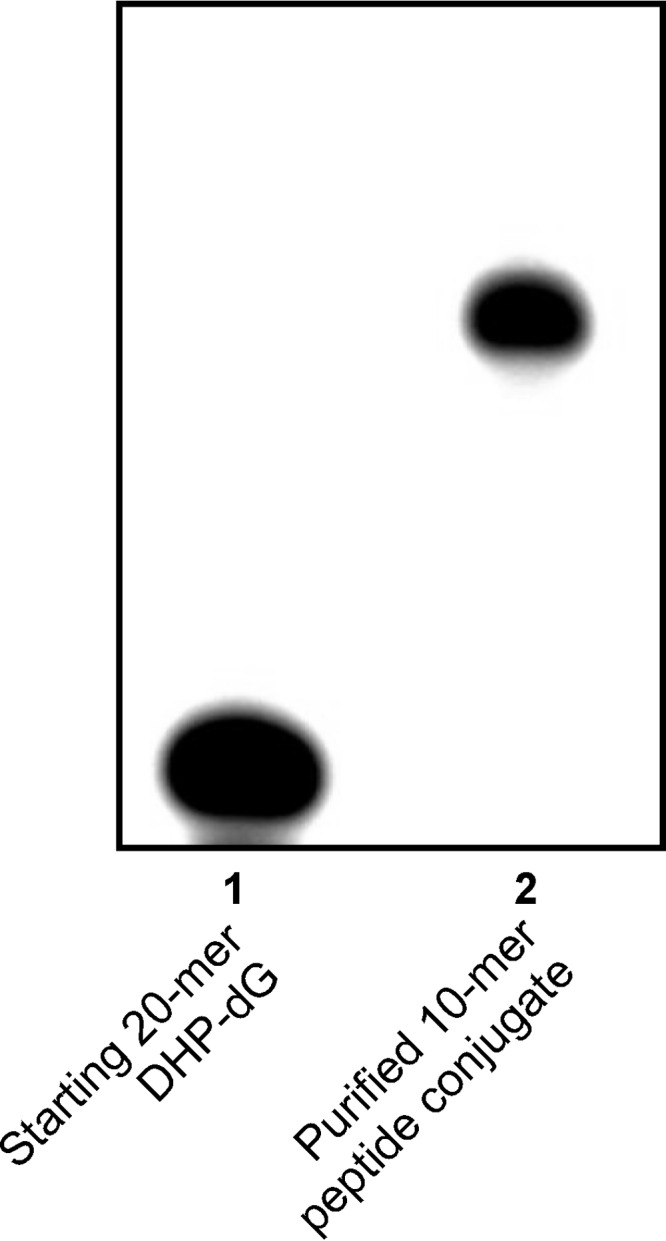
Representative purity check for a DNA-peptide conjugate (5′-G TCA CTG GTA XCA AGC ATT G-3′ where X is conjugated to EQKLISEEDL via the Lys residue.
Replication Bypass of DNA-Protein Cross-links
To examine the influence of DNA-protein cross-links conjugated to the C7 position of 7-deaza-G in DNA on DNA replication, template-primer complexes containing site-specific DPC lesions (10-mer peptide EQKLISEEDL derived from c-Myc protein (30, 31), 23-mer peptide PDAQLVPGINGKAIHLVNNESSE derived from tetanus toxoid, AlkB, histones H4 and H2A, and GFP protein) were subjected to standing start and running start primer extension in the presence of human translesion synthesis (TLS) polymerases η and κ. Templates containing native dG and deaza-DHP-dG served as negative controls.
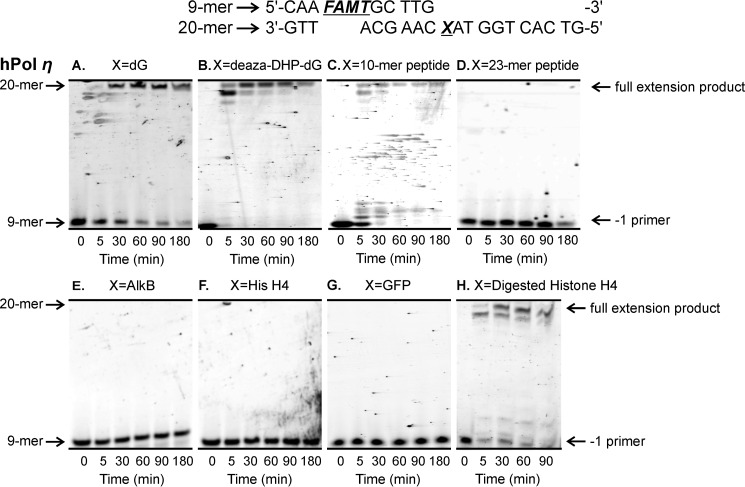
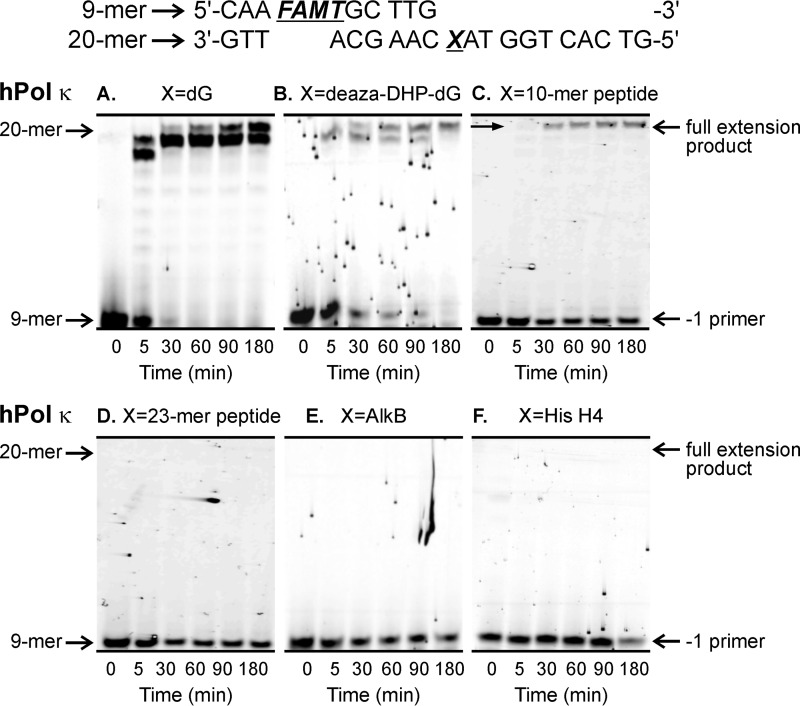
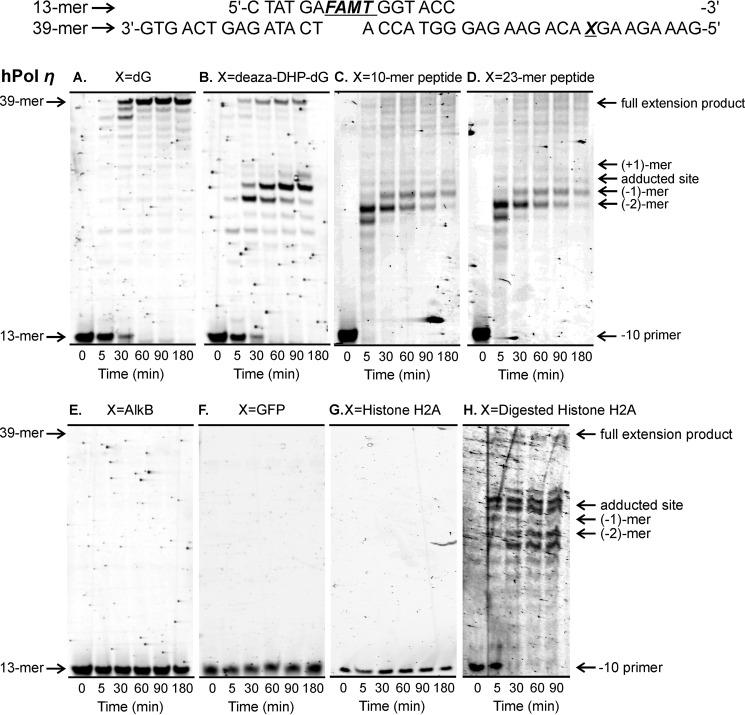
Our “standing start” assays used a 9-mer primer P1 (5′-CAA FAMTGC TTG-3′) extending to the penultimate position of the modified site (−1 primer) on the 20-mer templates T1–T9 (5′-G TCA CTG GTA XCA AGC ATT G-3′ where X is unmodified dG, deaza-DHP-dG, or a DPC adduct; Table 1 and Fig. 3A). Standing start primer extension past dG- and deaza-DHP-dG-containing templates resulted in complete extension products (20-mers) by hPols η and κ (Figs. 4, A and B, and 5, A and B). We found that the 10-mer peptide DPC was bypassed by hPols η and κ albeit with low efficiencies (Figs. 4C and 5C). In contrast, the 23-mer peptide cross-link and all three protein conjugates completely blocked primer extension (Figs. 4, D–G, and 5, D–F). The ability of polymerase η to copy the damaged strand was restored when the protein portion of DPC was digested with proteinase K (Fig. 4H). Taken together, these results indicate that the ability of TLS polymerases to bypass DPC lesions is a function of peptide size with lesions ≥23 amino acids long causing a complete replication block.
FIGURE 3.
Sequences of DNA oligomers used in this study. A, 9-mer FAMdT primer-20-mer template duplex used in standing start assays; B, 13-mer FAMdT primer-39-mer template DNA duplex used in running start assays; C, 9-mer primer-20-mer template duplex used for the single nucleotide incorporation and steady-state kinetic assays; D, biotinylated 19-mer primer-20-mer template duplexes used for the LC-MS/MS analyses of primer extension products. X = dG, 7-DHP-7-deaza-dG, or DPC.
FIGURE 4.
Standing start assays for replication bypass of DNA-peptide and DNA-protein cross-links by hPol η. 9-mer FAMdT primers were annealed with 20-mers containing unmodified dG (A), deaza-DHP-dG (B), or covalent cross-links to 10-mer peptide EQKLISEEDL (C), 23-mer peptide PDAQLVPGINGKAIHLVNNESSE (D), or full-sized protein AlkB (E), histone (His) H4 (F), or eGFP (G). The resulting primer-template duplexes (0.15 μm) were incubated in the presence of hPol η (0.30 μm). Reactions were initiated by the addition of a mixture of dNTPs (500 μm) and quenched at the indicated time points. Extension products were separated by 20% denaturing PAGE and visualized by fluorescence imaging. Panel H shows results for DPC to histone (His) H4 following digestion with Proteinase K.
FIGURE 5.
Standing start assays for replication bypass of DNA-protein cross-links by hPol κ. 9-mer FAMdT primers were annealed with 20-mers containing unmodified dG (A), deaza-DHP-dG (B), or covalent cross-links to 10-mer peptide EQKLISEEDL (C), 23-mer peptide PDAQLVPGINGKAIHLVNNESSE (D), or full-sized protein AlkB (E) or histone (His) H4 (F). The resulting primer-template duplexes (0.15 μm) were incubated in the presence of hPol κ (0.30 μm). Reactions were initiated by the addition of a mixture of dNTPs (500 μm) and quenched at the indicated time points. Extension products were separated by 20% denaturing PAGE and visualized by fluorescence imaging.
Because bulky DPC lesions may affect polymerase loading on the primer, in vitro replication experiments were repeated under “running start” conditions. Running start experiments were conducted using 13-mer primer P2 (5-CTA TGA FAMTGG TAC C-3) annealed to 39-mer templates T10–T16 (5′-GAA AGA AGX ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′ where X is unmodified dG, deaza-DHP-dG, or DNA-protein conjugate; Table 1 and Fig. 3B). In this primer-template complex, the 3′-end of the primer is placed 10 nucleotides upstream from the adducted site (−10 primer; Fig. 3B). We found that under running start conditions, both 10-mer and 23-mer peptide conjugates were partially bypassed by hPols η and κ to generate full-length products. For both peptide lesions, pronounced pausing was observed 1–2 nucleotides ahead of the adducted site and downstream from the lesion, suggesting inefficient polymerase translocation along the template (Figs. 6, C and D, and 7, C and D). Although the deaza-DHP-dG control caused some degree of polymerase pausing, it was less blocking than the peptide lesions (Figs. 6B and 7B). In contrast, all three DPC lesions completely blocked nucleotide addition, yielding no extension products (Figs. 6, E–G, and 7E). Similar results were observed when the primer was moved farther upstream from the lesion site (Fig. 8). As was the case with standing start experiments, proteinase K-digested cross-links were bypassed (Fig. 6H). Interestingly, replication past proteinase K-digested lesions and peptide cross-links generated large numbers of −3, −2, and −1 products (Fig. 6, C, D, and H). Taken together, our running start experiments are consistent with the standing start results (Figs. 4 and 5), indicating that although peptide DPCs can be bypassed by TLS polymerases η and κ DPCs to full-length proteins completely block DNA replication. In cells, such replication blockage may be relieved by proteolytic degradation of the protein component of DPC (20–22).
FIGURE 6.
Running start assays for replication bypass of DNA-protein cross-links by hPol η. 13-mer FAMdT primers were annealed with 39-mers containing unmodified dG (A), deaza-DHP-dG (B), or covalent cross-links to 10-mer peptide EQKLISEEDL (C), 23-mer peptide PDAQLVPGINGKAIHLVNNESSE (D), or full-sized protein AlkB (E), histone H2A (F), or eGFP (G). The resulting primer-template complexes (0.15 μm) were incubated in the presence of hPol η (0.30 μm). Reactions were initiated by the addition of the four dNTPs (500 μm) and quenched at the indicated time points. Extension products were separated by 20% denaturing PAGE and visualized by fluorescence imaging. Panel H shows results for DPC to histone H2A following digestion with Proteinase K.
FIGURE 7.
Running start assays for replication bypass of DNA-peptide and DNA-protein cross-links by hPol κ. 13-mer FAMdT primers were annealed with 39-mers containing unmodified dG (A), deaza-DHP-dG (B), or covalent cross-links to 10-mer peptide (C), 23-mer peptide (D), or AlkB protein (E). The resulting primer-template complexes (0.15 μm) were incubated in the presence of hPol κ (0.30 μm). Reactions were initiated by the addition of the four dNTPs (500 μm) and quenched at the indicated time points. Extension products were separated by 20% denaturing PAGE and visualized by fluorescence imaging.
FIGURE 8.
Running start assays for replication bypass of DNA-peptide and DNA-protein cross-links by hPol η. 12-mer FAMdT primers were annealed with 39-mers containing unmodified dG (negative control) or AlkB-DNA cross-link. The resulting primer-template complexes (0.15 μm) were incubated in the presence of hPol η (0.30 μm). Reactions were initiated by the addition of the four dNTPs (500 μm) and quenched at the indicated time points. Extension products were separated by 20% denaturing PAGE and visualized by fluorescence imaging.
Kinetics of Single Nucleotide Incorporation Opposite 10-mer Peptide Cross-link
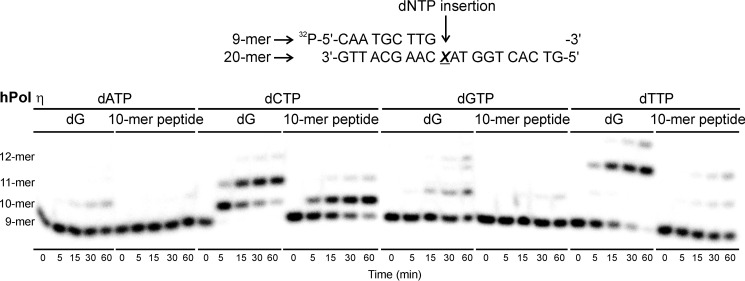
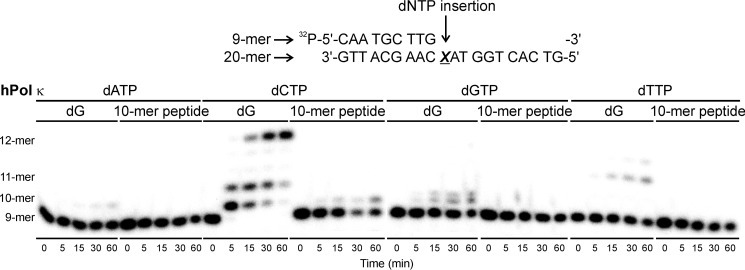
To examine the fidelity of nucleotide insertion opposite DNA-peptide lesions by TLS polymerases, single nucleotide insertion assays were conducted. Template-primer complexes created by annealing radiolabeled 9-mer primers (32P-5′-CAA TGC TTG-3′) to 20-mer templates (5′-G TCA CTG GTA XCA AGC ATT G-3′; Fig. 3C) were incubated with hPol η or κ in the presence of individual dNTPs (100 μm) for 0–60 min. Denaturing PAGE-phosphorimaging analyses revealed that both polymerases preferentially incorporated the correct base (dC) opposite native G as expected (see representative data in Figs. 9 and 10). Furthermore, hPol κ inserted a single nucleotide opposite the 10-mer peptide cross-link with high fidelity (Fig. 10). hPol η also favored the incorporation of the correct base (C), although a small amount of T was also incorporated (Fig. 9).
FIGURE 9.
Single nucleotide insertion opposite unmodified guanine (dG) and the 10-mer peptide cross-linked to C7 of 7-deazaguanine (10-mer peptide) by TLS polymerase hPol η. Primer-template duplexes (50 nm) were incubated with 50 nm polymerase in the presence of 100 μm individual dNTPs. The reactions were quenched at predetermined time points (0–60 min), and the products were separated by 20% denaturing PAGE and visualized by phosphorimaging.
FIGURE 10.
Single nucleotide insertion opposite unmodified guanine (dG) and the 10-mer peptide cross-linked to C7 of 7-deazaguanine (10-mer peptide) by TLS polymerase hPol κ. 50 nm primer-template duplexes were incubated with 150 nm polymerase in the presence 100 μm individual dNTPs. The reactions were quenched at predetermined time points (0–60 min), and the products were separated by 20% denaturing PAGE and visualized by phosphorimaging.
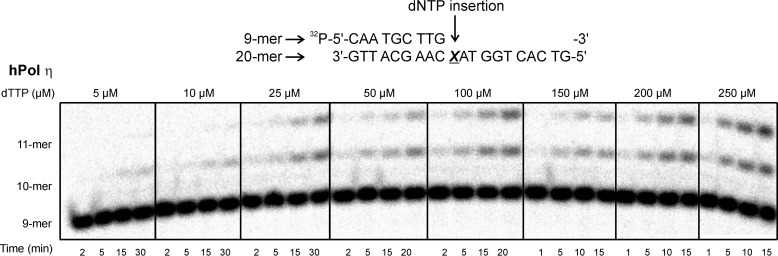
To determine the catalytic efficiency and to calculate the misinsertion frequency for incorporation of individual dNTPs opposite the 10-mer peptide cross-link, primer-template duplexes containing unmodified dG or 10-mer peptide cross-link (Fig. 3C) were incubated with hPol η or κ in the presence of increasing concentrations of individual dNTPs (0–500 μm), and the reactions were quenched at preselected time points (0–60 min). Polymerase concentrations and the time points were selected such that the extent of product formation was <35% of the starting substrate concentration (see representative gel image in Fig. 11). The specificity constant (kcat/Km) was calculated to evaluate the catalytic efficiency of incorporation of each dNTP and the frequency of incorporating an incorrect versus correct dNTP opposite the lesion (f) (32) were calculated by plotting the reaction velocity against dNTP concentration.
FIGURE 11.
Example of a steady-state kinetics experiment to determine the kinetics of single nucleotide insertion opposite the 10-mer peptide cross-linked to C7 of 7-deaza-G. Single nucleotide incorporation assays were performed using hPol η and 10-mer peptide-deaza-G cross-links in the presence of increasing concentrations of individual dTTPs. Polymerization reactions were quenched at preselected time points and loaded onto 20% (w/v) denaturing polyacrylamide gels.
Our steady-state kinetic studies have revealed that the specificity constants (kcat/Km) for nucleotide insertion by hPol η were 4–150-fold higher compared with those of hPol κ, indicating that replication of DNA-peptide lesions is more efficient in the presence of hPol η (Table 2). The specificity constants for the insertion of the correct base (dCTP) opposite the adduct were 0.03 and 0.0002 μm−1 min−1 for hPols η and κ, respectively (Table 2). These values were 49- and 1850-fold lower than those obtained for dCTP insertion opposite native G (1.5 and 0.37 μm−1 min−1, respectively, Table 2), reflecting the difficulty of accommodating the bulky peptide adduct in the polymerase active site.
TABLE 2.
Steady-state kinetic parameters for single nucleotide insertion opposite native dG (negative control) and the 10-mer peptide cross-linked to C7 of 7-deaza-G (10-mer peptide) by human TLS polymerases hPols κ and η
f, misinsertion frequency = (kcat/Km)incorrect dNTP/(kcat/Km)correct dNTP.
| Polymerase | Template | Incoming nucleotide | kcat | Km | kcat/Km | f |
|---|---|---|---|---|---|---|
| min−1 | μm | μm−1 min−1 | ||||
| hPol η | dG | dCTP | 3.7 ± 0.2 | 2.6 ± 1.0 | 1.5 | 1 |
| dTTP | 0.70 ± 0.08 | 120 ± 38 | 0.006 | 0.004 | ||
| 10-mer peptide | dCTP | 0.38 ± 0.04 | 13 ± 5 | 0.03 | 1 | |
| dTTP | 0.01 ± 0.001 | 32 ± 11 | 0.00005 | 0.002 | ||
| hPol κ | dG | dCTP | 0.53 ± 0.04 | 1.5 ± 1.1 | 0.37 | 1 |
| 10-mer peptide | dCTP | 0.02 ± 0.004 | 63 ± 24 | 0.0002 | 1 |
Nucleotide insertion opposite the 10-mer peptide conjugate by hPols η and κ was characterized by high fidelity, resulting in incorporation of C opposite the adduct. The insertion frequency of dTMP (f) was 500-fold lower compared with the frequency of incorporation of the correct nucleotide (dCMP) (Table 2). Overall, our steady-state kinetic data were indicative of low efficiency, high fidelity nucleotide insertion opposite the 10-mer peptide cross-linked to 7-deaza-G in DNA.
Sequence Analysis of Primer Extension Products by Liquid Chromatography-Tandem Mass Spectrometry
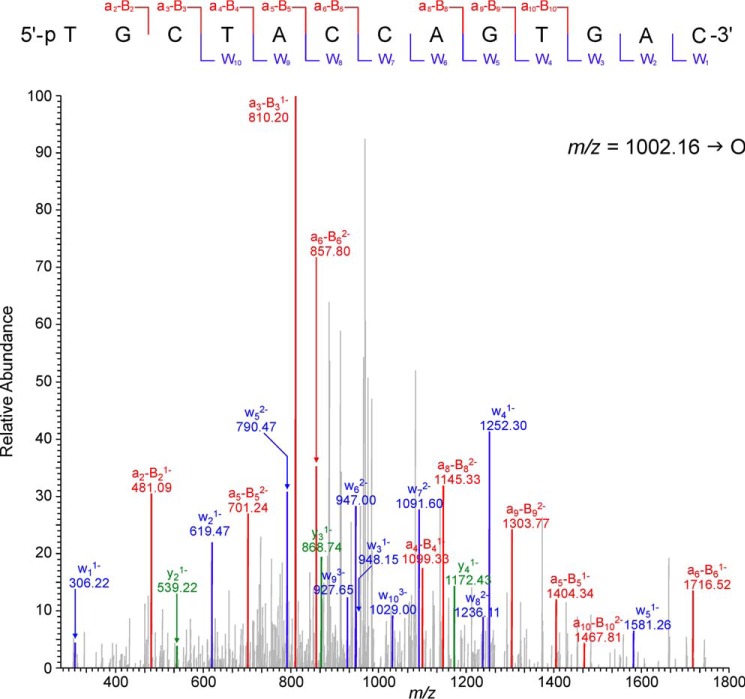
A mass spectrometry-based strategy reported previously (33–36) was used to sequence primer extension products of the translesion synthesis past peptide-dG conjugates. This method allows for detection of insertion, deletion, and point mutation products at the site of damage as well as any postlesion synthesis errors (34). Primer extension reactions were carried out with DNA duplexes containing native dG or the 10-mer peptide cross-link in the template strand (Fig. 3D). A uracil base was incorporated 3 nucleotides upstream from the primer end. Streptavidin capture of biotinylated primer extension products and treatment with UDG and hot piperidine allow for the release of short oligodeoxynucleotide products containing the region of interest that are amenable for sequencing by HPLC-ESI-MS/MS (Fig. 12).
FIGURE 12.
Collision-induced dissociation mass spectra of the error-free extension products observed following hPol η-catalyzed replication past the 10-mer peptide cross-linked to 7-deazaguanine.
The extension products from hPol η-catalyzed primer extension reactions were sequenced by HPLC-ESI-FTMS/MS using an Orbitrap Velos mass spectrometer. The samples were initially analyzed in the full scan mode to detect all extension products. HPLC-ESI−-FTMS peak areas in the extracted ion chromatograms corresponding to each product were compared with that of an internal standard to determine the relative quantities of each extension product.
Capillary HPLC-ESI−-FTMS analysis of in vitro replication products resulting from primer extension reactions of template T4 containing 10-mer dG-peptide conjugate revealed one main peak corresponding to the accurate replication product (m/z 1002.16, [M − 4H]4−). The MS/MS spectrum of this oligomer was consistent with the theoretical sequence (5′-pTG CTA CCA GTG AC-3′; Fig. 12). Targeted HPLC-ESI−-FTMS analyses failed to detect any point mutations, single nucleotide deletions, or postlesion synthesis errors. Taken together, our HPLC-ESI−-FTMS/MS sequencing results are in agreement with the data from steady-state kinetic experiments (Table 2), indicating that nucleotide insertion opposite the 10-mer peptide conjugate by hPol η is largely accurate.
Modeling and Simulation
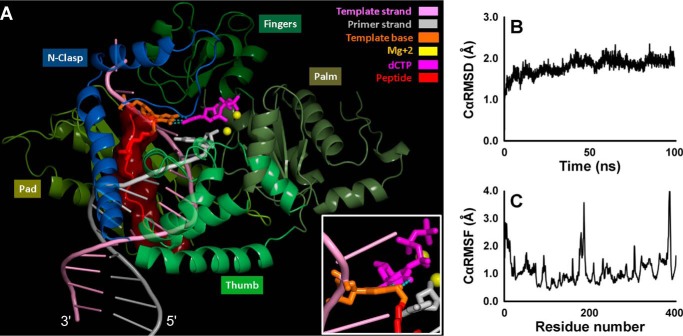
To better understand how the 10-mer peptide cross-link can be accommodated in the active site of TLS polymerases and copied with high fidelity, molecular dynamics simulations were carried out using a model for hPol κ developed earlier (37) based on the crystal structure of the hPol κ ternary complex by Lone et al. (38). The C7 atom of 7-deazaguanine is located in the DNA major groove. Modeling of the Ac-EQKLISEEDL-NH2 cross-link was achieved by extending the longer N-terminal end of the peptide toward the 3′-end of the replicating DNA template (Fig. 13A and supplemental movie). Inspection of the trajectory showed an average Cα RMSD of 1.9 Å over the course of the 100-ns MD simulation, suggesting that the presence of the 10-mer cross-link within the major groove does not significantly affect the overall structure of hPol κ (Fig. 13B). The largest Cα RMSF of 3.6 Å occurred at Lys-204 in a turn of the palm region more than 30 Å away from the active site (Fig. 13C). All the interatomic distances between the incoming dCTP and the two Mg2+ ions were found to be conserved in the final structure of the simulation. The three Watson-Crick hydrogen bonds between 7-deazaguanine and the dCTP were present throughout the simulation, which would allow hPol κ to catalyze high fidelity nucleotide addition opposite the DNA-peptide lesion. However, the simulation also revealed that the Watson-Crick pairing was distorted because of the rigidity imposed on the damaged base by the cross-link and that there was steric crowding between the peptide lesion and the N-clasp domain of hPol κ (Fig. 13A and supplemental movie). This may contribute to inefficient lesion bypass and polymerase stalling at the peptide cross-link (Figs. 6 and 7).
FIGURE 13.
A, MD snapshot at 100 ns of the hPol κ-DNA ternary complex containing the 10-mer peptide cross-linked to 7-deazaguanine. The complex is displayed in schematic mode except for the dCTP and the cross-link-containing template base, which are in stick. The peptide is shown in red surface. The three Watson-Crick hydrogen bonds are shown with dotted lines. The inset, rotated about 90° along the y axis, highlights the Watson-Crick pair distortion. A rotating view is given in the supplemental movie. Cα RMSD (B) and Cα RMSF (C) of the hPol κ over the course of the 100-ns MD simulation for the hPol κ-DNA ternary complex containing the lesion peptide.
Discussion
Lesion bypass is an important mechanism of cellular DNA damage tolerance as it allows for DNA replication past bulky nucleobase adducts that block DNA polymerases (39–43). Specialized TLS polymerases can be recruited to blocked replication forks and insert nucleotides opposite damaged bases (39–41, 43–49). TLS polymerases are catalytically less efficient than replicative enzymes and are much more error-prone because of their open and flexible active sites that accommodate large DNA adducts (43, 49–51) and their lack of intrinsic 3′ → 5′ proofreading activity (41, 46, 52–55). In humans, TLS polymerases include Y family polymerases hPols η, κ, and ι and Rev1, A family Pol ν, and B family Pol ζ (41, 43–49). Polymerase switching is thought to be regulated, at least in part, by posttranslational modifications of the proliferating cell nuclear antigen sliding clamp, which acts as a scaffold for polymerase enzymes (41, 43–49).
DPCs represent a special challenge to DNA replication because of their large size and their ability to distort DNA structure. DPCs had been hypothesized to completely block DNA polymerases, leading to stalling of replication forks (43, 56). In our earlier study, we found that model DPCs generated by copper-catalyzed [3 + 2] Huisgen cycloaddition (click reaction) between an alkyne group from 5-(octa-1,7-diynyl)-uracil in DNA and an azide group within engineered proteins/polypeptides completely blocked DNA replication, whereas the corresponding 10-mer peptide conjugate was bypassed in an error-prone manner (29, 36). In these model DPCs, peptides were conjugated to the C5 position of thymidine via a six-carbon linker (29). However, the majority of DNA-protein conjugates induced by antitumor nitrogen mustards such as mechlorethamine and chlorambucil, platinum compounds, dibromoethane, and diepoxides such as 1,2,3,4-diepoxybutane involve the N7 atom of G residues in DNA (3–5, 8, 10, 57), and the cross-linking site within DNA duplex is likely to modulate the effects of DPC lesions on DNA replication.
N7-Guanine-conjugated DPCs are induced by many DNA cross-linking drugs such as nitrogen mustards, haloethylnitrosoureas, and platinum antitumor agents commonly used in the clinic to treat leukemia; lymphoma; ovarian adenocarcinoma; and breast, lung, and testicular cancers (58–62). The majority of N7-G lesions are labile to hydrolysis (23), making it difficult to synthesize site-specific DNA substrates containing such adducts for polymerase bypass assays. We recently developed a reductive amination methodology to prepare hydrolytically stable model 7-deaza-G DPC substrates that resemble DPCs formed by nitrogen mustards and dibromoethane (25). In the present study, this new methodology was used to prepare DNA-peptide and DNA-protein substrates of increasing size (1.3–28 kDa; Fig. 1). The resulting template-primer complexes were subjected to steady-state kinetics and polymerization experiments to investigate replication bypass of these bulky lesions by human TLS polymerases η and κ.
We found that human TLS polymerases η and κ were completely blocked by full size DPCs conjugated to the 7-deazaguanine within DNA (histone H4, 11.5 kDa; histone H2A, 14 kDa, AlkB, 23 kDa, and eGFP, 28 kDa) but were able to bypass the corresponding small peptide lesions (Figs. 4–7). DPC-induced replication blocks were observed in both standing start and running start experiments but were removed when the protein component of DPCs was digested with proteinase K (Figs. 4H and 6H). Importantly, the presence of DPC lesions affects polymerization at least 18 nucleotides upstream from the lesion, probably by interfering with polymerase loading (Fig. 8). These results are in agreement with our earlier primer extension studies of DPCs conjugated to the C5 position of thymidine (29). In both cases, the presence of TLS polymerases cannot overcome the replication block imposed by bulky protein-DNA conjugates. It should be noted that the C7 of G and the C5 of T occupy similar positions in the major groove of DNA.
Primer extension results shown here are consistent with published reports of polymerase blockage by DPCs in vivo (56, 63, 64). For example, Kreuzer and co-workers (56) detected bubble and Y molecule accumulation in wild-type pBR322 plasmids containing methyltransferase (53.5 kDa) cross-linked to 5-azacytosine in Escherichia coli. Nakano et al. (63) found that the transformation efficiency of pGL3-CMV plasmids containing formaldehyde-induced cross-link to histone H1 (21.7 kDa) was <10% as compared with undamaged plasmid. In another study, the relative transformation efficiency of pMS2 plasmids containing UV-induced T4-pyrimidine dimer glycosylase (16 kDa) DPCs was found to be <5%, whereas that of UV-irradiated plasmids was ∼50% (64). These results suggest that DPCs significantly hinder DNA replication in vivo.
In living cells, the protein component of DPC lesions may undergo proteolytic cleavage to smaller DNA-peptide or DNA-amino acid adducts, which represent a lesser block to polymerase bypass (65–68). For example, yeast Wss1 protease cleaves the protein constituent of DPC at blocked replisomes (21). Furthermore, repair of a plasmid-embedded DPC in Xenopus egg extracts is coupled to DNA replication (22). We found that the DPC-induced replication block is removed if the protein constituent of DPC is digested to amino acids with proteinase K (Figs. 4H and 6H). This is consistent with an earlier observation by Nakano et al. (63) that the transformation efficiency of pGL3-CMV plasmids containing partially digested histone H1 DPCs increased to 58% from <10% for plasmids containing undigested DPC.
To examine the influence of 7-deaza-G DNA-peptide lesions on replication efficiency and accuracy, we investigated primer extension past 10-mer and 23-mer peptides (EQKLISEEDL and PDAQLVPINGKAIHLVNNESSE) conjugated to a 7-deaza-G in an oligonucleotide. We found that both peptide conjugates were bypassed under running start conditions (Figs. 6, C and D, and 7, C and D), although the 23-mer peptide cross-link blocked replication in standing start experiments (see Figs. 4, C and D, and 5, C and D). Polymerase pause sites were observed 1–3 nucleotides prior to the adducted site (Fig. 6, C and D). Furthermore, both nucleotide insertion opposite DPC and postlesion synthesis were extremely inefficient (Figs. 6 and 7).
To our knowledge, this is the first investigation of the effects of 7-guanine-conjugated peptides on DNA replication in the presence of TLS polymerases. Previous studies have shown that Pol ν was able to catalyze primer extension past tetrapeptides and dodecapeptides cross-linked to the N6 position of adenine (69), whereas the same peptides conjugated to the N2-guanine position were bypassed by hPol κ and E. coli Pol IV (70). In our earlier study, the same 10-mer peptide conjugated to C5 of thymine via a six-carbon linker was bypassed by hPol κ and hPol η (29). Taken together, these results indicate that unlike DPCs containing full size proteins peptide conjugates can be tolerated by human TLS polymerases.
Steady-state kinetic experiments have revealed that the efficiencies of dCMP insertion opposite 7-deaza-G DNA-peptide conjugates by hPol η and hPol κ were 49- and 1850-fold lower than for the template containing native G (Table 2). As observed previously for C5-thymine lesions (36), hPol η was more efficient than hPol κ. Higher catalytic efficiency of hPol η than hPol κ is consistent with previous reports for other bulky nucleobase lesions such as exocyclic adducts (35, 71–73) and DNA-glutathione conjugates (74).
Unlike C5-thymine-peptide lesions, which induce high numbers of frameshift and base substitution mutations upon primer extension in the presence of hPol κ and hPol η (36), C7-G conjugated DPCs were not miscoding. Our gel electrophoresis and mass spectrometry experiments have shown that replication bypass of the 10-mer peptide cross-linked to C7-G by hPol κ was essentially error-free, whereas hPol η incorporated the correct base (dC) with 500-fold preference over the mispair (dT) (Table 2 and Figs. 9 and 10). These results indicate that the biological outcomes of DNA-peptide lesions are largely defined by the cross-linking site on DNA (C7 of deazaguanine versus C5 of thymine).
Our model cross-links differ from lesions formed in cells by the absence of a positive charge on the guanine 7-position. The replacement of N7-guanine with carbon introduces changes in hydration and charge localization (75). However, 7-deazaguanine-containing DNA has essentially the same structure as native DNA (76), and 7-deazaguanine mimics have been successfully used in a number of previous studies of DNA lesion repair and polymerase bypass (27, 77–79).
Molecular dynamics simulations based on the published crystal structure of a hPol κ-DNA template-primer-dCTP ternary complex (37, 38) indicated that the 10-mer peptide conjugated to 7-deaza-G can be accommodated in the major groove of the DNA oriented toward the 3′-end of the template strand with minimal overall structural perturbations (Cα RMSD of 1.9 Å; Fig. 13B). The Watson-Crick base pairing between the 7-deazaguanine and the incoming dCTP explains the observed high fidelity bypass results (Table 2). However, the rigidity imposed on the damaged base by the cross-link causes the Watson-Crick base pair to be distorted, which explains the reduced nucleotide incorporation efficiency. Furthermore, the close contact of the peptide with the hPol κ N-clasp likely causes polymerase stalling by hindering translocation, which is expected to impede extension efficiency and hinder postlesion synthesis (Fig. 13A and supplemental movie). Other bulky major groove DNA lesions are also known to stall hPol κ (80, 81).
hPol η and hPol κ have very significant structural differences (82). One important difference in relation to our experimental results is the absence of the N-clasp in hPol η. Consequently, the DNA major groove of the primer-template complex with hPol η remains open, allowing for ready accommodation of the 7-deazaguanine 10-mer peptide cross-link. The cross-linked template is expected to retain Watson-Crick pairing at the 5′-templating base but distort the pairing and impede translocation due to the rigidity of the damaged base. Thus, the greater efficiency and fidelity of hPol η compared with hPol κ very likely stems from the absence of the N-clasp, whereas the overall high fidelity but low efficiency in the two polymerases has a common origin in the lesion structure.
In summary, we have examined the ability of human lesion bypass polymerases to catalyze replication past DNA-protein and DNA-peptide cross-links conjugated to the major groove of DNA via the C7 atom of deazaguanine. We found that large DNA-protein and DNA-peptide cross-links completely blocked human TLS polymerases, whereas smaller peptide cross-links were bypassed with low efficiency but with high fidelity. The 7-deazaguanine DNA substrates used here are structurally analogous to the major DNA-protein cross-links formed in vivo when cancer patients are treated with nitrogen mustard drugs (3). These results suggest that proteolytic degradation of toxic DPCs to smaller DNA-peptide cross-links allows for them to be bypassed in an accurate manner by human TLS polymerases.
Experimental Procedures
Materials
Fluorescein-dT phosphoramidite, protected 2′-deoxyribonucleoside 3′-phosphoramidites (dA-CE, Ac-dC-CE, dmf-dG-CE, and dT-CE), Ac-dC-CPG ABI, dmf-dG-CPG ABI columns, and all other reagents required for automated DNA synthesis were purchased from Glen Research (Sterling, VA). Recombinant human DNA polymerases hPols η and κ were expressed and purified as described elsewhere (84, 85). Recombinant AlkB protein was a generous donation from Prof. Chuan He (Department of Chemistry, University of Chicago). Expression of His6-eGFP protein (referred to as eGFP throughout) (86) and synthesis of 23-mer and 10-mer peptides (PDAQLVPINGKAIHLVNNESSE and EQKLISEEDL) (29) were carried out according to previously reported protocols. T4 polynucleotide kinase (PNK) and recombinant histones H2A and H4 were obtained from New England Biolabs (Beverly, MA); [γ-32P]ATP was purchased from PerkinElmer Life Sciences. 40% 19:1 acrylamide/bisacrylamide solution and Micro Bio-Spin 6 columns were purchased from Bio-Rad. Unlabeled dNTPs were obtained from Omega Bio-Tek (Norcross, GA). Size exclusion columns (NAP-5 and Microspin G25) and Sep-Pak C18 solid phase extraction cartridges were obtained from GE Healthcare and Waters (Milford, MA), respectively. All other chemicals and solvents were purchased from Sigma-Aldrich and were of the highest grade available.
Synthesis and Characterization of Oligodeoxynucleotides
Synthetic oligonucleotides containing site-specific 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine (5′-G TCA CTG GTA deaza-DHP-dGCA AGC ATT G-3′ and 5′-GAA AGA AGdeaza-DHP-dG ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′) were prepared as described previously (27). Primers internally labeled with 5′-[N-((fluoresceinyl)-aminohexyl)-3-acrylimido]-2′-deoxyuridine (5′-CAAFAMdTGC TTG-3′ and 5′-CTA TGA FAMdTGG TAC C-3′) and native DNA strands (5′-G TCA CTG GTA GCA AGC ATT G-3′, 5′-GAA AGA AGG ACA GAA GAG GGT ACC ATC ATA GAG TCA GTG-3′, and 5′-CAA TGC TTG-3′) were prepared by standard solid phase synthesis using an ABI 394 DNA synthesizer (Applied Biosystems, Foster City, CA). Biotinylated 19-mer primers (biotin-5′-T10 CAA TGC UTG-3′) were purchased from Integrated DNA Technologies (Coralville, IA). All synthetic oligodeoxynucleotides were purified by semipreparative HPLC, desalted by Illustra NAP-5 columns, and characterized by HPLC-ESI−-MS.
Synthesis and Characterization of DNA-Protein and DNA-Peptide Cross-links
Site-specific DNA-protein and DNA-peptide cross-links were generated by a postsynthetic reductive amination strategy developed in our laboratory (25). Briefly, synthetic DNA oligomers containing site-specific deaza-DHP-dG (1 nmol in 36 μl of water) were oxidized in the presence of 50 mm NaIO4 (70 μl) in 15 mm sodium phosphate buffer (pH 5.4; 70 μl) for 6 h at 4 °C in the dark to convert deaza-DHP-dG to 7-deaza-7-(2-oxoethyl)-2′-deoxyguanosine (Fig. 1). Excess NaIO4 was quenched with Na2SO3 (55 mm; 70 μl), and the resulting aldehyde-functionalized DNA was incubated with the protein of interest (AlkB, histone H4, histone H2A, or eGFP; 2–10-fold molar excess) in the presence of 25 mm NaCNBH3 at 37 °C overnight to generate DNA-protein cross-links (Fig. 1). The resulting DNA-protein conjugates (Table 1) were purified using 20% (w/v) denaturing polyacrylamide gels containing 7 m urea.
DNA-peptide conjugates were prepared analogously (Fig. 1) except that a 10–100-fold molar excess of the peptide was used, and the pH was adjusted to 7.0 prior to peptide addition. DNA-peptide conjugates were isolated using 20% (w/v) denaturing polyacrylamide gels containing 7 m urea. DNA-protein and DNA-peptide cross-links were desalted by Micro Bio-Spin 6 columns and Sep-Pak C18 solid phase extraction cartridges, respectively. The purified conjugates were characterized by HPLC-ESI+-MS/MS as described previously (25). To assess DPC purity, aliquots of the purified samples were radiolabeled with γ-32P and resolved on 20% (w/v) denaturing polyacrylamide gels containing 7 m urea followed by visualization using a Typhoon FLA 7000 phosphorimaging system (GE Healthcare). Depending on the purity of the cross-links obtained, additional gel purifications were carried out to achieve >98% purity.
Preparation of Primer-Template Duplexes
For primer extension assays, fluorescein-labeled primer P1 or P2 (Table 1; 50 pmol) were mixed with 2 eq of template strands containing dG (T1 or T10), deaza-DHP-dG (T2 or T11), or model DPCs (T3–T9 or T12–T16 in Table 1) in 10 mm Tris-HCl buffer (pH 8.0) containing 50 mm NaCl. The mixtures were heated at 95 °C for 10 min and allowed to cool overnight to afford the corresponding primer-template duplexes.
For steady-state experiments to determine the kinetics of single nucleotide insertion opposite the cross-links, the 9-mer 5′-CAA TGC TTG-3′ (1 nmol) was radiolabeled with T4 PNK (20 units) and [γ-32P]ATP (25 μCi) in the presence of T4 PNK reaction buffer (total volume, 20 μl) at 37 °C for 1 h to obtain primer P3 (Table 1). The mixtures were heated at 65 °C for 10 min to inactivate the enzyme and passed through Illustra Microspin G25 columns to remove excess [γ-32P]ATP. 5′-32P-Labeled primer P3 (Table 1; 50 pmol) was annealed to template strands (5′-G TCA CTG GTA XCA AGC ATT G-3′) containing unmodified dG (T1 in Table 1) or 7-deaza-dG-10-mer peptide cross-link (T4 in Table 1). For HPLC-ESI−-MS/MS sequencing of extension products, biotinylated primers P4 (Table 1; 100 pmol) were annealed to the template strands containing either unmodified dG or the dG-10-peptide conjugate at position X (T1 or T4 in Table 1; 200 pmol).
Primer Extension Assays
For standing start experiments, DNA complexes containing primer P1 annealed to templates T1–T9 in Table 1 (0.15 μm; Fig. 3A) were incubated with human DNA polymerase η or κ (0.30 μm) at room temperature in the presence of a buffered solution containing 50 mm Tris (pH 7.5), 50 mm NaCl, 5 mm DTT, 100 μg/ml BSA, 10% glycerol (v/v), and 5 mm MgCl2. Polymerization reactions were initiated by adding a solution containing all four dNTPs (0.5 mm each). Aliquots of the reaction mixtures (4 μl) were withdrawn at preselected time intervals (0, 5, 30, 60, 90, and 180 min) and quenched by the addition of 95% (v/v) formamide containing 10 mm EDTA (18 μl). Samples were loaded onto 20% (w/v) denaturing polyacrylamide gels and run at 80 watts for 3 h. The extension products were visualized using a Typhoon FLA 7000 in the fluorescence imaging mode. Running start experiments were conducted in a similar manner using 13-mer FAMdT primer-39-mer template DNA duplexes (Fig. 3B).
Single Nucleotide Incorporation Assays
32P-End-labeled primer-template duplexes containing native dG or 10-mer peptide-deaza-dG conjugate (50 nm) at position X (Fig. 3C) were incubated with human TLS polymerases (50 nm hPol η or 150 nm hPol κ) in 50 mm Tris-HCl (pH 7.5) buffer containing 50 mm NaCl, 5 mm DTT, 5 mm MgCl2, 100 μg/ml BSA, and 10% (v/v) glycerol at room temperature. Polymerization reactions were initiated by the addition of individual dNTPs (100 μm) in a final volume of 20 μl. Aliquots (4 μl) were withdrawn at preselected time points, and the reactions were quenched by the addition of a solution containing 10 mm EDTA, 0.03% (w/v) bromphenol blue, and 0.03% (w/v) xylene cyanol in 95% (v/v) formamide (8 μl). The extension products were resolved by 20% (w/v) denaturing PAGE containing 7 m urea and visualized using a Typhoon FLA 7000 phosphorimaging system.
Steady-state Kinetic Analyses
The steady-state kinetics for incorporation of individual nucleotides opposite the native dG or the 10-mer peptide-deaza-dG cross-link were investigated by performing single nucleotide incorporation assays with 0.5–50 nm hPol η or 3–55 nm hPol κ in the presence of increasing concentrations of individual dNTPs (0–500 μm). Polymerization reactions were quenched at preselected time points (0–60 min). The product bands were visualized using a Typhoon FLA 7000 phosphorimaging system and quantified by volume analysis using Image Quant TL 8.0 software (GE Healthcare). Steady-state kinetic parameters were calculated by nonlinear regression analysis using one-site hyperbolic fits in Prism 4.0 (GraphPad Software, La Jolla, CA).
Sequencing of hPol η Primer Extension Products by Liquid Chromatography-Tandem Mass Spectrometry
Biotinylated primer-template duplexes (150 pmol) prepared by annealing uracil-containing primer P4 with template T1 or T4 (Table 1 and Fig. 3D) were incubated with hPol η (60 pmol) in 50 mm Tris-HCl (pH 7.5) buffer containing 50 mm NaCl, 5 mm DTT, 5 mm MgCl2, 100 μg/ml BSA, and a 1 mm concentration each of the four dNTPs. Following a 6-h incubation at room temperature, biotinylated DNA strands were captured on streptavidin beads. Briefly, 20 mm sodium phosphate (pH 7.0) buffer containing 150 mm NaCl (400 μl) was added to each reaction. Streptavidin-Sepharose high performance beads (0.2 ml; GE Healthcare) were washed with 500 μl of 20 mm sodium phosphate (pH 7.0) buffer containing 150 mm NaCl and then added to the polymerase reaction mixture. The resulting suspension was incubated at room temperature for 2 h with mixing every 10 min. Following binding of the biotinylated DNA to the beads, the supernatant was removed, and the beads were washed three times with water (300 μl). A solution of 50 mm Tris-HCl buffer (pH 7.5) containing uracil-DNA glycosylase (20 units), 1 mm EDTA, and 1 mm DTT (500 μl) was added followed by incubation at 37 °C for 4 h. The beads were washed three times with H2O (300 μl) and then incubated with 250 mm piperidine (400 μl) at 95 °C for 1 h to cleave the abasic sites resulting from uracil-DNA glycosylase-mediated excision of U. The resulting samples were dried in vacuo, and the residue was reconstituted in 25 μl of water containing a 14-mer internal standard (5′-pCTT CAC GAG CCC CC-3′; 40 pmol).
Capillary HPLC-ESI−-MS/MS analyses were conducted as described previously (36). An Agilent 1100 HPLC system (Agilent Technologies, Wilmington, DE) coupled to a Thermo LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA) was operated in the negative ion ESI-FTMS/MS mode. HPLC separation was conducted using an Agilent Zorbax 300SB-C18 column (0.5 × 150 mm; 5 μm) eluted with a gradient of 15 mm NH4(CH3CO2) (buffer A) and CH3CN (buffer B) at a flow rate of 15 μl/min. The percentage of solvent B was changed linearly from 1 to 10% in 24 min, further to 75% in 1 min, held at 75% for 3 min, and finally brought back to 1% in 2 min (all v/v). Nucleotide sequences of the extension products were confirmed by comparing the observed MS/MS fragments to the expected collision-induced dissociation fragmentation patterns of oligodeoxynucleotides (Mongo Oligo mass calculator version 2.06, available online from The RNA Institute, College of Arts and Sciences, State University of New York at Albany).
Modeling and Simulation
All molecular modeling was carried out using the Schrödinger modeling package (Schrödinger LLC, New York, NY). The starting model of hPol κ in complex with unmodified DNA was based on our earlier publication (37). We placed the Ac-EQKLISEEDL-NH2 peptide lesion with its N terminus directed toward the 3′-end of the template strand; the opposite orientation, in the 5′-direction, is crowded, whereas the 3′-orientation places the peptide comfortably in the major groove. The templating G within the polymerase active site was replaced by a 7-deazaguanine and conjugated to the side chain of a single lysine residue via a two-carbon linker (Fig. 1). Individual amino acids were iteratively added and locally minimized with fixed protein backbone and DNA until the full sequence of the 10-mer peptide (Ac-QKLISEEDL-NH2 where lysine is conjugated to DNA) was assembled. The final model was solvated with a 15-Å buffer region of explicit TIP3 water layer containing 0.15 m NaCl as counterions inside a rectangular box. The long range electrostatic interactions were evaluated by the particle mesh Ewald method under periodic boundary conditions. MD simulation was carried out using Desmond (D. E. Shaw Research, New York, NY) with the default initialization protocol followed by a 100-ns production simulation run under isothermal isobaric (NPT) conditions at 300 K and 1 atm with the OPLS-AA 2005 force field. A force constant of 50 kcal/mol Å2 restraint was applied to the heavy atoms of DNA. The structural changes to hPol κ in the presence of the 10-mer peptide cross-link were assessed by evaluating the Cα RMSF and the Cα RMSD of the protein with respect to the minimized starting structure.
Author Contributions
N. T. conceived and directed the studies. Y. S. and M. G. P. prepared recombinant DNA polymerases. S. W. and S. J. conducted steady-state kinetics analyses and LC-MS analyses. S. M. and O. D. S. synthesized oligonucleotide containing deaza-DHP-dG. Y. Y. S. conducted molecular modeling. L. L.-H. and I. F. interpreted molecular modeling results and generated figures for publication. S. W. and N. T. wrote the paper with the help of Y. Y. S., M. D., S. B., O. D. S., and F. P. G.
Supplementary Material
Acknowledgments
We thank Dr. Chuan He (Department of Chemistry, University of Chicago) for the generous donation of recombinant AlkB protein, Dr. Dan Harki and Dr. Angela Perkins (Department of Medicinal Chemistry, University of Minnesota) for synthesizing the Ac-Myc peptide, Jeffrey Vervacke for synthesizing the tetanus toxoid peptide, and Bob Carlson (Masonic Cancer Center, University of Minnesota) for preparing the figures for this manuscript. We thank the University of Minnesota Supercomputing Institute for providing all the necessary computational resources.
This work was supported by National Institutes of Health NIEHS Grants R01 ES023350 (to N. T.), R01 ES010546 (to F. P. G.), and R01 ES025987 (to S. B.); NCI, National Institutes of Health Grants R01 CA100670 (to N. T.) and R01 CA75449 (to S. B.); and NIGMS, National Institutes of Health Grant R01 GM084152 (to M. D.) and a Brainstorm Award from the University of Minnesota Masonic Cancer Center. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains a supplemental movie.
A. Groehler and N. Tretyakova, unpublished observations.
- DPC
- DNA-protein cross-link
- TLS
- translesion synthesis
- ESI
- electrospray ionization
- eGFP
- enhanced GFP
- RMSD
- root mean square deviation
- RMSF
- root mean square fluctuation
- hPol
- human polymerase
- deaza-DHP-dG
- 7-deaza-7-(2,3-dihydroxypropan-1-yl)-2′-deoxyguanosine
- Pol
- polymerase
- dmf
- dimethylformadine
- CE
- cyanoethyl
- CPG
- controlled pore glass
- PNK
- polynucleotide kinase
- FAM
- fluorescein amidite
- MD
- molecular dynamics.
References
- 1. Barker S., Weinfeld M., and Murray D. (2005) DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 589, 111–135 [DOI] [PubMed] [Google Scholar]
- 2. Ide H., Shoulkamy M. I., Nakano T., Miyamoto-Matsubara M., and Salem A. M. (2011) Repair and biochemical effects of DNA-protein crosslinks. Mutat. Res. 711, 113–122 [DOI] [PubMed] [Google Scholar]
- 3. Loeber R. L., Michaelson-Richie E. D., Codreanu S. G., Liebler D. C., Campbell C. R., and Tretyakova N. Y. (2009) Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem. Res. Toxicol. 22, 1151–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michaelson-Richie E. D., Ming X., Codreanu S. G., Loeber R. L., Liebler D. C., Campbell C., and Tretyakova N. Y. (2011) Mechlorethamine-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J. Proteome. Res. 10, 2785–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michaelson-Richie E. D., Loeber R. L., Codreanu S. G., Ming X., Liebler D. C., Campbell C., and Tretyakova N. Y. (2010) DNA-protein cross-linking by 1,2,3,4-diepoxybutane. J. Proteome Res. 9, 4356–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barker S., Weinfeld M., Zheng J., Li L., and Murray D. (2005) Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J. Biol. Chem. 280, 33826–33838 [DOI] [PubMed] [Google Scholar]
- 7. Tretyakova N. Y., Groehler A. 4th, and Ji S. (2015) DNA-protein cross-links: formation, structural identities, and biological outcomes. Acc. Chem. Res. 48, 1631–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ming X. (2011) DNA-Protein Cross-linking by cis-1,1,2,2-Diamminedichloroplatinum(II) (Cisplatin). M.Sc. thesis, University of Minnesota [Google Scholar]
- 9. O'Connor P. M., and Fox B. W. (1989) Isolation and characterization of proteins cross-linked to DNA by the antitumor agent methylene dimethanesulfonate and its hydrolytic product formaldehyde. J. Biol. Chem. 264, 6391–6397 [PubMed] [Google Scholar]
- 10. Gherezghiher T. B., Ming X., Villalta P. W., Campbell C., and Tretyakova N. Y. (2013) 1,2,3,4-Diepoxybutane-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. J. Proteome Res. 12, 2151–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bau D. T., Wang T. S., Chung C. H., Wang A. S., Wang A.S., and Jan K. Y. (2002) Oxidative DNA adducts and DNA-protein cross-links are the major DNA lesions induced by arsenite. Environ. Health Perspect. 110, 753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhitkovich A., Voitkun V., Kluz T., and Costa M. (1998) Utilization of DNA-protein cross-links as a biomarker of chromium exposure. Environ. Health Perspect. 106, 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chakrabarti S. K., Bai C., and Subramanian K. S. (1999) DNA-protein crosslinks induced by nickel compounds in isolated rat renal cortical cells and its antagonism by specific amino acids and magnesium ion. Toxicol. Appl. Pharmacol. 154, 245–255 [DOI] [PubMed] [Google Scholar]
- 14. Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., and Ide H. (2003) DNA-protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 278, 25264–25272 [DOI] [PubMed] [Google Scholar]
- 15. Dizdaroglu M., and Gajewski E. (1989) Structure and mechanism of hydroxyl radical-induced formation of a DNA-protein cross-link involving thymine and lysine in nucleohistone. Cancer Res. 49, 3463–3467 [PubMed] [Google Scholar]
- 16. Gajewski E., and Dizdaroglu M. (1990) Hydroxyl radical induced cross-linking of cytosine and tyrosine in nucleohistone. Biochemistry 29, 977–980 [DOI] [PubMed] [Google Scholar]
- 17. Gueranger Q., Kia A., Frith D., and Karran P. (2011) Crosslinking of DNA repair and replication proteins to DNA in cells treated with 6-thioguanine and UVA. Nucleic Acids Res. 39, 5057–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tretyakova N. Y., Michaelson-Richie E. D., Gherezghiher T. B., Kurtz J., Ming X., Wickramaratne S., Campion M., Kanugula S., Pegg A. E., and Campbell C. (2013) DNA-reactive protein monoepoxides induce cell death and mutagenesis in mammalian cells. Biochemistry 52, 3171–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groehler A. 4th, Villalta P. W., Campbell C., and Tretyakova N. (2016) Covalent DNA-protein cross-linking by phosphoramide mustard and nornitrogen mustard in human cells. Chem. Res. Toxicol. 29, 190–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stingele J., Schwarz M. S., Bloemeke N., Wolf P. G., and Jentsch S. (2014) A DNA-dependent protease involved in DNA-protein crosslink repair. Cell 158, 327–338 [DOI] [PubMed] [Google Scholar]
- 21. Stingele J., Habermann B., and Jentsch S. (2015) DNA-protein crosslink repair: proteases as DNA repair enzymes. Trends Biochem. Sci. 40, 67–71 [DOI] [PubMed] [Google Scholar]
- 22. Duxin J. P., Dewar J. M., Yardimci H., and Walter J. C. (2014) Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singer B., and Grunberger D. (1983) Molecular Biology of Mutagens and Carcinogens, Plenum Press, New York [Google Scholar]
- 24. Gates K. S., Nooner T., and Dutta S. (2004) Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol. 17, 839–856 [DOI] [PubMed] [Google Scholar]
- 25. Wickramaratne S., Mukherjee S., Villalta P. W., Schärer O. D., and Tretyakova N. Y. (2013) Synthesis of sequence-specific DNA-protein conjugates via a reductive amination strategy. Bioconjug. Chem. 24, 1496–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loecken E. M., and Guengerich F. P. (2008) Reactions of glyceraldehyde 3-phosphate dehydrogenase sulfhydryl groups with bis-electrophiles produce DNA-protein cross-links but not mutations. Chem. Res. Toxicol. 21, 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Angelov T., Guainazzi A., and Schärer O. D. (2009) Generation of DNA interstrand cross-links by post-synthetic reductive amination. Org. Lett. 11, 661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ho T. V., Guainazzi A., Derkunt S. B., Enoiu M., and Schärer O. D. (2011) Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 39, 7455–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeo J. E., Wickramaratne S., Khatwani S., Wang Y. C., Vervacke J., Distefano M. D., and Tretyakova N. Y. (2014) Synthesis of site-specific DNA-protein conjugates and their effects on DNA replication. ACS Chem. Biol. 9, 1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nilsson J. A., and Cleveland J. L. (2003) Myc pathways provoking cell suicide and cancer. Oncogene 22, 9007–9021 [DOI] [PubMed] [Google Scholar]
- 31. Hoffman B., and Liebermann D. A. (2008) Apoptotic signaling by c-MYC. Oncogene 27, 6462–6472 [DOI] [PubMed] [Google Scholar]
- 32. Geacintov N. E., and Broyde S. (2010) The Chemical Biology of DNA Damage, Wiley, Hoboken, NJ [Google Scholar]
- 33. Zang H., Goodenough A. K., Choi J. Y., Irimia A., Loukachevitch L. V., Kozekov I. D., Angel K. C., Rizzo C. J., Egli M., and Guengerich F. P. (2005) DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 280, 29750–29764 [DOI] [PubMed] [Google Scholar]
- 34. Maddukuri L., Eoff R. L., Choi J. Y., Rizzo C. J., Guengerich F. P., and Marnett L. J. (2010) In vitro bypass of the major malondialdehyde- and base propenal-derived DNA adduct by human Y-family DNA polymerases κ, ι, and Rev1. Biochemistry 49, 8415–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kotapati S., Maddukuri L., Wickramaratne S., Seneviratne U., Goggin M., Pence M. G., Villalta P., Guengerich F. P., Marnett L., and Tretyakova N. (2012) Translesion synthesis across 1,N6-(2-hydroxy-3-hydroxymethylpropan-1,3-diyl)-2′-deoxyadenosine (1,N6-γ-HMHP-dA) adducts by human and archebacterial DNA polymerases. J. Biol. Chem. 287, 38800–38811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wickramaratne S., Boldry E. J., Buehler C., Wang Y. C., Distefano M. D., and Tretyakova N. Y. (2015) Error-prone translesion synthesis past DNA-peptide cross-links conjugated to the major groove of DNA via C5 of thymidine. J. Biol. Chem. 290, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lior-Hoffmann L., Ding S., Geacintov N. E., Zhang Y., and Broyde S. (2014) Structural and dynamic characterization of polymerase κ's minor groove lesion processing reveals how adduct topology impacts fidelity. Biochemistry 53, 5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lone S., Townson S. A., Uljon S. N., Johnson R. E., Brahma A., Nair D. T., Prakash S., Prakash L., and Aggarwal A. K. (2007) Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell 25, 601–614 [DOI] [PubMed] [Google Scholar]
- 39. Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev. 13, 2191–2195 [DOI] [PubMed] [Google Scholar]
- 40. Friedberg E. C., Lehmann A. R., and Fuchs R. P. (2005) Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell 18, 499–505 [DOI] [PubMed] [Google Scholar]
- 41. Lehmann A. R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J. F., Kannouche P. L., and Green C. M. (2007) Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 6, 891–899 [DOI] [PubMed] [Google Scholar]
- 42. Huen M. S., and Chen J. (2008) The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 18, 8–16 [DOI] [PubMed] [Google Scholar]
- 43. Sale J. E., Lehmann A. R., and Woodgate R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burgers P. M., Koonin E. V., Bruford E., Blanco L., Burtis K. C., Christman M. F., Copeland W. C., Friedberg E. C., Hanaoka F., Hinkle D. C., Lawrence C. W., Nakanishi M., Ohmori H., Prakash L., Prakash S., Reynaud C. A., Sugino A., Todo T., Wang Z., Weill J. C., and Woodgate R. (2001) Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 276, 43487–43490 [DOI] [PubMed] [Google Scholar]
- 45. Ohmori H., Friedberg E. C., Fuchs R. P., Goodman M. F., Hanaoka F., Hinkle D., Kunkel T. A., Lawrence C. W., Livneh Z., Nohmi T., Prakash L., Prakash S., Todo T., Walker G. C., Wang Z., and Woodgate R. (2001) The Y-family of DNA polymerases. Mol. Cell 8, 7–8 [DOI] [PubMed] [Google Scholar]
- 46. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., and Walker G. C. (2009) Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marini F., Kim N., Schuffert A., and Wood R. D. (2003) POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 278, 32014–32019 [DOI] [PubMed] [Google Scholar]
- 48. McCulloch S. D., and Kunkel T. A. (2008) The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Friedberg E. C. (2005) Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 6, 943–953 [DOI] [PubMed] [Google Scholar]
- 50. Yang W. (2005) Portraits of a Y-family DNA polymerase. FEBS Lett. 579, 868–872 [DOI] [PubMed] [Google Scholar]
- 51. Yang W., and Woodgate R. (2007) What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mailand N., Gibbs-Seymour I., and Bekker-Jensen S. (2013) Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14, 269–282 [DOI] [PubMed] [Google Scholar]
- 53. Matsuda T., Bebenek K., Masutani C., Hanaoka F., and Kunkel T. A. (2000) Low fidelity DNA synthesis by human DNA polymerase η. Nature 404, 1011–1013 [DOI] [PubMed] [Google Scholar]
- 54. Goodman M. F. (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71, 17–50 [DOI] [PubMed] [Google Scholar]
- 55. Prakash S., Johnson R. E., and Prakash L. (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 56. Kuo H. K., Griffith J. D., and Kreuzer K. N. (2007) 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 67, 8248–8254 [DOI] [PubMed] [Google Scholar]
- 57. Loeber R., Michaelson E., Fang Q., Campbell C., Pegg A. E., and Tretyakova N. (2008) Cross-linking of the DNA repair protein O6-alkylguanine DNA alkyltransferase to DNA in the presence of antitumor nitrogen mustards. Chem. Res. Toxicol. 21, 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kelland L. (2007) The resurgence of platinum-based cancer chemotherapy. Nat. Rev. Cancer 7, 573–584 [DOI] [PubMed] [Google Scholar]
- 59. Wheate N. J., Walker S., Craig G. E., and Oun R. (2010) The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 39, 8113–8127 [DOI] [PubMed] [Google Scholar]
- 60. Hall A. G., and Tilby M. J. (1992) Mechanisms of action of, and modes of resistance to, alkylating agents used in the treatment of haematological malignancies. Blood Rev. 6, 163–173 [DOI] [PubMed] [Google Scholar]
- 61. Wang C. C., Li J., Teo C. S., and Lee T. (1999) The delivery of BCNU to brain tumors. J. Control Release 61, 21–41 [DOI] [PubMed] [Google Scholar]
- 62. Gilman A., and Philips F. S. (1946) The biological actions and therapeutic applications of the β-chloroethyl amines and sulfides. Science 103, 409–436 [DOI] [PubMed] [Google Scholar]
- 63. Nakano T., Morishita S., Katafuchi A., Matsubara M., Horikawa Y., Terato H., Salem A. M., Izumi S., Pack S. P., Makino K., and Ide H. (2007) Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell 28, 147–158 [DOI] [PubMed] [Google Scholar]
- 64. Kumari A., Minko I. G., Smith R. L., Lloyd R. S., and McCullough A. K. (2010) Modulation of UvrD helicase activity by covalent DNA-protein cross-links. J. Biol. Chem. 285, 21313–21322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Minko I. G., Zou Y., and Lloyd R. S. (2002) Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 99, 1905–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Quievryn G., and Zhitkovich A. (2000) Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 21, 1573–1580 [PubMed] [Google Scholar]
- 67. Baker D. J., Wuenschell G., Xia L., Termini J., Bates S. E., Riggs A. D., and O'Connor T. R. (2007) Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 282, 22592–22604 [DOI] [PubMed] [Google Scholar]
- 68. Reardon J. T., Cheng Y., and Sancar A. (2006) Repair of DNA-protein cross-links in mammalian cells. Cell Cycle 5, 1366–1370 [DOI] [PubMed] [Google Scholar]
- 69. Yamanaka K., Minko I. G., Takata K., Kolbanovskiy A., Kozekov I. D., Wood R. D., Rizzo C. J., and Lloyd R. S. (2010) Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem. Res. Toxicol. 23, 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Minko I. G., Yamanaka K., Kozekov I. D., Kozekova A., Indiani C., O'Donnell M. E., Jiang Q., Goodman M. F., Rizzo C. J., and Lloyd R. S. (2008) Replication bypass of the acrolein-mediated deoxyguanine DNA-peptide cross-links by DNA polymerases of the DinB family. Chem. Res. Toxicol. 21, 1983–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Levine R. L., Miller H., Grollman A., Ohashi E., Ohmori H., Masutani C., Hanaoka F., and Moriya M. (2001) Translesion DNA synthesis catalyzed by human pol η and pol κ across 1,N6-ethenodeoxyadenosine. J. Biol. Chem. 276, 18717–18721 [DOI] [PubMed] [Google Scholar]
- 72. Zhao L., Pence M. G., Christov P. P., Wawrzak Z., Choi J. Y., Rizzo C. J., Egli M., and Guengerich F. P. (2012) Basis of miscoding of the DNA adduct N2,3-ethenoguanine by human Y-family DNA polymerases. J. Biol. Chem. 287, 35516–35526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Patra A., Su Y., Zhang Q., Johnson K. M., Guengerich F. P., and Egli M. (2016) Structural and kinetic analysis of miscoding opposite the DNA adduct 1,N6-ethenodeoxyadenosine by human translesion polymerase η. J. Biol. Chem. 291, 14134–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cho S. H., and Guengerich F. P. (2013) Replication past the butadiene diepoxide-derived DNA adduct S-[4-(N6-deoxyadenosinyl)-2,3-dihydroxybutyl]glutathione by DNA polymerases. Chem. Res. Toxicol. 26, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ganguly M., Wang F., Kaushik M., Stone M. P., Marky L. A., and Gold B. (2007) A study of 7-deaza-2′-deoxyguanosine 2′-deoxycytidine base pairing in DNA. Nucleic Acids Res. 35, 6181–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kowal E. A., Ganguly M., Pallan P. S., Marky L. A., Gold B., Egli M., and Stone M. P. (2011) Altering the electrostatic potential in the major groove: thermodynamic and structural characterization of 7-deaza-2′-deoxyadenosine:dT base pairing in DNA. J. Phys. Chem. B 115, 13925–13934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Räschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., and Walter J. C. (2008) Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Enoiu M., Ho T. V., Long D. T., Walter J. C., and Schärer O. D. (2012) Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol. Biol. 920, 203–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Roy U., and Schärer O. D. (2016) Involvement of translesion synthesis DNA polymerases in DNA interstrand crosslink repair. DNA Repair 44, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rechkoblit O., Zhang Y., Guo D., Wang Z., Amin S., Krzeminsky J., Louneva N., and Geacintov N. E. (2002) trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 277, 30488–30494 [DOI] [PubMed] [Google Scholar]
- 81. Jia L., Geacintov N. E., and Broyde S. (2008) The N-clasp of human DNA polymerase κ promotes blockage or error-free bypass of adenine- or guanine-benzo[a]pyrenyl lesions. Nucleic Acids Res. 36, 6571–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yang W. (2014) An overview of Y-family DNA polymerases and a case study of human DNA polymerase η. Biochemistry 53, 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fedtke N., Boucheron J. A., Walker V. E., and Swenberg J. A. (1990) Vinyl chloride-induced DNA adducts. II: Formation and persistence of 7-(2′-oxoethyl)guanine and N2,3-ethenoguanine in rat tissue DNA. Carcinogenesis 11, 1287–1292 [DOI] [PubMed] [Google Scholar]
- 84. Patra A., Nagy L. D., Zhang Q., Su Y., Müller L., Guengerich F. P., and Egli M. (2014) Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase η. J. Biol. Chem. 289, 16867–16882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Irimia A., Eoff R. L., Guengerich F. P., and Egli M. (2009) Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J. Biol. Chem. 284, 22467–22480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chung J. A., Wollack J. W., Hovlid M. L., Okesli A., Chen Y., Mueller J. D., Distefano M. D., and Taton T. A. (2009) Purification of prenylated proteins by affinity chromatography on cyclodextrin-modified agarose. Anal. Biochem. 386, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.