Abstract
The mitochondrial electron transport chain consists of individual protein complexes arranged into large macromolecular structures, termed respiratory chain supercomplexes or respirasomes. In the yeast Saccharomyces cerevisiae, respiratory chain supercomplexes form by association of the bc1 complex with the cytochrome c oxidase. Formation and maintenance of these assemblies are promoted by specific respiratory supercomplex factors, the Rcf proteins. For these proteins a regulatory function in bridging the electron transfer within supercomplexes has been proposed. Here we report on the maturation of Rcf2 into an N- and C-terminal peptide. We show that the previously uncharacterized Rcf3 (YBR255c-A) is a homolog of the N-terminal Rcf2 peptide, whereas Rcf1 is homologous to the C-terminal portion. Both Rcf3 and the C-terminal fragment of Rcf2 associate with monomeric cytochrome c oxidase and respiratory chain supercomplexes. A lack of Rcf2 and Rcf3 increases oxygen flux through the respiratory chain by up-regulation of the cytochrome c oxidase activity. A double gene deletion of RCF2 and RCF3 affects cellular survival under non-fermentable growth conditions, suggesting an overlapping role for both proteins in the regulation of the OXPHOS activity. Furthermore, our data suggest an association of all three Rcf proteins with the bc1 complex in the absence of a functional cytochrome c oxidase and identify a supercomplex independent interaction network of the Rcf proteins.
Keywords: cytochrome c oxidase (complex IV), membrane protein, mitochondria, protein assembly, respiratory chain
Introduction
Eukaryotic cells cover their energy demands mainly with ATP generated by oxidative phosphorylation in mitochondria. The complexes of the respiratory chain in the inner mitochondrial membrane establish a proton motif force, which is used by the F1Fo-ATP synthase to generate ATP from ADP and inorganic phosphate. In the yeast Saccharomyces cerevisiae, cytosolic substrates like NADH or glycerol-3-phosphate can be oxidized via distinct mitochondrial enzymes like NADH-ubiquinone oxidoreductase proteins or glycerol-3-phosphate dehydrogenase located either on the intermembrane space site of the mitochondrial inner membrane (Nde1, Nde2, or Gut2) or facing the matrix (Ndi1) (1). The mitochondrial respiratory chain is further formed by multiprotein containing enzymes like the succinate dehydrogenase (complex II), the ubiquinol-cytochrome c oxidoreductase (complex III), and the cytochrome c oxidase (complex IV) (2–4). The latter two complexes are composed of proteins with dual genetic origin. The mitochondrial genome encodes for the hydrophobic core subunits of complex III and complex IV. Nuclear encoded structural and regulatory subunits are post-translationally transported into mitochondria by defined import routes and incorporated with the help of assembly factors into the growing enzymes (5–7). The respiratory chain complexes III and IV form homo- and hetero-oligomers in the inner membrane. A dimer of complex III (III2) associates with one (III2/IV) or two (III2/IV2) copies of complex IV to form respiratory chain supercomplexes (8–10). Although the function and formation of supercomplexes has been extensively studied in the past decade, it is not fully understood which physiological changes drive supercomplex formation and which of the factors are required to mediated formation and maintenance of these complexes. Suggested functions of supercomplexes are an optimized electron transport between the respiratory chain complexes, reduction of reactive oxygen species leakage, and a general stabilizing effect on the individual complexes (11–13). Cardiolipin and a small number of proteins participate in supercomplex formation (14–18). Recent studies identified Rcf1 as a protein that promotes efficient assembly of III2IV2 supercomplexes and is further proposed to regulate supercomplex function. Rcf2 was identified together with Rcf1 as complex IV-associated protein (19–22). Rcf2 displays sequence similarity to Rcf1 but is not required for supercomplex organization, and its molecular function has remained enigmatic.
Here we analyzed assembly of Rcf2 into supercomplexes and identified an unexpected modification of the protein by proteolytic processing into a labile N-terminal fragment and a stable C-terminal fragment. Using in silico analysis, we mapped the Rcf1 homology region of Rcf2 to the C terminus, whereas the N terminus resembles an uncharacterized protein encoded by the open reading frame YBR255c-A. The YBR255c-A gene product, later termed Rcf3, encodes a mitochondrial protein that is located in the inner membrane. Rcf3 associates with supercomplexes predominantly via complex IV. Single gene deletions of RCF2 and RCF3 showed an increased oxygen flux via complex IV but no growth defect on non-fermentable medium. Contrary to the single mutants, the rcf2Δ/rcf3Δ double deletion results in a decreased oxygen flux and loses the capability of the cells to growth on non-fermentable media, revealing an overlapping role of both proteins in mitochondrial respiration. In addition, our analyses on the Rcf3 interaction environment reveal that the Rcf proteins associate with complex III in the absence of complex IV. The observed affinity of Rcf proteins for complex III provides new insight into the molecular basis of supercomplexes.
Results and Discussion
Respiratory chain supercomplexes assemble from individual complexes. Rcf1 and Rcf2 have been identified as supercomplex-associated proteins (19–21). In S. cerevisiae, Rcf1 facilitates the oligomerization and stability of the complex III (cytochrome c reductase) dimer with complex IV (cytochrome c oxidase). Although Rcf1 is conserved from yeast to human and shows a direct role in the association of the complex III/complex IV into supercomplexes, the function of Rcf2 is still unknown.
A Processed Form of Rcf2 Associates with Respiratory Chain Supercomplexes
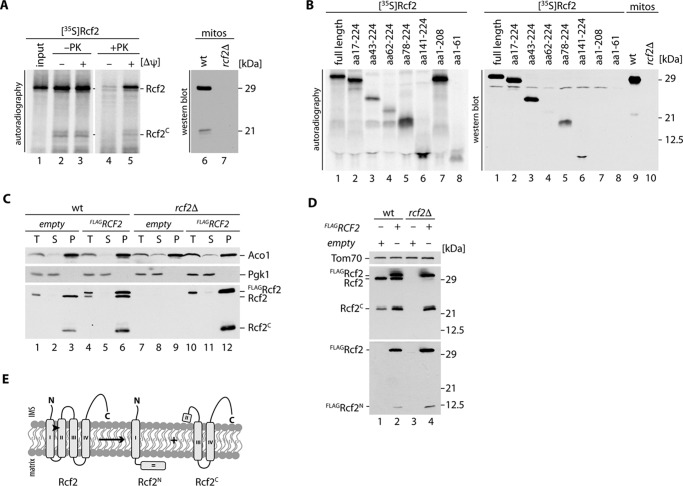
Rcf2 lacks a classical mitochondrial targeting sequence but can be efficiently imported into mitochondria in vitro. When we imported radiolabeled Rcf2 into purified wild-type mitochondria, we observed formation of an Rcf2 fragment with an estimated mass of 21 kDa. The fragment was only protected against protease treatment when a membrane potential (Δψ) was present (Fig. 1A). A corresponding Rcf2 fragment was also detected with lower intensity of the full-length protein in Western blot analysis of mitochondrial extracts using Rcf2-specific antiserum (Fig. 1A). Because the Rcf2 antiserum was raised against a C-terminal peptide of Rcf2, we concluded that the detected fragment contained the C terminus of Rcf2. To define the size of the Rcf2 fragment, we created a library of Rcf2 truncation constructs in vitro. Seven truncations of Rcf2, as well as the full-length protein, were translated and radiolabeled in reticulocyte lysate and separated by SDS-PAGE. For comparison, mitochondrial extracts from wild-type and rcf2Δ cells were separated in parallel. Compared with the Western blot analysis, the autoradiogram of the radiolabeled Rcf2 truncations revealed two fragments, amino acids 62–224 and 78–224 (Fig. 1B, lanes 4 and 5), that displayed a migration pattern in the size range of the endogenous C-terminal fragment of Rcf2 (Fig. 1B, lane 9). This finding led us to investigate the fate of the cleaved N- and C-terminal fragments. Hence, we cloned N-terminally FLAG tagged Rcf2 (FLAGRcf2) for expression in wild-type and rcf2Δ yeast cells. We performed subcellular fractionation analyses and could show that FLAGRcf2 was expressed and migrated slightly slower than the untagged protein. FLAGRcf2 and the C-terminal fragment could be detected in the organellar fraction like the mitochondrial matrix protein aconitase 1 (Aco1), whereas cytosolic proteins like phosphoglycerate kinase 1 (Pgk1) stayed in the supernatant (Fig. 1C). However, the N-terminal fragment was not detected because of its low abundance. We prepared mitochondrial extracts from both strains. The FLAGRcf2 obviously localized to mitochondria and similar to the untagged protein underwent proteolytic processing. Hence, the C-terminal fragment was efficiently generated from the tagged construct. Only when we expressed FLAGRcf2, was a weak signal at the expected size of 12 kDa corresponding to the N-terminal fragment plus FLAG tag detected with FLAG antibodies (Fig. 1D). To evaluate the physiological relevance of the Rcf2 proteolytic processing, we investigated in the steady state levels of full-length and processed Rcf2 in wild-type mitochondria under different growth conditions. The amount of processed Rcf2 ranged from 55% of the full-length protein under fermentable growth conditions to 40–50% under growth on non-fermentable carbon sources, whereas the cell growth in the presence of galactose leads to lower levels (20–30%) of detectable processed Rcf2 fragment (supplemental Fig. S1A).
FIGURE 1.
After import a part of Rcf2 is processed into Rcf2C and an unstable Rcf2N fragment. A, radiolabeled Rcf2 was imported into isolated mitochondria for 15 min in the presence or absence of membrane potential (Δψ). Samples were treated with proteinase K where indicated and analyzed by SDS-PAGE and digital autoradiography. For comparison with endogenous Rcf2C, mitochondria were analyzed by SDS-PAGE followed by Western blotting and immunodecoration. B, radiolabeled Rcf2 and N- and C-terminally truncated constructs (ΔN and ΔC) were analyzed by SDS-PAGE and compared with endogenous Rcf2C. C, subcellular fractionation of wild-type and rcf2Δ cells expressing FLAGRcf2. Samples of homogenized cells (T), post mitochondrial supernatant (S), and mitochondrial pellet (P) were analyzed by SDS-PAGE and Western blotting. Cytosolic proteins are represented by Pgk1, and mitochondrial proteins are represented by Aco1. D, Isolated mitochondria of the strains used in C were analyzed by SDS-PAGE and Western blotting. Rcf2 variants were detected using FLAG and Rcf2 antibodies. E, model for Rcf2 processing in the inner mitochondrial membrane. The data shown in A–D represent the results of at least two individual experiments.
Rcf2 is predicted to possess four transmembrane segments (TMpred). Taking the predicted transmembrane spans into consideration, we conclude that the observed processing of Rcf2 occurs between amino acids 62 and 78 within the second transmembrane segment of Rcf2 (Fig. 1E). The processing generates a labile N-terminal and a stable C-terminal fragment of Rcf2.
Rcf2 Associates with Respiratory Chain Complexes through Its C-terminal Portion
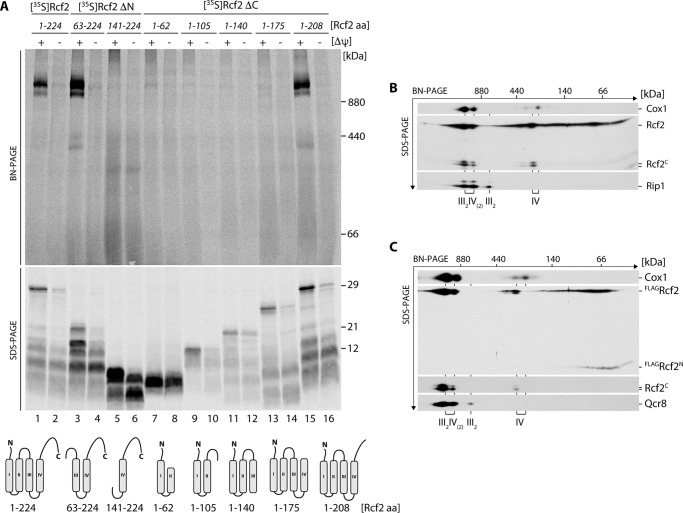
To address whether the labile N-terminal or the stable C-terminal fragment of Rcf2 were capable to associate with supercomplexes, we imported radiolabeled Rcf2 truncations and for comparison with the full-length Rcf2 into wild-type mitochondria. After import, mitochondria were solubilized in digitonin buffer. Solubilized protein complexes were separated by blue native (BN)-PAGE2 to assess assembly into supercomplexes. In parallel, the individual import reactions were analyzed by SDS-PAGE to assess whether the Rcf2 versions were imported into mitochondria. Membrane potential-dependent import into mitochondria was observed for all tested fragments, albeit with different efficiencies. However, assembly into supercomplexes could only be observed for fragments that contained at least the third and fourth transmembrane segment and a region of the C-terminal IMS tail of Rcf2 (Fig. 2A). Accordingly, the N-terminal Rcf2 fragment was unable to assemble into supercomplexes in vitro. At steady state, it is not possible to distinguish between the full-length Rcf2 protein and the N-terminal fragment within supercomplexes by BN-PAGE and Western blotting. Therefore, we carried out a second dimension SDS-PAGE after separation of the supercomplexes by BN-PAGE. In agreement with previous work, full-length Rcf2 co-migrated with complex IV-containing supercomplexes and monomeric complex IV and to a lower extent as a 66-kDa complex (Fig. 2B) (19). The stable C-terminal fragment behaves like the full-length protein and could be found associated with complex IV, either in its monomeric form or assembled into supercomplexes with complex III2 (Fig. 2B). In contrast, the labile N-terminal fragment was detected via the FLAG tag and no longer displayed an association with complex IV and supercomplexes (Fig. 2C). Based on this finding, we hypothesized that the processing of Rcf2 takes place after its association with complex IV and processing is physiologically relevant.
FIGURE 2.
Rcf2C is low abundant in the oxidase, whereas Rcf2N is free and not assembled into respiratory supercomplexes. A, radiolabeled N- and C-terminally truncated Rcf2 constructs (ΔN and ΔC) were imported into isolated mitochondria for 45 min in the presence or absence of membrane potential (Δψ). Proteinase K-treated samples were split for solubilization in 1% digitonin buffer and SDS sample buffer. The samples were analyzed by BN-PAGE or SDS-PAGE and digital autoradiography. B, isolated wild-type mitochondria were solubilized in 1% digitonin and analyzed by BN-PAGE followed by a second dimension SDS-PAGE and Western blotting. Cox1 and Rip1 indicate the positions of respiratory supercomplexes (III2IV2 and III2IV), complex III dimer (III2) and complex IV. Rcf2 and Rcf2C were detected using Rcf2 antibody. C, isolated mitochondria of FLAGRcf2-expressing rcf2Δ were analyzed as in B. Qcr8 was used as marker for complex III. Rcf2 variants were detected using a FLAG and Rcf2 antibodies. The data shown in A–C represent the results of at least two individual experiments.
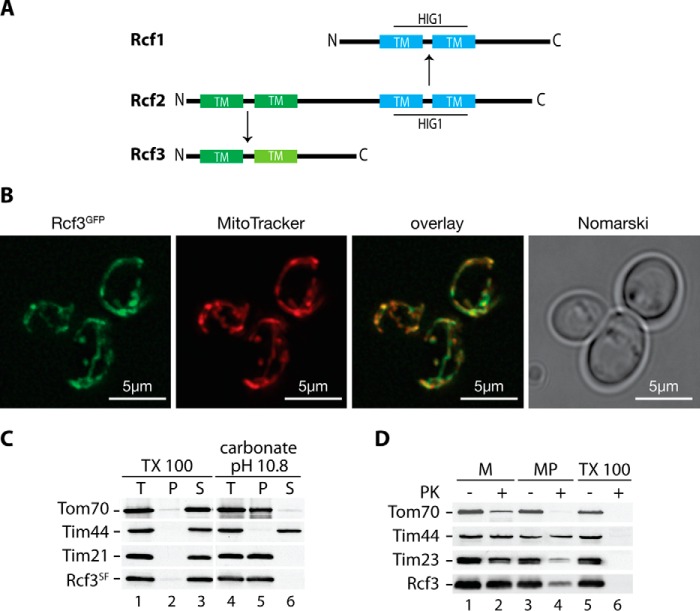
Rcf3 Is an Integral Protein of the Mitochondrial Inner Membrane
The C-terminal fragment of Rcf2 shares two major features with the Rcf1 protein. Both proteins display similar topology with regard to their predicted membrane topology. Moreover, Rcf1 and Rcf2 comprise a conserved HIG1 domain, the function of which is unknown (21). Interestingly, the N-terminal fragment of Rcf2 displays significant sequence similarity to an uncharacterized protein encoded by the yeast open reading frame YBR255C-A, later called Rcf3 (Fig. 3A and supplemental Fig. S2). Rcf3 was reported in a proteomic analysis by Helbig et al. (34) as a yeast mitochondrial membrane protein associated with respiratory supercomplexes but was not further analyzed in detail (34). The similarity between Rcf2 and Rcf3 led us to assess the mitochondrial localization of Rcf3. We expressed a GFP fusion of the protein from its authentic chromosomal locus. Fluorescence microscopic analyses of living cells expressing Rcf3GFP showed co-localization of the GFP signal with mitochondria as visualized by MitoTracker staining (Fig. 3B), revealing an exclusive localization of Rcf3 in mitochondria. Computer-based analysis of the primary sequence of Rcf3 predict two transmembrane segments within the protein (Fig. 3A and supplemental Fig. S2). To confirm this prediction, we performed carbonate extraction analyses using isolated yeast mitochondria. Rcf3 was resistant to alkaline treatment at pH 10.8 and remained in the sediment together with the integral membrane proteins Tom70 and Tim21, whereas the peripheral membrane protein Tim44 was released to the supernatant. All tested proteins including Rcf3 were released from the membrane fraction upon solubilization with Triton X-100 (Fig. 3C). These analyses supported the prediction that Rcf3 is an integral membrane protein. To determine the submitochondrial localization of Rcf3, we performed protease protection experiments with isolated mitochondria. The Rcf3 protein was protected against protease treatment in mitochondria but became accessible to the protease in mitoplasts (Fig. 3D). We conclude that Rcf3 is an integral protein of the mitochondrial inner membrane. Because we used an antibody directed against the C terminus of Rcf3, the protease sensitivity in mitoplasts indicates that Rcf3 exposes its C terminus to the intermembrane space.
FIGURE 3.
Rcf3 (YBR255C-A) is an integral inner mitochondrial membrane protein. A, alignment of Rcf1, Rcf2, and Rcf3 (YBR255C-A). Dark green and cyan indicate transmembrane spans (TM); light green indicates a possible transmembrane span or hydrophobic patch. B, live cell fluorescence microscopy of Rcf3GFP expressing cells with stained mitochondria by MitoTracker treatment. Scale bar, 5 μm. C, mitochondria of Rcf3SF expressing cells were subjected to carbonate extraction or lysed with 1% Triton X-100 and were separated by ultracentrifugation into pellet (P), supernatant (S), and total (T). Samples were analyzed by SDS-PAGE and Western blot analysis. D, wild-type mitochondria were left untreated (M), swollen (MP), or lysed with 1% Triton X-100, treated with proteinase K where indicated, and subjected to SDS-PAGE and Western blotting. The data shown in B–D represent the results of at least two individual experiments.
Phenotypic Characterization of rcf3Δ Mutant Mitochondria
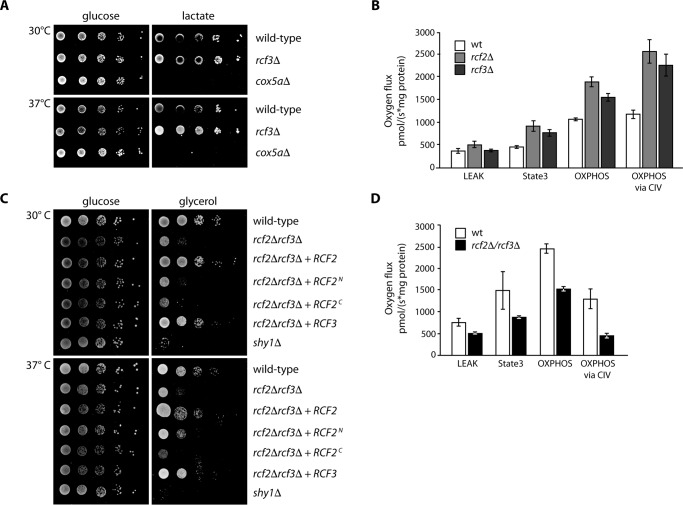
To assess Rcf3 function, we generated an RCF3 gene deletion mutant to analyze respiratory chain function and organization. When we compared the growth of rcf3Δ to cox5aΔ cells, we found that RCF3 was dispensable for the growth of yeast cells on fermentable carbon sources. Although the deletion of COX5a displayed a strong growth defect on non-fermentable media such as lactate, this was not the case for the rcf3Δ mutant (Fig. 4A). Next we addressed the organization of respiratory chain supercomplexes at steady state. Therefore, we solubilized wild-type and rcf3Δ mitochondria in digitonin containing buffer and performed BN-PAGE analysis. The mutant mitochondria displayed no changes in the amount or the organization of the respiratory chain complexes III, IV, and V compared with wild-type mitochondria (supplemental Fig. S3A). The observation that the deletion of Cox13, a subunit of the yeast complex IV, results in a significant decrease in complex IV enzymatic activity and O2 consumption but does not lead to a growth phenotype on non-fermentable medium or change in the oligomerization of complex IV (19, 35), led us to measure the oxygen flux in rcf3Δ and rcf2Δ mutant mitochondria compared with wild type. Based on the used substrate in combination with the inhibition of specific respiratory chain complexes, we could distinguish between a leak, state3 oxphos, and oxphos via CIV-dependent oxygen flux (Fig. 4B). Deletion of rcf3Δ and rcf2Δ maintains an increased oxygen flux specifically related to complex IV activity. The boost in the complex IV-dependent oxygen flux was not related to an altered organization of respiratory chain supercomplexes at steady state (supplemental Fig. S3A) and approve a regulatory role for Rcf3 and Rcf2 in supercomplex function.
FIGURE 4.
Deletion of RCF3 and RCF2 influence mitochondrial respiration via complex IV. A, rcf3Δ cells were tested for growth on minimal media supplemented with glucose or lactate at indicated temperatures in comparison to wild-type and respiratory deficient cox5aΔ. B, oxygen flux rates of mitochondria derived from wild-type and rcf3Δ cells were analyzed in an Oxygraph 2 k (Oroboros) at 30 °C (n = 4). C, rcf2Δ/rcf3Δ cells with additional expression of Rcf2 derivates or Rcf3 were tested for growth on minimal medium supplemented with glucose or glycerol at the indicated temperatures in comparison to wild-type and respiratory deficient shy1Δ. D, oxygen flux rates were measured in isolated mitochondria of wild type and rcf2Δ/rcf3Δ in an Oxygraph 2 k (Oroboros) at 30 °C (n = 4). The data shown in A–D represent the results of at least three individual experiments.
Overlapping Role of Rcf2 and Rcf3
The primary amino acid sequence of Rcf2 seems to harboring the combined coding sequences of Rcf1 and Rcf3. In that regard, the C-terminal fragment of Rcf2 could overcome a possible mitochondrial defect caused by the deletion of the RCF3 gene. To prove this possibility, we generated a rcf2Δ/rcf3Δ double deletion strain and performed a growth test (Fig. 4C). The deletion of RCF3 together with RCF2 results in a drastic reduced growth on the non-fermentable carbon source glycerol. This growth phenotype could be reverted to wild-type level by re-expressing either Rcf2 or Rcf3. In addition, we tested whether the previously observed Rcf2 fragments could restore the growth on glycerol. None of the expressed Rcf2 fragments led to a reversion of the observed phenotype (Fig. 4C). Given that we do not know the exact position where the Rcf2 processing occurs, it is possible that the construct used in this study, which should resemble the N-terminal Rcf2 fragment, is too short for successful complementation of the double deletion strain. To investigate the reason for the growth defect, we analyzed the oxygen flux of mitochondria from the rcf2Δ/rcf3Δ mutant compared with wild type. In contrast to the rcf3Δ and rcf2Δ single mutants, we observed a decrease in the oxygen flux during our measurements of the rcf2Δ/rcf3Δ double deletion mitochondria (Fig. 4D). The use of N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) as direct complex IV substrate in combination with the inhibition of complex III by antimycin A showed us the specific down-regulation of respiration in the rcf2Δ/rcf3Δ double mutant mitochiondria via complex IV. To avoid the possibility that this effect is based on an altered organization of supercomplexes, as shown for Rcf1 (19–21), we performed BN-PAGE and Western blot analyses of the rcf2Δ/rcf3Δ mutant mitochondria compared with wild type and could not observe any changes in supercomplex formation (supplemental Fig. S3B). The growth defect of the rcf2Δ/rcf3Δ double deletion is most likely caused by the drop of complex IV activity. In combination with the results of the increased oxygen flux of the RCF3 and RCF2 single deletions, we suggest an overlapping role of Rcf3 and Rcf2 in the regulation of respiratory chain supercomplexes.
Rcf3 Associates with Respiratory Chain Supercomplexes
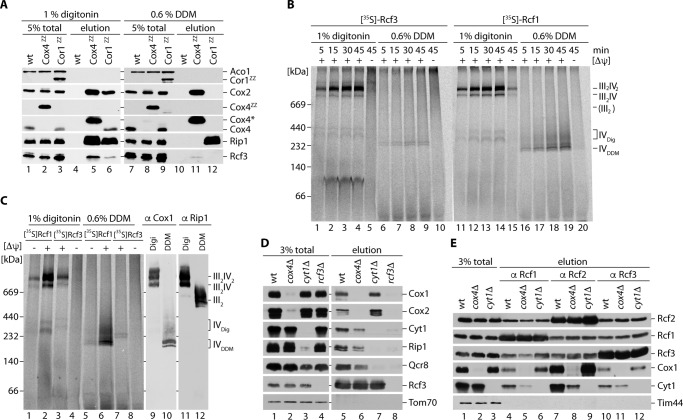
Rcf1 and Rcf2 are associated with complex III/IV supercomplexes. Detailed analysis by different groups showed that Rcf1 and Rcf2 assemble into supercomplexes via an association to complex IV. To address whether Rcf3 assembles into supercomplexes, we first analyzed Rcf3 interaction with complex III and IV. Therefore, we affinity-purified complex III and complex IV by IgG chromatography utilizing the ZZ-tagged structural subunits Cor1ZZ and Cox4ZZ (19, 27). Using the mild detergent digitonin for solubilization, we were able to efficiently purify complex III/IV supercomplexes. Rcf3 was co-isolated with Cor1ZZ and even more with Cox4ZZ (Fig. 5A, lanes 5 and 6). To assess whether Rcf3 was bound to complex III or IV, we dissociated supercomplexes into the complex III dimer and complex IV monomer by solubilizing mitochondria in DDM-containing buffer prior to isolation via Cor1ZZ and Cox4ZZ. Under these conditions, significantly less Rcf3 co-purified with Cox4ZZ. Moreover, Cor1ZZ failed to co-purify Rcf3 (Fig. 5A, lanes 11 and 12). These analyses suggested that the association of Rcf3 was sensitive to the DDM. To address respiratory chain association of Rcf3 by alternative means, we imported radiolabeled Rcf3 into purified mitochondria. After import, mitochondria were reisolated and solubilized in either digitonin or DDM containing buffer, and the protein complexes were separated by BN-PAGE. Upon solubilization in digitonin buffer, Rcf3 and Rcf1 assembled predominantly into two high molecular weight protein complexes, reminiscent of complex IV-containing supercomplexes. No complex corresponding to the size of a complex III dimer could be observed (Fig. 5B). After solubilization with DDM, Rcf3 and Rcf1 migrated at the size of monomeric complex IV populations. However, the amount of Rcf3 was drastically reduced in agreement with dissociation during the solubilization process (Fig. 5B). We repeated the import experiment and loaded solubilized mitochondria next to the import reaction samples on BN-PAGE for Western blot analysis of complex III and complex IV. Immunodecoration showed a co-migration of the imported Rcf3 and Rcf1 protein with the signal for Cox1, whereas a co-migration with Rip1 was only observed in cases where association between complex III and complex IV were maintained (Fig. 5C). Compared with the total amount of Rcf3 that is bound to supercomplexes under digitonin conditions, significantly less Rcf3 association to respiratory chain complexes could be observed upon DDM treatment. Because the Rcf3 antibody failed to detect the protein in Western blot analyses of separated supercomplexes by BN-PAGE after digitonin solubilization, we performed additional two-dimensional SDS-PAGE to detect the Rcf3 protein. Rcf3 co-migrates with complexes containing cytochrome c oxidase like Rcf1 and Rcf2 (supplemental Fig. S1B). By quantitative Western blot analyses of all three Rcf proteins, compared with Cox1 and Qcr8, we could confirm an overall equal tendency to associate either with supercomplexes (50–60%) or free complex IV (40–50%) (supplemental Fig. S1B). To assess Rcf3 association with complexes III or IV under milder conditions, we performed immunoprecipitations of Rcf3 from digitonin-solubilized wild type, cyt1Δ (lacking complex III), and cox4Δ (lacking complex IV) mitochondria. As a control for unspecific binding of mitochondrial proteins to the Rcf3 antibody, we included digitonin-solubilized rcf3Δ mitochondria. Supercomplex constituents were co-purified with Rcf3 in wild-type mitochondria. In the absence of Rcf3 (rcf3Δ), the Rcf3 antibody sample did not display unspecific binding of complex III or complex IV components (Fig. 5D, lane 8). The deletion of CYT1 leads to the loss of complex III formation leaving complex IV in its monomeric form (19, 27). Under these conditions Rcf3 is still associated with the complex IV components (Fig. 5D, lane 7). To our surprise, we could co-isolate the complex III structural subunits Cyt1 and Rip1 in the absence of an assembled complex IV, although in reduced amounts (Fig. 5D, lane 6). To this end, we found that the complex IV subunit Rcf3 associates with complex III in the absence of complex IV and independent of supercomplex formation. Based on this observation, we addressed whether this was also the case for Rcf1 and Rcf2. We performed immunoprecipitations with Rcf1, Rcf2, and Rcf3 antibodies using digitonin-solubilized wild-type, cyt1Δ, and cox4Δ mitochondria. The individual Rcf-antibodies precipitated their bait with different efficiencies. Nevertheless, all three Rcf proteins could co-isolate complex III and complex IV components and the two other Rcf proteins from wild-type mitochondria. In the absence of a mature complex III, the co-isolation of complex IV subunits with Rcf1, Rcf2, and Rcf3 were slightly increased compared with the wild-type sample (Fig. 5E). Finally, all three Rcf proteins co-purified complex III subunits in the absence of mature complex IV. At the same time the amount of co-isolated Rcf proteins was not altered. We conclude that Rcf1, Rcf2, and Rcf3 associate independent of complex III and complex IV (Fig. 5E).
FIGURE 5.
Rcf3 associates with complex III and IV and assembles into respiratory supercomplexes. A, mitochondria isolated from wild-type, Cox4ZZ, and Cor1ZZ strains were solubilized in 1% digitonin or 0.6% DDM and subjected to IgG chromatography. Upon TEV protease cleavage, eluates were analyzed by SDS-PAGE and Western blotting. Cox4* marks Cox4 after removal of ZZ by TEV cleavage. B, radiolabeled Rcf3 or Rcf1 were imported into isolated rcf3Δ mitochondria for indicated times in the presence or absence of membrane potential (ΔΨ). Proteinase K-treated samples were solubilized in 1% digitonin or 0.6% DDM and analyzed by BN-PAGE and digital autoradiography. IVDig and IVDDM indicate the size of the Rcf1-containing pool of monomeric complex IV when solubilized in the respective detergent. C, radiolabeled Rcf3 or Rcf1 were imported into isolated rcf3Δ mitochondria for 45 min and analyzed as specified in B. For comparison, wild-type mitochondria were solubilized in 1% digitonin or 0.6% DDM and analyzed by BN-PAGE and Western blotting. Complex III and IV containing assemblies were visualized by immunodetection of Rip1 and Cox1, respectively. D, digitonin-solubilized mitochondria of wild type, cox4Δ, cyt1Δ, or rcf3Δ were used for immunoprecipitation of Rcf3. Totals and glycine eluates were analyzed by SDS-PAGE and Western blotting. E, digitonin-solubilized mitochondria of wild type, cox4Δ, or cyt1Δ were used for immunoprecipitation of Rcf1, Rcf2, or Rcf3. Totals and glycine eluates were analyzed by SDS-PAGE and Western blotting. The data shown in A–E represent the results of at least two individual experiments.
Conclusions
Several studies showed that Rcf1 is involved in supercomplex formation by binding and priming complex IV for its association with complex III (19–21). The physiological role for supercomplex formation is still unknown, but it is hypothesized to promote electron flux, prevent the generation of reactive oxygen species, and stabilize respiratory chain complexes (12, 13). We could show that the respiratory supercomplex factor 2 is partially processed into a labile N-terminal and a stable C-terminal fragment. The C-terminal fragment remains associated with respiratory chain supercomplexes via complex IV. This Rcf2 fragment includes all necessary targeting information for import into mitochondria and assembly into complex IV-containing supercomplexes. Sequence comparison allowed us to identify Rcf3, encoded by the open reading frame YBR255C-A, as a protein that displays significant sequence similarity to the Rcf2 N-terminal half. Rcf3 localizes to the inner mitochondrial membrane where it associates predominantly with complex IV and complex IV-containing supercomplexes. In the absence of either Rcf2 or Rcf3, complex IV activity is increased, promoting oxygen consumption. An rcf2Δ/rcf3Δ double mutant displays the opposite effect, a decreased complex IV activity that results in the loss of respiratory growth. These results suggest an overlapping function of both proteins in the regulation of complex IV containing supercomplexes without altering supercomplex organization, in contrast to Rcf1. So far it is not clear under which physiological conditions Rcf2 and Rcf3 regulate the respiratory activity of supercomplexes, and neither is it clear whether they compete or work synergistically. In addition, considering its short half-life the N-terminal half of Rcf2, it appears that this fragment is dispensable in a wild-type situation but becomes crucial in the absence of Rcf3. Therefore, it will be a critical issue to identify the exact processing site within Rcf2 and the related protease. By immunoprecipitation, a fraction of Rcf3 is found to be bound to complex III, Rcf1 and Rcf2. However, we did not observe co-migration of Rcf3 with individual complex III by BN-PAGE followed by two-dimensional SDS-PAGE and Western blotting. This discrepancy could be caused by a dissociation of weakly bound proteins during BN-PAGE. The complex IV-independent association of Rcf3 with Rcf1/Rcf2 and complex III emphasize Rcf3 as a genuine subunit of the Rcf interaction network. It will be of a great interest to investigate the functional interplay of the three Rcf proteins in the regulation of supercomplex formation and function through their association to each other and with complex III and complex IV.
Experimental Procedures
Yeast Strains and Molecular Cloning
Yeast strains used in this study are derivatives of S. cerevisiae strain YPH499 except for Rcf3GFP, which is derived from BY4741 (Table 1). Deletion of RCF3 (YBR255C-A) and COX5A was achieved by homologous recombination of a HIS3MX6 cassette into the corresponding locus (23, 24). Generation of GFP- and StrepFLAG (SF)-tagged strains was performed by chromosomal integration. Cox4ZZ, Cor1ZZ, cox4Δ, cox5aΔ, cyt1Δ, rcf2Δ, and shy1Δ were described previously (19, 25–28). For expression of N-terminally FLAG-tagged Rcf2, the rcf2Δ strain was transformed with the centromeric yeast plasmid pRS416-FLAGRCF2. The RCF2 open reading frame including promoter and terminator was amplified from yeast genomic DNA by PCR and cloned SalI/SacI into pRS416. The coding sequence for the tag (GAC TAC AAG GAC GAC GAT GAC AAA) was introduced downstream of the start codon using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) on pRS416-RCF2. For complemental expression in yeast, the genomic fragments of RCF2N, RCF2C, and RCF3 were subcloned under their endogenous promoter into pRS416.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MATa, his3-Δ1, leu2-Δ0, met15Δ0, ura3Δ0 | Euroscarf |
| Rcf3GFP | MATa, his3-Δ1, leu2-Δ0, met15Δ0, ura3Δ0, rcf3::rcf3-EGFP-KANMX4 | This study |
| YPH499 | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801 | Ref. 19 |
| Rcf3SF | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, rcf3::rcf3-SF-HIS3MX6 | This study |
| rcf3Δ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, rcf3::HISMX6 | This study |
| rcf2Δ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, rcf2::HISMX6 | Ref. 19 |
| rcf2Δ/rcf3Δ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, rcf2::KANMX4, rcf3::HISMX6 | This study |
| Cox4ZZ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, cox4::cox4-TEV-ProA-7HIS-HIS3MX6 | Ref. 19 |
| Cor1ZZ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, cor1::cor1-TEV-ProA-7HIS-HIS3MX6 | Ref. 19 |
| cyt1Δ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, cyt1::HISMX6 | Ref. 19 |
| cox4Δ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, cox4::HISMX6 | Ref. 25 |
| cox5aΔ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, cox5a::HISMX6 | This study |
| shy1Δ | MATa, ade2-101, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, lys2-801, shy1::HISMX6 | Ref. 28 |
Yeast Growth Conditions
Yeast cells were grown in liquid medium containing 1% yeast extract, 2% peptone and 2% glucose, 2% galactose, 3% lactate or 3% glycerol (YPD, YPGal, YPLac, or YPG). Strains containing derivatives of pRS416 or pRS426 were grown on synthetic medium containing 0.67% yeast nitrogen base and 0.07% CSM lacking uracil (MP Biomedicals) and 2% glucose or 3% glycerol (SD-Ura, SG-Ura) and 2.8% agar. Mitochondria were isolated essentially as previously described (29). Subcellular fractionations were performed according to Horazdovsky and Emr (30).
Mitochondrial Oxygen Consumption
Mitochondria were isolated from cells grown on YPL according to the protocol of Onishi et al. (31). Oxygen consumption was assessed using high resolution respirometry (Oxygraph-2k; Oroboros Instruments, Innsbruck, Austria) in 2 ml of respiration buffer (225 mm sucrose, 75 mm mannitol, 10 mm Tris, 10 mm KH2PO4, 5 mm MgCl2, 10 mm KCl, pH 7.4) at 30 °C. Non-phosphorylating respiration (LEAK) was addressed using pyruvate (10 mm) and malate (2 mm). Adding ADP in a saturating concentration (1 mm) (State3) followed by succinate (10 mm) determines the maximal capacity for oxidative phosphorylation (OXPHOS). Respiration was killed by addition of antimycin A (500 μm) to prevent electron transfer from complex III to complex IV, and ascorbate (1 mm) and TMPD (50 μm) were added to address OXPHOS capacity via complex IV. To distinguish between respiration and auto-oxidation of TMPD/ascorbate, NaN3 (100 mm) was added to block the O2 binding site of complex IV, and the values were subtracted from the values after TMP/ascorbate addition.
In Vitro Import into Mitochondria
RCF1, RCF2, and RCF3 and RCF2 truncations were amplified using appropriate primers including a 3′ overhang for SP6 recognition. PCR products were used for in vitro transcription (Ambion®). Respective proteins were synthesized in rabbit reticulocyte lysate (Promega) in the presence of [35S]methionine. For import analysis, isolated mitochondria were incubated with radiolabeled proteins in import buffer (32) in the presence of 2 mm ATP and 2 mm NADH for the indicated times. The reactions were stopped on ice by dissipation of the membrane potential with 8 mm antimycin A, 1 mm valinomycin, and 10 mm oligomycin. For assembly analyses of imported proteins, the buffer was further supplemented with 5 mm creatine phosphate and 0.1 mg/ml creatine kinase, and samples were lysed in 1% digitonin or 0.6% DDM buffer for BN-PAGE.
Protein Localization Analysis
Protein localization analyses were assayed as described (27). For fluorescence microscopy analyses, yeast cells expressing Rcf3GFP were grown to a mid-log phase in minimal medium supplemented with 2% galactose at 30 °C. The cells were stained with 500 nm MitoTracker® Orange CMTMRos probe (Invitrogen) for 20 min at 30 °C in the dark and used for fluorescence microscopy. The images were collected by using a DeltaVision microscope (Olympus IX71; Applied Precision, Issaquah, WA) and deconvoluted using Softworx, version 3.5.1 (Softworx, Great Falls, MT). For protease protection assay, isolated mitochondria were converted to mitoplasts by hypotonic swelling in EM buffer (1 mm EDTA, 10 mm MOPS, pH 7.2, with KOH), kept intact in SE buffer (EM buffer supplemented with 250 mm sucrose) or lysed in 1% Triton X-100 and then treated with proteinase K. For carbonate extraction of proteins, mitochondria were incubated in 0.1 m Na2CO3 (pH 10.8) or 1% Triton X-100 supplemented with 150 mm NaCl for 20 min and centrifuged at 45,000 rpm at 4 °C for 45 min. Samples of both assays were precipitated with TCA and analyzed by SDS-PAGE and Western blotting.
Isolation of Protein Complexes
For native isolation of cytochrome c oxidase and bc1 complex, Cox4ZZ and Cor1ZZ expressing strains were used, respectively. Cox4- or Cor1-containing complexes were bound to Sepharose-coupled human IgGs and eluted by TEV protease cleavage as described previously (28) with the exception that buffers contained 0.5 mm EDTA. Samples were solubilized in 1% digitonin or 0.6% DDM. Eluates were analyzed by SDS-PAGE and Western blotting. For co-immunoprecipitiation, Rcf1, Rcf2, or Rcf3-specific antisera were bound to protein A-Sepharose (GE Healthcare) as described (28). Mitochondria were lysed in 20 mm Tris (pH 7.4), 80 mm NaCl, 0.5 mm EDTA, 1% digitonin, 10% glycerol, and 1 mm PMSF for 40 min under agitation. Lysates were cleared at 20,000 × g for 10 min at 4 °C, and input samples (total) were taken.
Miscellaneous
Standard techniques were used for SDS-PAGE and Western blotting on PVDF membranes. Proteins were detected with polyclonal antibodies raised in rabbit, except for α-FLAG (Sigma-Aldrich) and α-Pgk1 (Invitrogen) (monoclonal; mouse). For visualization of antibody-protein complexes, peroxidase-conjugated goat anti-rabbit (or anti-mouse) IgG (Jackson ImmunoResearch) and enhanced chemiluminescence reagent (GE Healthcare) were used. BN-PAGE protocols followed published procedures (33). In brief, mitochondria were solubilized in solubilization buffer (20 mm Tris/HCl, pH 7.4, 5 mm EDTA, 50 mm NaCl, 10% glycerol, and 1 mm PMSF supplemented with either 1% digitonin or 0.6% DDM) for 20 min at 4 °C. The lysate was cleared at 20,000 × g at 4 °C for 10 min. The supernatants were mixed with 10× loading dye (5% Coomassie G-250, 500 mm 6-amino-hexanoic acid and 0.1 m Bis-Tris, pH 7.0) and separated on a 4–13% or 4–10% polyacrylamide gradient gel. For two-dimensional analyses, whole BN-PAGE lanes were cut, incubated in SDS buffer (250 mm Tris, 1.91 m glycin, 10% SDS) for 30 min, and subjected to a second dimension SDS-PAGE, followed by Western blotting. Quantification of Western blots were done by the use of a LAS3000 imaging system (FujiFilm) and analyzed with the AIDA software (Raytest).
Author Contributions
K. R. performed the experiments and prepared the figures. T. M. performed the OROBOROS measurements. L. J. did the microscopic analysis, and M. W. provided technical assistance and contributed to the experiments. M. V. initially characterized Rcf2 and identified Rcf3. K. H. performed sequence analysis. M. D. conceived the study and wrote the paper.
Supplementary Material
Acknowledgments
We thank P. Rehling, D. M. Katschinski, H. Krebber, and F. N. Vögtle for helpful discussions.
This work was supported by Deutsche Forschungsgemeinschaft Grant DE 2277/1-1. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S3.
- BN
- blue native
- DDM
- n-dodecyl-β-d-maltopyranoside
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- TMPD
- N,N,N′,N′-tetramethyl-p-phenylenediamine.
References
- 1. Grandier-Vazeille X., Bathany K., Chaignepain S., Camougrand N., Manon S., and Schmitter J. M. (2001) Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry 40, 9758–9769 [DOI] [PubMed] [Google Scholar]
- 2. Rutter J., Winge D. R., and Schiffman J. D. (2010) Succinate dehydrogenase: assembly, regulation and role in human disease. Mitochondrion 10, 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zara V., Conte L., and Trumpower B. L. (2009) Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta. 1793, 89–96 [DOI] [PubMed] [Google Scholar]
- 4. Maréchal A., Meunier B., Lee D., Orengo C., and Rich P. R. (2012) Yeast cytochrome c oxidase: a model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochim. Biophys. Acta. 1817, 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mick D. U., Fox T. D., and Rehling P. (2011) Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat. Rev. Mol. Cell Biol. 12, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soto I. C., Fontanesi F., Liu J., and Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta. 1817, 883–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McStay G. P., Su C. H., and Tzagoloff A. (2013) Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell 24, 440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Althoff T., Mills D. J., Popot J.-L., and Kühlbrandt W. (2011) Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 30, 4652–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schägger H., and Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stuart R. A. (2008) Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40, 411–417 [DOI] [PubMed] [Google Scholar]
- 11. Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., and Enriquez J. A. (2008) Respiratory active mitochondrial supercomplexes. Mol. Cell 32, 529–539 [DOI] [PubMed] [Google Scholar]
- 12. Maranzana E., Barbero G., Falasca A. I., Lenaz G., and Genova M. L. (2013) Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 19, 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Genova M. L., and Lenaz G. (2014) Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta. 1837, 427–443 [DOI] [PubMed] [Google Scholar]
- 14. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., and Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 15. Zhang M., Mileykovskaya E., and Dowhan W. (2005) Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brandner K., Mick D. U., Frazier A. E., Taylor R. D., Meisinger C., and Rehling P. (2005) Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth syndrome. Mol. Biol. Cell 16, 5202–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenzie M., Lazarou M., Thorburn D. R., and Ryan M. T. (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J. Mol. Biol. 361, 462–469 [DOI] [PubMed] [Google Scholar]
- 18. Deckers M., Balleininger M., Vukotic M., Römpler K., Bareth B., Juris L., and Dudek J. (2014) Aim24 stabilizes respiratory chain supercomplexes and is required for efficient respiration. FEBS Lett. 588, 2985–2992 [DOI] [PubMed] [Google Scholar]
- 19. Vukotic M., Oeljeklaus S., Wiese S., Vögtle F. N., Meisinger C., Meyer H. E., Zieseniss A., Katschinski D. M., Jans D. C., Jakobs S., Warscheid B., Rehling P., and Deckers M. (2012) Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 15, 336–347 [DOI] [PubMed] [Google Scholar]
- 20. Chen Y.-C., Taylor E. B., Dephoure N., Heo J.-M., Tonhato A., Papandreou I., Nath N., Denko N. C., Gygi S. P., and Rutter J. (2012) Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 15, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Strogolova V., Furness A., Robb-McGrath M., Garlich J., and Stuart R. A. (2012) Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 32, 1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rydström Lundin C., von Ballmoos C., Ott M., Ädelroth P., and Brzezinski P. (2016) Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 113, E4476–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 24. Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., and Schiebel E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15, 963–972 [DOI] [PubMed] [Google Scholar]
- 25. Frazier A. E., Taylor R. D., Mick D. U., Warscheid B., Stoepel N., Meyer H. E., Ryan M. T., Guiard B., and Rehling P. (2006) Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 172, 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reinhold R., Bareth B., Balleininger M., Wissel M., Rehling P., and Mick D. U. (2011) Mimicking a SURF1 allele reveals uncoupling of cytochrome c oxidase assembly from translational regulation in yeast. Hum. Mol. Genet. 20, 2379–2393 [DOI] [PubMed] [Google Scholar]
- 27. Levchenko M., Wuttke J.-M., Römpler K., Schmidt B., Neifer K., Juris L., Wissel M., Rehling P., and Deckers M. (2016) Cox26 is a novel stoichiometric subunit of the yeast cytochrome c oxidase. Biochim. Biophys. Acta. 1863, 1624–1632 [DOI] [PubMed] [Google Scholar]
- 28. Bareth B., Dennerlein S., Mick D. U., Nikolov M., Urlaub H., and Rehling P. (2013) The heme a synthase Cox15 associates with cytochrome c oxidase assembly intermediates during Cox1 maturation. Mol. Cell. Biol. 33, 4128–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meisinger C., Pfanner N., and Truscott K. N. (2006) Isolation of yeast mitochondria. Methods Mol. Biol. 313, 33–39 [DOI] [PubMed] [Google Scholar]
- 30. Horazdovsky B. F., and Emr S. D. (1993) The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268, 4953–4962 [PubMed] [Google Scholar]
- 31. Onishi T., Kröger A., Heldt H. W., Pfaff E., and Klingenberg M. (1967) The response of the respiratory chain and adenine nucleotide system to oxidative phosphorylation in yeast mitochondria. Eur. J. Biochem. 1, 301–311 [DOI] [PubMed] [Google Scholar]
- 32. Ryan M. T., and Pfanner N. (2001) Protein Translocation across Membranes, John Wiley & Sons, Ltd., Chichester, UK [Google Scholar]
- 33. Dekker P. J., Martin F., Maarse A. C., Bömer U., Müller H., Guiard B., Meijer M., Rassow J., and Pfanner N. (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16, 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Helbig A. O., de Groot M. J., van Gestel R. A., Mohammed S., de Hulster E. A., Luttik M. A., Daran-Lapujade P., Pronk J. T., Heck A. J., and Slijper M. (2009) A three-way proteomics strategy allows differential analysis of yeast mitochondrial membrane protein complexes under anaerobic and aerobic conditions. Proteomics 9, 4787–4798 [DOI] [PubMed] [Google Scholar]
- 35. Taanman J. W., and Capaldi R. A. (1993) Subunit VIa of yeast cytochrome c oxidase is not necessary for assembly of the enzyme complex but modulates the enzyme activity. Isolation and characterization of the nuclear-coded gene. J. Biol. Chem. 268, 18754–18761 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.