Abstract
α-Conotoxins represent a large group of pharmacologically active peptides that antagonize nicotinic acetylcholine receptors (nAChRs). The α3β4 nAChR, a predominant subtype in the peripheral nervous system, has been implicated in various pathophysiological conditions. As many α-conotoxins have multiple pharmacological targets, compounds specifically targeting individual nAChR subtypes are needed. In this study, we performed mutational analyses to evaluate the key structural components of human β2 and β4 nAChR subunits that determine α-conotoxin selectivity for α3β4 nAChR. α-Conotoxin RegIIA was used to evaluate the impact of non-conserved human β2 and β4 residues on peptide affinity. Two mutations, α3β2[T59K] and α3β2[S113R], strongly enhanced RegIIA affinity compared with wild-type α3β2, as seen by substantially increased inhibitory potency and slower off-rate kinetics. Opposite point mutations in α3β4 had the contrary effect, emphasizing the importance of loop D residue 59 and loop E residue 113 as determinants for RegIIA affinity. Molecular dynamics simulation revealed the side chains of β4 Lys59 and β4 Arg113 formed hydrogen bonds with RegIIA loop 2 atoms, whereas the β2 Thr59 and β2 Ser113 side chains were not long enough to form such interactions. Residue β4 Arg113 has been identified for the first time as a crucial component facilitating antagonist binding. Another α-conotoxin, AuIB, exhibited low activity at human α3β2 and α3β4 nAChRs. Molecular dynamics simulation indicated the key interactions with the β subunit are different to RegIIA. Taken together, these data elucidate the interactions with specific individual β subunit residues that critically determine affinity and pharmacological activity of α-conotoxins RegIIA and AuIB at human nAChRs.
Keywords: electrophysiology, homology modeling, molecular dynamics, nicotinic acetylcholine receptors (nAChR), oocyte, site-directed mutagenesis, toxin, Xenopus
Introduction
Neuronal nicotinic acetylcholine receptors (nAChR)3 are ligand-gated ion channels endogenously activated by acetylcholine (ACh). Presynaptic nAChRs in the central nervous system play an important role in synaptic transmission, as they mediate the release of various neurotransmitters (1). Postsynaptic nAChRs are involved in fast excitatory transmission, and non-synaptic nAChRs modulate many neurotransmitter systems by influencing neuronal excitability (2). The widespread expression and functional diversity makes nAChRs key components in numerous physiological functions of the central and peripheral nervous systems such as learning, memory, attention, motor control, synaptic plasticity, and analgesia (2, 3). Based on these versatile physiological roles, any dysfunction of nAChR subtypes contributes to various disease states, including epilepsy, schizophrenia, Parkinson's disease, depression, autism, Alzheimer's disease, and addiction (2–5).
Functional neuronal nAChRs are composed of five transmembrane-spanning subunits combining α (α2–α10) and β (β2–β4) subunits. Combinations of different α and β subunits yields a variety of heteromeric receptors with individual physiological roles and pharmacological profiles (6, 7). Functional neuronal nAChRs containing an α2, α3, α4, or α6 subunit also require a β2 or β4 subunit. The β subunit in these nAChR subtypes influences pharmacological properties including agonist efficacy, desensitization kinetics, and Ca2+ permeability (3).
The ligand binding site of nAChRs lies at the extracellular interface between α and β subunits, with the α subunit contributing the principal (+) face and the β subunit contributing the complementary (−) face. The ligand binding sites of nAChR subtypes are structurally very similar and hence the residues involved in agonist binding are considerably conserved. Therefore, the development of highly subtype-specific competitive ligands is a difficult ongoing task (6).
α-Conotoxins are a large group of disulfide-bonded peptides isolated from the venom of carnivorous marine Conus snails. They have been found to be specific competitive ligands of nAChRs, inhibiting ACh-evoked currents in a concentration-dependent manner. All α-conotoxins contain two conserved disulfide bridges and can be subclassified by the number of residues between the cysteines forming the disulfide bonds (e.g. α4/3, α4/4, and α4/7) (8). Individual α-conotoxins have their own selectivity profile and can discriminate between different nAChR subunit combinations and stoichiometries (9, 10).
To date, a number of highly selective α-conotoxins potently inhibiting the α3β2* subtype have been identified (* indicates the presence of potential other subunits) (8). However, few probes specifically inhibiting single α3-containing nAChR subtypes exist (6). The α3β4 nAChR is the predominant subtype in the peripheral nervous system and medial habenula of the brain and it has been shown to be involved in several pathophysiological disease conditions such as lung cancer, nicotine addiction, and drug abuse (3, 11, 12). Therefore, α3β4 is a promising therapeutic target and pharmacologically active substances modulating this nAChR subtype are sought-after.
The first α3β4 selective α-conotoxin described was α4/6-conotoxin AuIB, which inhibits rat α3β4 with an IC50 of 0.75 μm, whereas activity at other subtypes was reported as >100-fold lower (13). Another α4/6-conotoxin, TxID from Conus textile, was recently described as the most potent α3β4* nAChR-inhibiting α-conotoxin, however, besides α3β4 it also inhibits α6/α3β4 with 7.5-fold less potency (14).
α4/7-Conotoxin RegIIA from Conus regius has been reported to potently inhibit α3β4 besides α3β2 and α7 nAChRs (15). Interestingly, a non-natural analogue of RegIIA in which two residues in loop 2 were exchanged by alanines exhibited an enhanced selectivity for the α3β4 subtype compared with the wild-type toxin (16). Furthermore, we have previously shown that the selectivity profile of RegIIA differs between nAChRs of different species, as RegIIA was equipotent at the rat (r) and human (h) α3β4 subtypes but significantly less active at hα3β2 compared with rα3β2. This potency difference could be mapped to a single non-conserved Glu198 on the rα3 subunit (proline in human) (17).
Here, we investigated the molecular determinants for nAChR subtype selectivity of α-conotoxins, especially for the hα3β4 subtype. We tested residues, which are non-conserved between the homologous β2 and β4 subunits, for their impact on α-conotoxin affinity and activity using β subunit chimeras and point mutations. α-Conotoxin RegIIA was used as a probe for a major part of this study. We aimed to exhibit key β subunit residues of the ligand-binding interface that profoundly and subtype specifically affect both RegIIA sensitivity and wash-off kinetics.
Experimental Procedures
Peptide Synthesis
α-Conotoxin RegIIA was synthesized as described previously (15). The α-conotoxin AuIB was assembled on Rink amide methylbenzhydrylamine resin using solid-phase peptide synthesis with a neutralization/2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate activation procedure for Fmoc (N-(9-fluorenyl)-methoxycarbonyl) chemistry. Cleavage was achieved by treatment with 88:2:5:5 (v:v) ratio of trifluoroacetic acid/triisopropylsilane/phenol/water at room temperature (20–25 °C) for 2 h. Trifluoroacetic acid was evaporated at low pressure in a rotary evaporator. Peptides were precipitated with ice-cold ether, filtered, dissolved in 50% buffer A/B (buffer A consists of 99.95% H2O, 0.05% trifluoroacetic acid and buffer B consists of 90% CH3CN, 10% H2O, 0.045% trifluoroacetic acid), and lyophilized. Crude peptides were purified by RP-HPLC on a Phenomenex C18 column using a gradient of 0–50% of buffer B for 50 min, with the eluent monitored at 214/280 nm. Electrospray mass spectrometry confirmed the molecular mass of the peptides before they were pooled and lyophilized for oxidation.
The four cysteines in the peptides were selectively oxidized in two steps to yield the globular conformation. This was achieved by incorporating Fmoc-Cys-acetamidomethyl (Acm)–OH at positions 2 and 8 of the amino acid sequence. In the first step, the two non-protected cysteines were oxidized in 0.1 m NH4HCO3 (pH 8–8.5) at a concentration of 0.5 mg/ml, and the mixture was stirred at room temperature for 48 h. The oxidized peptides were then purified and lyophilized as previously outlined. In the second step, the Acm-protected cysteines were oxidized by dissolving the peptides in iodine solution filled at 1 mg/ml, and the mixture was stirred for 35 min. After two rounds of oxidation, peptides were purified by RP-HPLC using a gradient of 0–80% buffer B over 180 min. Analytical RP-HPLC and electrospray mass spectrometry were used to confirm the purity and molecular mass of the synthesized peptides.
Protein Sequence Alignment
Protein sequences of hβ2 and hβ4 nAChR subunit ECDs (RefSeq accession numbers NP_000739 and NP_000741, respectively) were aligned using CLC Viewer 7 software (CLC bio, Aarhus, Denmark). Residues were numbered according to the mature protein sequences.
Site-directed Mutagenesis
Plasmid DNAs encoding human and rat α3, β2, and β4 nAChR subunits were subcloned into the pT7TS Xenopus expression vector (Addgene plasmid 17091) as described previously (17). Two chimeric receptors consisting of hβ2 backbone and portions of the N-terminal extracellular domain (ECD) (N-terminal T1 to loop D Glu63 and a lesser conserved Lys70-His86 segment between loops D and E) replaced with the corresponding hβ4 sequences, were generated by overlap PCR and molecular cloning. Additional mutants of human β subunit loops D, E and F (Table 1), as well as rat β2[T59K] were engineered using the Geneart Site-directed Mutagenesis System (Invitrogen). All point mutations were confirmed by DNA sequencing (Australian Genome Research Facility, Melbourne, Australia).
TABLE 1.
RegIIA inhibition of α3-containing nAChR subtypes and subtype mutants
IC50 values (nm) with 95% CI. Hill slope (nH) was obtained from concentration–response curves for RegIIA at wild-type and mutant human α3β2 and α3β4 nAChR subtypes. Human α3β2 nAChR mutations hα3β2[T59K] and hα3β2[S113R] notably decrease the IC50 of RegIIA towards the human α3β4 subtype value, whereas the opposite human α3β4[K59T] and α3β4[R113S] result in lower potency of RegIIA and significantly increased IC50 values. Data from wild-type nAChRs and the mutants mentioned are highlighted in bold font. All data represent mean of n = 3–9 experiments.
| nAChR/mutant | IC50 | 95% CI | nH |
|---|---|---|---|
| nm | |||
| hα3β2 | 132.4 | 109.7–159.7 | 1.5 |
| hα3β4 | 45.6 | 31.5–65.9 | 0.9 |
| hα3β2[1–63β4] | 159.2 | 113.0–224.4 | 1.4 |
| hα3β2[70–86β4] | 1268.1 | 935.7–1718.5 | 0.9 |
| hα3β2[T59K] | 49.4 | 33.4–73.1 | 1.2 |
| hα3β4[K59T] | 2795.1 | 2346.6–3329.3 | 1.4 |
| hα3β4[113–115β2] | 1056.5 | 924.9–1206.9 | 1.1 |
| hα3β2[S113R] | 23.7 | 19.0–29.4 | 1.5 |
| hα3β4[R113S] | 805.3 | 554.7–1169.0 | 0.9 |
| hα3β4[S114Y] | 133.1 | 105.0–168.6 | 1.2 |
| hα3β4[N115D] | 275.0 | 221.3–341.5 | 1.6 |
| hα3β4[M163K+T164S] | 138.6 | 113.2–169.8 | 2.3 |
| hα3β4[P165E] | 217.9 | 172.9–274.6 | 1.2 |
| hα3β4[T166V+M169L] | 130.2 | 100.1–169.5 | 1.9 |
Electrophysiological Recordings in Xenopus Oocytes and Data Analysis
RNA preparation, Xenopus laevis oocyte preparation, and expression of nAChR subunits in oocytes were performed as described previously (18).
Membrane currents from Xenopus oocytes were recorded using a single channel two-electrode voltage clamp setup (virtual ground circuit) with a GeneClamp 500B amplifier (Molecular Devices, Sunnyvale, CA) as described previously (17). Briefly, oocytes were continuously perfused with ND96 solution containing (in mm) 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES (pH 7.4) at 2 ml/min. ACh or ACh plus toxin was applied for ∼2 s using a manual HPLC injection module. The effect of the peptides on ACh-evoked currents was defined as ACh plus peptide peak current amplitude relative to the average peak current amplitude of 3–5 control ACh applications (300 μm for hα3β4 and 50 μm for all other nAChR subtypes) recorded before preincubation with the peptides (technical replicates of ACh application). Perfusion was switched off during preincubation with the peptide. Concentration-response curves were fitted by unweighted nonlinear regression to the logistic equation,
where Ex is the response, X is the antagonist concentration, Emax is the maximal response, nH is the Hill coefficient, and IC50 is the antagonist concentration that gives 50% inhibition of the agonist response.
Measurement of the recovery from block (koff) by the peptides was carried out by incubating the oocyte with the respective peptide for 5 min followed by repeated ACh applications at the indicated time points while continuously perfusing the oocyte with ND96 solution at 2 ml/min. Approximate half-maximal excitatory concentrations (EC50) of ACh were determined and used in the wash-off kinetic experiments.
All electrophysiological data were pooled (n = 3–9 for each data point) and represent arithmetic mean ± 95% confidence intervals (95% CI) of the fit. Each oocyte was tested only once with a given peptide concentration, therefore n indicates the number of oocytes used for generating each data point (biological replicates). Curves were fitted and statistics calculated using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA).
Homology Modeling
Models of RegIIA/AuIB-bound α3β2 and α3β4 nAChRs were built using Modeler (version 9v12), as described previously (19). The sequences of human α1, α3, α4, α6, α7, α9, α10, β2, β3, and β4 nAChR subunits were retrieved from the Uniprot database (20). The crystal structures ofAplysia californica AChBP in complex with α-conotoxin PnIA[A10L,D14K] (PDB code 2BR8) (21), and the ECDs of mouse α1 (PDB code 2QC1) (22) and human α9 (PDB code 4D01) (23) subunits were used as templates to model the α-conotoxin-bound nAChR complexes (200 models each). Models with the lowest DOPE score (24) were selected for further structural refinement using molecular dynamics simulations.
Protonation State Predictions
The protonation states of amino acids His, Asp, and Glu were predicted using the PropKa 3.1 method (25). The predictions were made for all 200 homology models generated by Modeler and the protonation states predicted for the majority of models were considered in the starting models for MD simulations.
Molecular Dynamics (MD) Simulations
MD simulations were performed using the AMBER 12 package (26) and ff12SB force field (27). The above α-conotoxin·nAChR complexes were solvated in a truncated octahedral periodic box with TIP3P water molecules, and neutralized using sodium ions. Prior to MD simulations, 2000 steps of steepest descent minimization and 3000 steps of conjugate gradient minimization were performed with the solute restrained using a harmonic force with a 100 kcal/mol·Å2 spring constant. After the first round of minimization, the entire system was minimized without position restraints. The minimized systems were gradually heated from 50 to 300 K over 100 ps using NVT ensemble with the solute atoms restrained to their positions by harmonic forces with a spring constant of 5 kcal/mol·Å2. Simulations were then switched to NPT ensemble, and spring constants of the restraints were gradually decreased from 5 to 0 kcal/mol·Å2 over 100 ps. In the production phase, 50-ns MD simulations were carried out with the temperature and pressure maintained at 300 K and 1 bar, respectively.
In all simulations, all bonds involving hydrogen atoms were constrained with the SHAKE algorithm and the time step was 2 fs (28). The particle-mesh Ewald method was used to treat long-range electrostatic interactions (29).
Results
Two Single Non-conserved Residues in Loops D and E of the Human β2 and β4 nAChR Subunit ECDs Determine the Differential Sensitivity of α3β2 and α3β4 to α-Conotoxin RegIIA
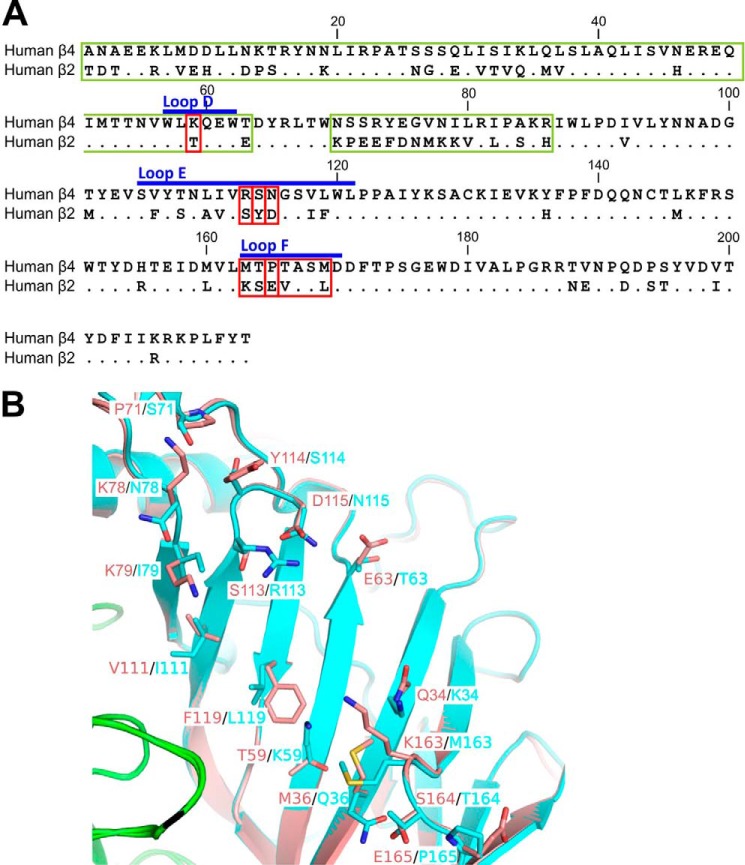
To identify potential structural determinants responsible for the lower potency of α-conotoxin RegIIA at the hα3β2 subtype relative to hα3β4, we compared the amino acid sequences of the N-terminal ECDs containing the agonist binding domains of each subtype. Pairwise alignments of mature protein sequences of hβ2 and hβ4 lacking the signal peptide revealed an overall homology of 70% (149 of the 213 residues in the ECD are homologous) (Fig. 1A). Interestingly, the amino acids contributing to the complementary face of the ACh-binding loops that correspond to the previously identified homologous loops of the α7 subunit (30) are among the lesser conserved regions in the sequence. Other less conserved regions include the N terminus preceding loop D and the region from amino acids 70 to 86 (Fig. 1A). Homology modeling and overlay of the hα3β2 and hα3β4 intersubunit interfaces revealed the side chain differences between these non-conserved residues on the β2 and β4 subunits (Fig. 1B). These differences could account for the α-conotoxin nAChR subtype selectivity.
FIGURE 1.
Sequence and structural comparison between human β2 and β4 nAChR ECDs. A, amino acid sequence alignment of the N-terminal ECDs of human β2 and β4 nAChR subunits shows 70% identity between the sequences (149 of the 213 residues homologous) and reveals several non-conserved residues in the ACh binding loops. Conserved residues are indicated with dots. Residues that were mutated to the opposite β subunit residue in this study are framed with red lines. Longer sequences that were replaced in hβ2 with the sequences of hβ4 are framed with green lines. Blue bars indicate ACh binding domain loops D, E, and F of the complementary interface (34). B, overlay of the hα3β2 and hα3β4 inter-subunit interfaces to emphasize the overall high structural similarity between them. The α3(+) interface is shown in green, β2(−) in pink, and β4(−) in cyan. Non-conserved residues from the complementary β2(−) and β4(−) subunits, respectively, are shown as licorice models and labeled.
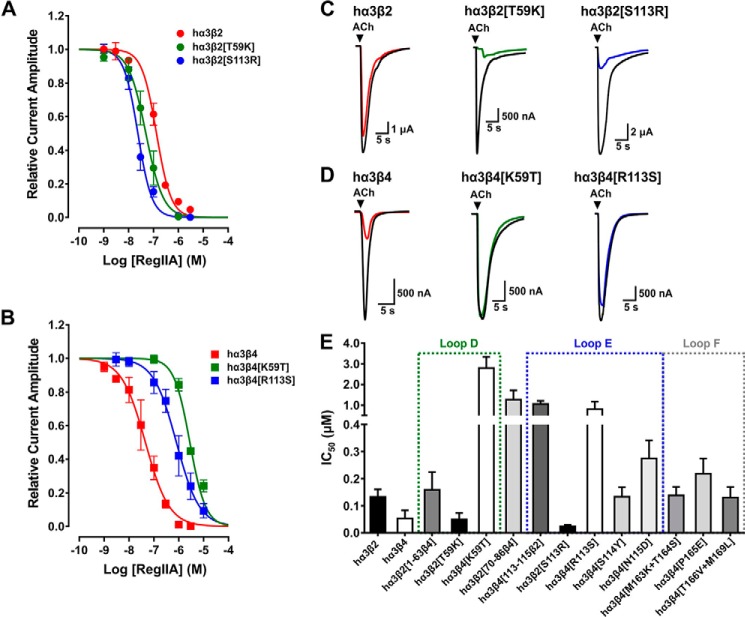
From electrophysiological recordings of oocyte-expressed nAChRs, RegIIA exhibited an IC50 of 45.6 nm (95% CI 31.5–65.9 nm; nH = 0.9) at the hα3β4 subtype, whereas an IC50 of 132.4 nm (95% CI 109.8–159.7 nm; nH = 1.5) was observed at the hα3β2 subtype (Fig. 2, A and B, and Table 1). To investigate if the less conserved sequences in the ECDs of the β-subunits determine the specificity of the peptide, we generated chimeric hβ2-β4 subunits, co-expressed them with wild-type hα3, and tested the effect of the peptide on ACh-evoked current amplitudes. The first chimeric subunit consisted of hβ2 in which the whole N-terminal region including loop D was replaced by the respective sequence of hβ4 (mutant hα3β2[1–63β4]) (Fig. 1A). Although the N-terminal region that precedes loop D is overall less conserved between the receptors, the loop itself only differs in one amino acid, the residue at position 59 (Lys in hβ4, Thr in hβ2). Mutant hα3β2[1–63β4] exhibited a similar sensitivity for RegIIA compared with wild-type hα3β2 with an IC50 of 159.2 nm (95% CI 113.0--224.4; nH = 1.4) (Fig. 2E, Table 1).
FIGURE 2.
The non-homologous residues at position 59 (Lys in hβ4, Thr in hβ2) and 113 (Arg in hβ4, Ser in hβ2) of the human β subunits are the key residues determining the selectivity profile in inhibitory potency of α-conotoxin RegIIA. A and B, concentration-response curves for RegIIA inhibition of wild-type and mutant human α3β2 (A) and α3β4 nAChRs (B). The two hα3β2 mutants exhibited a shift of the curve to the left compared with wild-type hα3β2 nAChR, indicating an increase in affinity to the peptide, whereas the opposite mutants of hα3β4 exhibited a lower affinity to RegIIA compared with wild-type hα3β4 nAChR. C, representative superimposed ACh-evoked currents obtained in the absence (control, black line) and presence of 100 nm RegIIA at wild-type hα3β2 (red), loop D mutant hα3β2[T59K] (green), and loop E mutant hα3β2[S113R] (blue). D, representative superimposed ACh-evoked currents obtained in the absence (control, black line) and presence of 100 nm RegIIA at wild-type (red) and mutant hα3β4 nAChRs (green and blue, respectively. E, bar graph summarizing the IC50 values with 95% CI obtained from concentration-response curves for RegIIA at wild-type hα3β2, hα3β4, and the mutants and chimeric subtypes analyzed. A gain in sensitivity was observed at mutants hα3β2[T59K] and hα3β2[S113R], whereas the most prominent reductions in sensitivity can be mapped to the opposite mutations hα3β4[K59T] and hα3β4[R113S]. Dotted squares indicate the agonist binding loops in which the respective mutants are located. All data points represent mean ± 95% CI. The IC50, 95% CI, and Hill slope (nH) values are summarized in Table 1.
A refined mutation in which only the single non-conserved Thr residue in loop D was exchanged to the corresponding Lys residue in hβ4 (mutant hα3β2[T59K]) exhibited a concentration-response curve with an IC50 of 49.4 nm (95% CI 33.4–73.1; nH = 1.2), which was significantly shifted to the left compared with wild-type hα3β2 and resembled the hα3β4 concentration-response curve (Fig. 2, A and C, and Table 1). Thus, the higher RegIIA sensitivity observed with hα3β4 can be (majorly) attributed to loop D. Conversely, the opposite mutation in hβ4 (mutant hα3β4[K59T]) led to a severe loss in sensitivity to RegIIA, represented by an IC50 of 2795.1 nm (95% CI 2346.6–3329.3; nH = 1.4) (Fig. 2, B and D, and Table 1) hence, confirming Lys59 as a key determinant for human nAChR β subunit sensitivity to RegIIA.
Similarly, the lesser conserved region between loops D and E (residues 70 to 86) of hβ2 was replaced with the β4 sequence (mutant hα3β2[70–86β4]) and tested in oocytes. However, this mutation did not decrease the IC50 compared with wild-type hα3β2 but rather resulted in considerably lowered potency of the peptide possibly because of structural effects (Fig. 2E, Table 1). Therefore, it was excluded as a factor for the β2- and β4-specific differences in sensitivity to RegIIA.
The non-conserved residues Arg113, Ser114, and Asn115 of hβ4 were replaced with the respective residues of hβ2 (mutant hα3β4[113–115β2]) to analyze the contribution of loop E for RegIIA affinity. This mutant considerably shifted the concentration-response curve for RegIIA to the right (IC50 = 1056.5 nm, 95% CI 924.9–1206.9 nm; nH = 1.1). Contribution of any of the three residues to the shift was further dissected with point mutations in hβ4. Of these, only mutant hα3β4[R113S] profoundly shifted RegIIA IC50 to the high nanomolar range (805.3 nm, 95% CI 554.7–1169.0 nm; nH = 0.9) (Fig. 2, B, D, and E, and Table 1) compared with little contribution of mutations S114Y and N115D (IC50 of 131.2 nm, 95% CI 112.7–152.7 nm, nH = 1.3, and IC50 of 275.0 nm, 95% CI 221.3–341.5 nm, nH = 1.6, respectively). The opposite mutation, hα3β2[S113R], exhibited a considerable shift of the concentration-response curve to the left compared with wild-type hα3β2 with an IC50 of 23.7 nm (95% CI 19.0–29.4 nm; nH = 1.5) (Fig. 2, A, C, and E, and Table 1), further confirming the key role of residue 113 in loop E in determining sensitivity toward RegIIA.
To probe the contribution of loop F to the sensitivity difference of RegIIA at human hα3β4 and hα3β2 we engineered three hβ4 receptor mutants: hα3β4[M163K+T164S], hα3β4[P165E], and hα3β4[T166V+M169L]. When tested in oocytes, none of these three mutants revealed a notable loss of sensitivity to RegIIA (Fig. 2E, Table 1) indicating loop F does not play a key role in determining RegIIA sensitivity. Taken together, the mutational analysis revealed the non-conserved residue in loop D at position 59 (Lys in hβ4, Thr in hβ2) and residue Arg113 in hβ4 (Ser in hβ2) of loop E as the two major determinants for the subtype selectivity of RegIIA at human nAChRs.
Non-conserved Loop D and E Residues Differentially Affect the ACh Half-maximal Excitatory Concentrations (EC50) at Human α3β4 and α3β2 nAChRs
We observed an ∼18-fold difference in the EC50 for ACh at human α3β4 and α3β2 nAChRs. The EC50 obtained at hα3β2 was 15.6 μm (95% CI 10.9–22.2 μm; n = 6), whereas an EC50 of 287.6 μm (95% CI 274.1–301.8 μm, n = 8) was observed at hα3β4 (Table 2 and supplemental Fig. S1). In addition, the approximate EC50 values of the mutants at positions 59 and 113 as well as other hα3β4 mutants were determined. Mutant hα3β2[T59K] had a similar ACh EC50 to the wild-type hα3β2 (14.3 μm, 95% CI 12.5–16.5 μm, n = 4). In contrast the opposite mutation hα3β4[K59T] exhibited a considerably lower EC50 than wild-type hα3β4 (29.8 μm, 95% CI 27.9–31.9 μm, n = 4). Mutant hα3β4[R113S] also resulted in a considerably lower than wild-type hα3β4 EC50 value of 42.9 μm (95% CI 36.4–50.5 μm, n = 4). Surprisingly, however, mutant hα3β2[S113R] revealed an EC50 in the low micromolar range (6.0 μm, 95% CI 5.0–7.2 μm, n = 5), which is comparable with wild-type hα3β2 (Table 2 and supplemental Fig. S1). Overall, both β2 mutants retained the high affinity to ACh as seen in wild-type hα3β2, whereas the opposite mutations in hα3β4 (hα3β4[K59T] and hα3β4[R113S]) considerably increased the affinity for ACh compared with hα3β4. Other hα3β4 loop E and F mutants that exhibited a less pronounced increase in RegIIA IC50 also had lower than wild-type ACh EC50 (Table 2). This is not unexpected given that these mutations are within the agonist binding loops.
TABLE 2.
ACh EC50 values for wild-type and mutant human α3β2 and α3β4 nAChRs
EC50 values (μm) with 95% CI. The Hill slope (nH) was obtained from concentration–response curves for ACh at wild-type and mutant human α3β2 and α3β4 nAChR subtypes. ACh concentrations from 0.01 μm to 10 mm (the highest applicable concentration) were tested. All data represent mean of n = 4–8 experiments.
| nAChR/mutant | EC50 | 95% CI | nH |
|---|---|---|---|
| μm | |||
| hα3β2 | 15.6 | 10.9–22.2 | 0.7 |
| hα3β4 | 287.6 | 274.1–301.8 | 2.3 |
| hα3β2[T59K] | 14.3 | 12.5–16.5 | 1.0 |
| hα3β4[K59T] | 29.8 | 27.9–31.9 | 1.1 |
| hα3β2[S113R] | 6.0 | 5.0–7.2 | 0.9 |
| hα3β4[R113S] | 42.9 | 36.4–50.5 | 1.1 |
| hα3β4[S114Y] | 65.1 | 56.3–75.3 | 1.1 |
| hα3β4[N115D] | 140.8 | 123.8–160.0 | 1.0 |
| hα3β4[M163K+T164S] | 63.1 | 58.6–67.9 | 1.0 |
| hα3β4[P165E] | 43.9 | 38.6–50.0 | 1.2 |
| hα3β4[T166V+M169L] | 17.5 | 15.8–19.4 | 1.1 |
The Non-conserved Residues hβ4-Lys59 and hβ4-Arg113 Are the Key Determinants for Recovery from Block by RegIIA
Human α3β4 and α3β2 nAChRs not only exhibited a difference in sensitivity to α-conotoxin RegIIA with considerably different IC50 values, but also differences in the time required to recover from block. By repeatedly applying ACh under constant perfusion after incubation with the peptide, we measured the time required for the ACh-evoked current amplitude to recover.
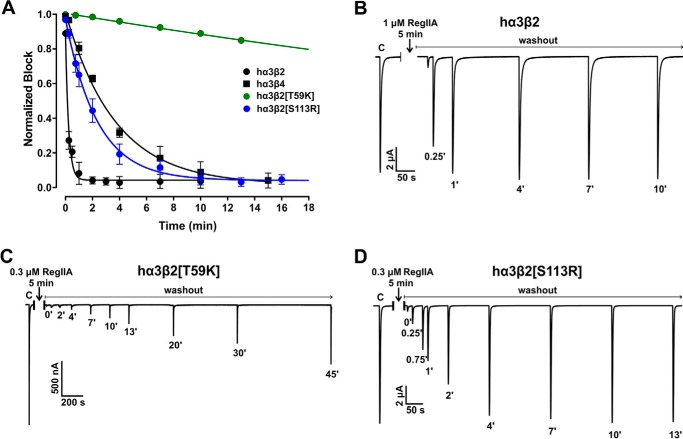
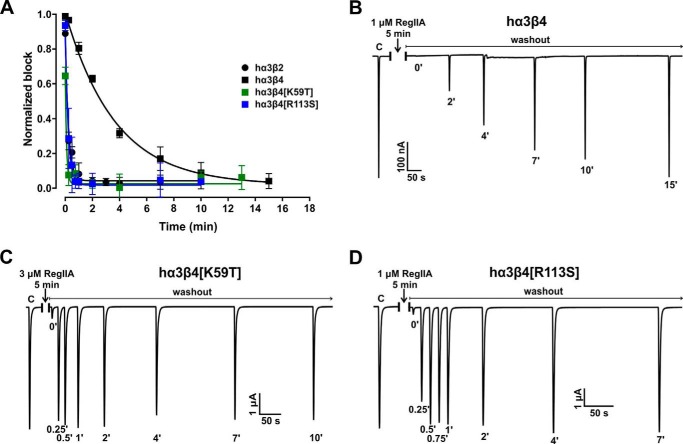
Interestingly, RegIIA revealed notably different wash-off kinetics at hα3β2 compared with hα3β4. At hα3β2, 95% recovery from block by RegIIA was reached within a minute (Fig. 3, A and B, and Table 3). In contrast, 95% wash-off at hα3β4 took ∼13–14 min (Fig. 4, A and B, and Table 3). A similar difference was observed at rat nAChRs, with faster recovery for rα3β2 (1–2 min) compared with a considerably slower rate for rα3β4 (16–18 min) (Table 3).
FIGURE 3.
Wild-type human α3β2 and α3β4 nAChRs display different wash-off kinetics from block by α-conotoxin RegIIA. Single residue β2 mutations in hα3β2 are sufficient to switch off-rates to those of the opposite subtype. A, graph summarizing the wash-off kinetics data. The hα3β2 nAChR subtype (black circles) exhibits fast recovery of ACh-evoked currents from block by RegIIA, with full recovery achieved in less than 2 min. In contrast, at hα3β4 (black squares) currents recovered from RegIIA block after a 13–14-min washout. When loop D residue 59 in hβ2 was replaced with the respective residue of hβ4 (mutant hα3β2[T59K], green circles) the off-rate of RegIIA was dramatically slowed. A similar replacement of loop E residue 113 in hβ2 (mutant hα3β2[S113R], blue circles) slowed the recovery rate similar to hα3β4. Representative ACh-evoked currents of hα3β2 (B), hα3β2[T59K] (C), and hα3β2[S113R] (D) illustrate the recovery from the RegIIA block differs between wild-type and the two mutant hα3β2 nAChR subtypes. Numbers at the respective ACh-evoked current peaks indicate the duration (in min) of washout and C indicates a representative control ACh application before incubation with the peptide. Oocytes were incubated with RegIIA for 5 min followed by repetitive application of ACh under continuous perfusion with ND96 solution. Approximate EC50 values for ACh and RegIIA concentrations giving major to full block of ACh-evoked currents under these conditions were used for each subtype tested. All data points in A represent mean ± 95% CI, n = 3–7. The times required to reach 95% recovery from block are summarized in Table 3.
TABLE 3.
Recovery time from block by α-conotoxin RegIIA
| nAChR/mutant | t95a |
|---|---|
| min | |
| hα3β2 | 1 |
| hα3β4 | 13–14 |
| rα3β2 | 1–2 |
| rα3β4 | 16–18 |
| hα3β2[T59K] | >45 |
| hα3β4[K59T] | <1 |
| rα3β2[T59K] | >45 |
| hα3β4[R113S] | <1 |
| hα3β2[S113R] | 9–3 |
| hα3β4[P165E] | 3–5 |
| hα3β4[T166V+M169L] | 5–7 |
a Time to reach 95% recovery after continuous peptide washout.
FIGURE 4.
Single residue β4 mutations in hα3β4 increase the α-conotoxin off-rate of RegIIA to values similar to wild-type hα3β2. A, graph summarizing the wash-off kinetics data. Wild-type hα3β2 and hα3β4 data are the same as described in the legend to Fig. 3 and shown for comparison. Replacing loop D residue 59 in hβ4 with the respective residue in hβ2 (mutant hα3β4[K59T], green squares) is sufficient to shift the off-rate to the opposite subtype. A mutant in which residue Arg113 of hβ4 is replaced with Ser, as in hβ2, (hα3β4[R113S], blue squares) similarly exhibits a fast wash-off rate resembling hα3β2. Representative ACh-evoked currents of hα3β4 (B), hα3β4[K59T] (C), and hα3β4[R113S] (D) illustrate the recovery from the RegIIA block differs between wild-type and the two mutant hα3β4 nAChR subtypes. Experimental conditions were the same as those described in the legend to Fig. 3. All data points in A represent mean ± 95% CI, n = 3–7. The times required to reach 95% recovery from block are summarized in Table 3.
We next tested if any of the mutations that affected the IC50 of RegIIA also had an impact on the rate of recovery from block. Mutant hα3β4[K59T] exhibited a rapid off-rate, similar to that observed for wild-type hα3β2 (95% recovery in <1 min) (Fig. 4, A and C). Conversely, peptide washout at the opposite mutant hα3β2[T59K] was very slow with less than 50% washout achieved after 45 min of perfusion (Fig. 3, A and C, and Table 3). In rat β2 and β4 subunits, the residue at position 59 is homologous with human (Thr in β2 and Lys in β4). A similar slowing in wash-off was observed with rat α3β2[T59K] compared with wild-type rα3β2 although less severe (Table 3), confirming the key role of residue 59 in loop D for the binding affinity of RegIIA.
Residue 113 mutants (hα3β2[S113R] and hα3β4[R113S]) also exhibited considerably different wash-off rates compared with the respective wild-type receptors. Peptide wash-off at hα3β4[R113S] was very fast, similar to the hα3β2 subtype (95% wash-off in <1 min), whereas at hα3β2[S113R] it was almost as slow as at the opposite wild-type hα3β4 nAChR (95% wash-off in 9–13 min) (Fig. 3A, D; Fig. 4, A and D, and Table 3).
In addition, mutants hα3β4[P165E] and hα3β4[T166V+M169L], which also showed a considerable higher affinity to ACh than wild-type, were tested for their RegIIA off-rate. Both mutants exhibited a faster off-rate compared with wild-type, which is consistent with the higher IC50 and lower EC50 observed. However, the wash-off was not as fast as observed for hα3β4[K59T] or hα3β4[R113S], placing these mutants between hα3β2 and hα3β4 with respect to their off-rates (Table 3).
In summary, these findings show that the residues at positions 59 and 113 not only affect the sensitivity of each receptor subtype to RegIIA, but they are also key determinants for the differences observed in wash-off kinetics. Other residues in the agonist binding loops also affected the affinities for ACh and the peptide, but to a lesser extent than the aforementioned positions and they appear to play an auxiliary role for peptide binding.
Molecular Modeling Shows the Interacting Mechanisms between the Non-conserved Residues of Human α3β4 and α3β2 nAChRs and RegIIA
The interactions between RegIIA and the binding sites of hα3β2 and hα3β4 nAChRs are very similar, with most of the residues in the first loop forming contacts with the (+) subunit and most of the second loop residues interacting with the (−) subunit.
The non-conserved residues at positions 59, 111, 113, and 119 of the β subunits form direct interactions with residues Asn9, Val10, and Asn11 of the second loop of RegIIA (Fig. 5 and Table 4). The residues at positions 111 and 119 of hβ2 and hβ4 are hydrophobic and possess similar biophysical properties thus, explaining their minor effects on the sensitivity of RegIIA to hα3β2 and hα3β4 nAChRs.
FIGURE 5.
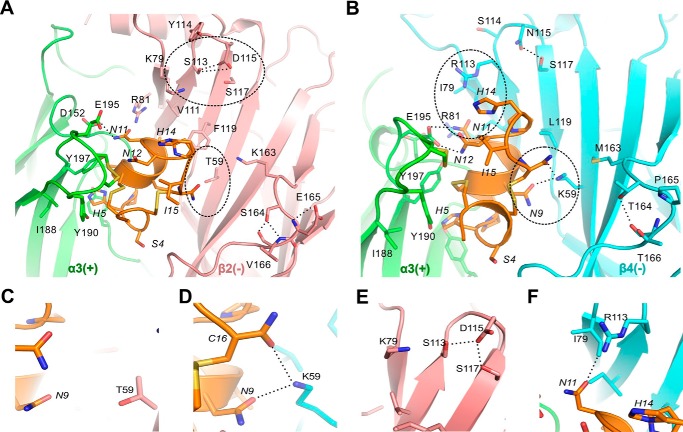
Molecular docking models illustrate the binding modes of RegIIA to hα3β2 (A) and hα3β4 (B), respectively. Several hydrogen bonds (dashed lines) are formed between pairwise interacting residues of different loops or β-sheets, thereby affecting their local conformation or dynamics, which in turn affects the binding of RegIIA. Note that hydrogen bonds are formed by several non-conserved β2-subunit (Ser113, Asp115, Ser164, Glu165) and β4-subunit residues (Asn115, Thr164). The α3(+) interface is shown in green, β2(−) in pink, β4(−) in cyan, and RegIIA in orange. Residues near the agonist binding site that affect RegIIA binding are shown as licorice models. Residues from the receptor and RegIIA are labeled using normal and italic fonts, respectively. Key interaction sites responsible for the differences in α-conotoxin RegIIA binding are highlighted with dashed circles. C–F, magnification of the key sites in hαβ2 (C and E) and hαβ4 (D and F) highlighted with circles in A and B.
TABLE 4.
Contacts of α-conotoxin RegIIA with hα3β2 and hα3β4 nAChR, respectively
Contacts between nAChR and RegIIA are defined as van der Waals interactions if the distance between heavy atoms of RegIIA and nAChR is between 2.6 and 4 Å. Residues of the nAChR forming hydrogen bonds with RegIIA are underlined. Residues that are non-conserved between α3β2 and α3β4 nAChRs, as well as RegIIA residues making contact with them are shown in bold.
| Residuea | α3β2 nAChR |
α3β4 nAChR |
||
|---|---|---|---|---|
| +b | −c | +b | −c | |
| Ser4 | Asp171 | Asp171 | ||
| His5 | Tyr93, Tyr190, Tyr197 | Tyr93, Tyr190, Tyr197 | ||
| Pro6 | Tyr93, Trp149 | Trp57 | Tyr93, Trp149 | Leu121 |
| Ala7 | Ser150, Tyr197 | Ser150, Tyr197 | ||
| Asn9 | Trp57, Thr59, Phe119, Leu121 | Lys59, Leu121 | ||
| Val10 | Ser150 | Arg81, Val111, Phe119, Leu121 | Ser150 | Arg81, Ile111, Leu119, Leu121 |
| Asn11 | Asp152, Glu195, Tyr197 | Lys79, Arg81, Val111 | Asp152, Glu195, Tyr197 | Ile79, Arg81, Ile111 |
| Asn12 | Cys192, Cys193, Glu195 | Cys192, Cys193, Glu195 | ||
| His14 | Cys192, Cys193 | Cys192, Cys193 | ||
| Ile15 | Cys192, Cys193 | Cys192, Cys193 | ||
a Only RegIIA residues making direct contact with the nAChRs are listed.
b Residues of the principal side of the binding site.
c Residues of the complementary side of the binding site.
In contrast, biophysical differences for the side chains at positions 59 and 113 are substantial, with two long and positively charged side chains contributed by residues Lys59 and Arg113 at hβ4, whereas at the hβ2 subunit two neutral and short side chains from residues Thr59 and Ser113 are present (Fig. 5, C–F). In our MD simulations, the side chains of β4-Lys59 and Arg113 formed hydrogen bonds with backbone atoms of Asn9 and Cys16 (for β4-K59), and Asn11 (for β4 Arg113) in the loop 2 of RegIIA, whereas the side chains of β2 Thr59 and β2 Ser113 are not long enough to form such interactions with the corresponding residues of RegIIA (Figs. 5, C–F, and 6). Residue β2 Ser113 is instead forming hydrogen bonds with the adjacent β2 Asp115 and β2 Ser117 residues, which stabilize it in an orientation facing away from the peptide binding site (Fig. 5E).
FIGURE 6.
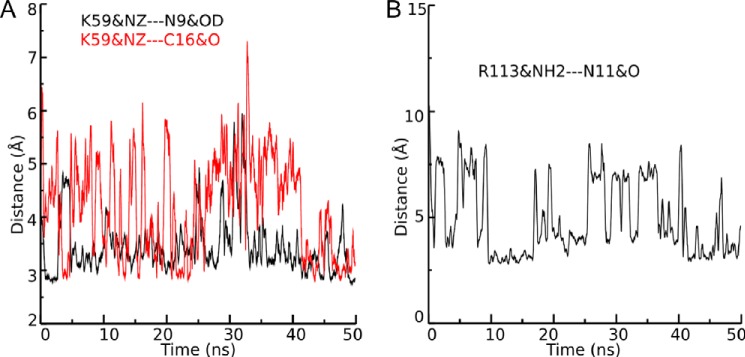
MD simulations of 50-ns duration show the evolution of distances between β4 Lys59 heavy atoms and RegIIA residues Asn9 as well as Cys16 (A), and between β4 Arg113 and RegIIA Asn11 (B), respectively. The strengthening of the hydrogen bond is reversely proportional to the distance between hydrogen bond donor and hydrogen bond acceptor. When the distance between the heavy atoms is more than 3.5 Å, the strength of the hydrogen bond is believed to be weak. Red and black colors are used to discriminate the two simulations performed with β4 Lys59. The labels K59&NZ-N9&OD and K59&NZ-C16&O designate the distance between the heavy atom NA of residue β4 Lys59 and the heavy atom OD of residue RegIIA Asn9, and the distance between heavy atom K59&NZ and the heavy atom O of residue RegIIA C16, respectively. The label R113&NH2-N11&O refers to the distance between the heavy atoms NH2 of β4 Arg113 and O of RegIIA Asn11.
MD simulation of a RegIIA-bound mutant hα3β4[K59T] model revealed that the β4 Lys59 to Thr mutation removed the hydrogen bond between the RegIIA amide terminus and the position 59 side chain, thereby significantly decreasing the affinity of RegIIA compared with wild-type hα3β4. Although the conformation of residues near position 59 was not significantly perturbed by the mutation, the non-conserved residues near the binding site have an unfavorable effect for RegIIA binding at hα3β4[K59T]. MD simulation indicates the opposite mutant hα3β2[T59K] exhibits higher affinity to RegIIA compared with wild-type hα3β2, because RegIIA is stabilized by additional hydrogen bonds formed between β2 Lys59, Asp170, and the amide group of RegIIA. However, this mutation also introduced significant local conformation changes of the β2 binding interface, thereby decreasing contacts between RegIIA and the β2 residue side chains Phe119 and Lys163. These effects counteract the affinity gain resulting from introduction of Lys59, and are likely responsible for the overall only mildly increased RegIIA activity at mutant hα3β2[T59K].
Computational studies on a RegIIA-bound mutant hα3β2[S113R] model revealed that residue β2 Arg113 forms salt bridges with Asp115, similar to Ser113 in wild-type hα3β2 (see Fig. 5E), instead of directly interacting with Asn11 of RegIIA. Interestingly we observed that the backbone oxygen atom of Asn11 forms a hydrogen bond with Lys79 rather than Arg113 due to the relocation of the latter. Thus, our MD simulation indicates that Arg113 of hα3β2[S113R] strengthens the binding of RegIIA through stabilization of the local conformation near the binding site, thereby making mutant hα3β2[S113R] even more RegIIA-sensitive than wild-type hα3β4.
Furthermore, we computationally analyzed RegIIA binding to other hα3β4 mutants that exhibited a major loss in RegIIA activity. The loop F mutation in hα3β4[P165E] created a local conformation change to the entire agonist binding loop F. We identified that the angle defined by the Cα atoms at 164, 165, and 166 positions became significantly larger (right shift). We conclude that this local conformation change might affect RegIIA binding to the α3β4[P165E], but that Pro165, or its neighboring residues, are not determinants for RegIIA binding in the wild-type nAChR subtypes.
The hα3β4[N115D] mutant, which also showed significantly decreased sensitivity to RegIIA compared with wild-type hα3β4, was analyzed further. Based on MD simulation data we suggest β4 Asp115 might compete with residues of RegIIA to interact with Arg113. Similar to the interaction of β2 Ser113 in hα3β2 (Fig. 5, A and E) residue β4 Arg113 forms salt bridges with Asp115 rather than with residues of RegIIA in hα3β4[N115D]. This explains the mutational effects of [N115D] to the binding affinity of RegIIA. Although the opposite mutation in β2 (mutant hα3β2[D115N]) would weaken the H-bond with Ser113, the overall effect would not be a reciprocal gain in RegIIA affinity, because the Ser113 residue is too short to directly interact with RegIIA.
Overall, our computationally directed mutational studies suggest that mutants hα3β4[P165E] and hα3β4[N115D] negatively affect RegIIA binding through mildly altering the local conformation of either the backbone or the side chains of the residues near the binding site. But those non-conserved positions do not appear to determine the difference in RegIIA sensitivity between wild-type hα3β2 and hα3β4 to a similar extent as do residues 59 and 113.
The non-conserved β subunit residues at positions 34, 36, 71, 78, 79, 114, 115, 163, 164, 165, and 166 are far from the binding site, and their side chains merely form weak interactions with RegIIA. Their effects to the binding affinity of RegIIA are mostly indirect, by forming hydrogen bonds with neighboring residues to affect the local conformation of the binding site (Fig. 5).
Taken together, MD simulations indicate the non-conserved residues 59 and 113 of the (−) interface are crucial for the observed differences in RegIIA activity, due to their considerably different mode of interaction with the peptide. In hα3β4 nAChRs, hydrogen bonds are formed between the respective residues and RegIIA loop 2, but they are absent in hα3β2.
The α3β4 nAChR-selective α-Conotoxin AuIB Is Less Active at the Homologous Human nAChR Subtype
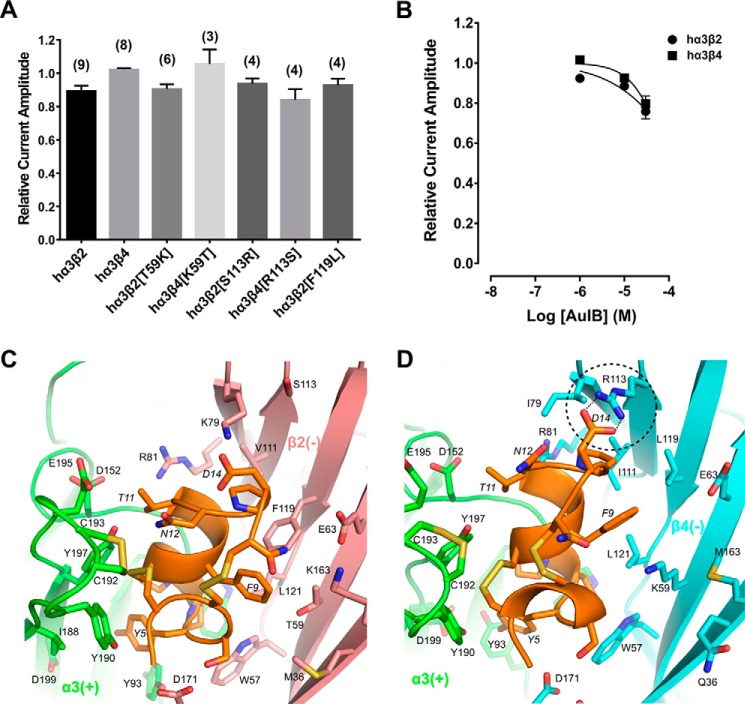
The 4/6 α-conotoxin AuIB has been shown to selectively inhibit the rat α3β4 nAChR with an IC50 of 0.75 μm (13). Activity at human nAChRs has not yet been reported. Therefore, we determined the activity of AuIB at wild-type hα3β2, hα3β4, and relevant β subunit mutants. Interestingly, 1 μm AuIB was inactive at hα3β4 and exhibited a minor block of ACh-evoked currents (10.7 ± 3.2%, n = 9) at hα3β2 nAChR. The nAChR β subunit mutants that distinctly affected the RegIIA activity profile (positions 59 and 113) were all relatively insensitive to 1 μm AuIB (Fig. 7A). Concentration-response analyses with wild-type hα3β2 and hα3β4 nAChRs confirmed the low sensitivity of both nAChR subtypes to AuIB. α-Conotoxin AuIB at 30 μm, reduced ACh-evoked currents mediated by hα3β2 to 75.9 ± 3.7% of control (n = 10) and hα3β4-mediated currents to only 79.6 ± 4.0% (n = 10) of control, indicating that the IC50 is considerably higher than 30 μm at either nAChR subtype (Fig. 7B).
FIGURE 7.
α-Conotoxin AuIB has minor activity at human α3β2 and α3β4 nAChRs. A, bar graph representing potency of block by α-conotoxin AuIB (1 μm) at wild-type and mutant human α3β2 and α3β4 nAChRs. Data represent mean ± S.E., n = 3–9. B, concentration-response analysis of AuIB at wild-type α3β2 and α3β4 nAChRs indicated the IC50 is considerably higher than 30 μm at both nAChR subtypes. AuIB (30 μm) reduced ACh-evoked current amplitude mediated by α3β2 to 75.9 ± 3.7% of control (n = 10) and α3β4 currents to 79.6 ± 4.0% (n = 10), respectively. C and D, molecular dynamics simulation predicted binding modes of AuIB to α3β2 (C) and α3β4 (D). Several hydrogen bonds are formed between pairwise interacting residues of different loops and between toxin and receptor, e.g. AuIB Asp14 with β4 Arg113 (dashed circle, hydrogen bonds as dotted lines). The α3(+) interface is shown in green, β2(−) in pink, β4(−) in cyan, and AuIB in orange. Non-conserved residues are shown as licorice models and labeled. Residues from the receptor and AuIB are labeled using normal and italic fonts, respectively.
MD Simulation of α-Conotoxin AuIB Binding to hα3β2 and hα3β4 nAChRs, Compared with RegIIA, Indicates Other Key Interactions Affect Binding
In MD simulations, the α4/6-conotoxin AuIB binds at the interface between the subunits and overlaps with the agonist binding sites of hα3β2 and hα3β4 nAChRs. Overall molecular interactions are relatively similar to those with RegIIA, as most of AuIB loop 1 residues form contacts with the (+)-subunit and most of the residues at loop 2 interact with residues at the (−)-subunit (Fig. 7, C and D, and Table 5). However, despite the general similarity of RegIIA and AuIB binding to hα3β2 and hα3β4 nAChRs, several side chains, including the non-conserved residues at positions 59 and 113 of both nAChR β subunits, formed remarkably different interactions with residues in loop 2 of AuIB (Fig. 7, C and D, and Table 5). These differences are a result of a significant shift in orientation between the two α-conotoxins. Direct comparison of the orientation of AuIB with RegIIA at the α3(+)β4(−) binding interface revealed that the shifted AuIB backbone places AuIB Phe9 far away from the β4 Trp57 side chain. Thereby no direct interaction between the aromatic ring of AuIB Phe9 and the tryptophan can occur. Instead AuIB Phe9 is in relatively close proximity to β4 Leu119 and β4 Lys59 (Fig. 7D). It is likely the missing interaction between the AuIB Phe9 aromatic ring and β4 Trp57 is a significant factor for the lack of activity of AuIB at hα3β4, whereas for hα3β2 it is the insufficient stabilization of AuIB Phe9 by residue β2 Thr59.
TABLE 5.
Contacts of α-conotoxin AuIB with hα3β2 and hα3β4 nAChRs, respectively
Contacts between nAChR and AuIB are defined as van der Waals interactions if the distance between heavy atoms of AuIB and nAChR is between 2.6 and 4 Å. Residues of the nAChR forming hydrogen bonds with AuIB are underlined. Residues that are non-conserved between α3β2 and α3β4 nAChRs, as well as AuIB residues making contact with them are shown in bold.
| Residuea | α3β2 nAChR |
α3β4 nAChR |
||
|---|---|---|---|---|
| +b | −c | +b | −c | |
| Ser4 | Asp171 | Asp170 | ||
| Tyr5 | Tyr93, Tyr190, Asp199 | Tyr93, Tyr190, Asp199 | ||
| Pro6 | Trp149 | Trp57, Leu121 | Trp149 | Trp57, Leu121 |
| Pro7 | Trp149, Ser150, Tyr197 | Trp149, Ser150, Tyr197 | ||
| Phe9 | Met36, Trp57, Thr59, Phe119, Leu121 | Lys59, Glu61, Leu119, Leu121 | ||
| Ala10 | Ser150 | Arg81, Val111, Phe119 | Ser150 | Arg81, Ile111 |
| Thr11 | Asp152, Glu195, Tyr197 | Asp152, Glu195, Tyr197 | ||
| Asn12 | Cys192, Cys193 | Cys192, Cys193 | ||
| Pro13 | Lys79, Val111, Phe119 | Ile111, Arg113 | ||
| Asp14 | Lys79 | Arg113 | ||
a Only AuIB residues making direct contact with the nAChRs are listed.
b Residues of the principal side of the binding site.
c Residues of the complementary side of the binding site.
Discussion
The α3β4 nAChR subtype represents an important pharmacological target as it is involved in several pathophysiological disease conditions such as lung cancer, nicotine addiction, and drug abuse (3, 11, 12). To date, only a few α-conotoxins have been found to be active at the α3β4 nAChR subtype and even fewer are highly specific for it. A problem complicating the task of developing specific α3β4 receptor antagonists is that the ligand binding sites are structurally very similar among nAChR subtypes. However, the small structural differences between subtypes can cause profound pharmacological effects, as seen with the selectivity profiles of various α-conotoxins.
α-Conotoxin RegIIA has previously been shown to exhibit a species-specific difference in activity between human and rat α3β2 nAChRs, whereas this species difference was not observed at the α3β4 nAChR subtype (17). Given that both the species and subtype selectivity profiles are well described for RegIIA, it represents a suitable probe to investigate pharmacological differences between α3β2 and α3β4 nAChR subtypes in detail.
RegIIA inhibits ACh-evoked currents competitively, like most α-conotoxins, indicating that its binding site overlaps with the agonist binding site (16). Sequence alignment between human β4 and β2 subunits shows their extracellular agonist binding regions are moderately conserved on the amino acid level between subtypes (70% homology, Fig. 1A). A mutational approach was chosen to elucidate how non-conserved structural elements account for functional differences in α-conotoxin binding.
Using receptor chimeras and point mutations to exchange individual residues to those of the opposite subtype, we identified two key residues (at positions 59 and 113, respectively) that crucially determine the selectivity of RegIIA. Exchange of β2 Thr59 to Lys increased RegIIA sensitivity similar to that observed at hα3β4, whereas the opposite exchange of β4 Lys59 to Thr resulted in a 61-fold loss of sensitivity compared with wild-type hα3β4. Furthermore, the rate of recovery from block by RegIIA differs considerably between hα3β2 and hα3β4. By exchanging the single residue at position 59 of the β subunit, we were able to swap the wash-off rate of subtypes.
Residue 113 on the β subunit has a similarly important role for RegIIA selectivity. Exchange of β2 Ser113 to Arg made the mutant hα3β2[S113R] receptor twice as sensitive to RegIIA than wild-type α3β4. In contrast, the opposite mutation in β4 made the mutant receptor (hα3β4[R113S]) 18-fold less sensitive to RegIIA than wild-type hα3β4 and 34-fold less sensitive than mutant hα3β2[S113R]. In addition, the single exchange of residue 113 between β2 and β4 was sufficient to switch the off-rate of RegIIA to that of the opposite subtype. To our knowledge, this is the first time Arg113 at the complementary agonist binding interface of nAChRs has been identified as a key determinant in antagonist binding. Computational modeling of RegIIA-bound mutant hα3β2[S113R] provided insights about why this mutant is even more sensitive to RegIIA than hα3β4. When Arg113 is introduced into the β2 backbone it forms salt bridges with Asp115, similar to Ser113 in wild-type hα3β2 (see Fig. 5E). However, in the case of Arg113 this interaction also affects the conformations of residues near the binding site in such way that residue β2 Lys79 can form a hydrogen bond with the backbone oxygen atom of RegIIA Asn11.
It is important to note that the ACh EC50 at hα3β2 is ∼18-fold lower than at hα3β4. As the aforementioned β-subunit residues are both within the agonist binding region at the interface between the α and β subunits, it can be assumed that point mutations would affect the EC50 of ACh. Indeed mutant hα3β4[R113S] exhibited ∼7-fold lower EC50 than wild-type hα3β4. However, unlike the IC50 of RegIIA, there was no reciprocal effect on ACh EC50 with the opposite mutant hα3β2[S113R]. This mutant exhibited an even slightly lower EC50 than hα3β2. Comparably, both hα3β2[T59K] and hα3β4[K59T] revealed an EC50 similar to hα3β2 (Table 2). This discrepancy suggests the mutations affecting ACh EC50 most likely impact the observed RegIIA potencies as well, as ACh competes with RegIIA for the agonist binding site. Mutants hα3β2[T59K] and hα3β2[S113R] are already more RegIIA sensitive than wild-type hα3β2, but it can be assumed they would be even more sensitive if their ACh EC50 values were not considerably lower than the EC50 seen with hα3β4.
Other hα3β4 mutants, primarily within loop F, also decreased RegIIA sensitivity and increased ACh affinity. Compared with the aforementioned hα3β4[K59T] and hα3β4[R113S] mutants, we observed a less extensive increase in RegIIA IC50 and RegIIA wash-off kinetics. We hypothesize the other mutants play an auxiliary role for the subtype selectivity of RegIIA.
Residue 59 on the β subunits of rat nAChRs has previously been identified as a determinant for α-conotoxin LvIA potency and wash-off kinetics (31). α-Conotoxin LvIA exhibited a 17-fold higher selectivity for rat α3β2 over α3β4, and the rat α3β2[T59K] mutation further increased the the activity of the peptide by ∼10-fold. Furthermore, similar to our findings with RegIIA, mutant rα3β2[T59K] considerably slowed the recovery from block by LvIA compared with wild-type α3β2 (31).
In our MD simulations, the evolution of distances between key RegIIA and receptor residues over 50 ns revealed the involvement of the β4 Lys59 residue side chain in forming hydrogen bonds with main chain atoms of RegIIA Asn9 and Cys16 residues, and similarly, β4 Arg113 formed a hydrogen bond with the Asn11 in loop 2 of RegIIA (Figs. 5 and 6). Both RegIIA and LvIA have an asparagine at position 9. Therefore, it is likely the Asn9 residue interacts with β4 Lys59 via a direct hydrogen bond in both peptides. The increased potency and slower wash-off caused by the point mutation of β2 Thr59 to Lys could then be explained by gain of these additional hydrogen bonds, resulting in tighter binding of the peptide to the mutant nAChR. The side chain of β2 Thr59, however, is not long enough to form such interactions with the corresponding residues in loop 2 of RegIIA (Fig. 5C), which would explain the faster wash-off at wild-type α3β2 and mutant hα3β4[K59T], as well as lower potency of the peptide compared with hα3β4 and mutant hα3β2[T59K], respectively.
Furthermore, MD simulation of RegIIA binding to mutant hα3β4[K59T] revealed that the mutation also removed a H-bond between the RegIIA amide terminus and the position 59 side chain, thereby significantly decreasing RegIIA affinity compared with wild-type hα3β4. In addition, non-conserved residues near the RegIIA binding site cannot compensate the affinity decrease of RegIIA with the β4 Lys59 to Thr mutation, which explains the observation that hα3β4[K59T] is even significantly less sensitive to RegIIA than hα3β2.
Our hypothesis that the RegIIA Asn9/β4 Lys59 contact is critical for the activity of RegIIA at α3β4 is also corroborated by RegIIA alanine-scanning mutagenesis data. It has been shown that the [N9A]RegIIA analogue was inactive at rα3β4 and α7, but maintained inhibitory activity at rα3β2 (16). This study also noted that alanine analogues [N11A]RegIIA, [N12A]RegIIA, and [N11A,N12A]RegIIA had improved selectivity for α3β4 compared with the native RegIIA. However, these analogues were less active than RegIIA at α3β4 and the overall higher selectivity for α3β4 resulted from considerable loss of activity at the α3β2 and α7 nAChR subtypes. MD simulation also showed that for α3β2, this was mainly due to destabilization of peptide contacts with multiple residues at the plus (+) and minus (−) interfaces (16).
α-Conotoxin LvIA remarkably revealed a considerable specificity for α3β2 over α6/α3β2β3 rat and human nAChRs, a rare feature not shared by other α-conotoxins such as RegIIA (32). As loops 1 of the two peptides are identical, these specificity differences must be conferred by the second loop in which three residues (11, 12, and 14) are different. Indeed, in our modeling most of the residues in the first loop of RegIIA form contacts with the principal β3 subunit and most of the residues of the second loop form contacts with the complementary subunits (Table 4). At α3β2 and α3β4 nAChRs, we identified RegIIA residues Asn9, Asn11, and Asn12 as main determinants for interaction with the β subunit. We speculate that the residues at positions 11 and 12 primarily account for the differences in α3β2* subtype sensitivity between LvIA and RegIIA.
Zhangsun et al. (31) also identified residues 111 and 119 in loop E of β2 and β4 subunits as determinants for LvIA potency, whereas for RegIIA, only residue 113 was responsible for the potency of the peptide in loop E. As mentioned previously, our modeling suggests that that the β subunit specificity of RegIIA is conferred via interactions of its residues Asn11 and Asn12 with loop E of the receptor, specifically β4 residue Arg113. As LvIA has different residues at these positions, the data on LvIA regarding key interacting residues with loop E (31) cannot be directly compared with RegIIA. However, as Asn9 is present in both RegIIA and LvIA, the interaction with β4 Lys59 likely occurs via the same mechanism leading to the similar observations in mutational analyses.
α-Conotoxin AuIB has been characterized as a selective inhibitor of the rat α3β4 nAChR subtype (13), however, to our knowledge, its specificity profile at human nAChRs had not been evaluated. We tested AuIB at hα3β2 and hα3β4 nAChRs and selected mutants. AuIB appeared to be considerably less active at hα3β4, as 1 μm AuIB did not inhibit ACh-evoked currents at this nAChR subtype and higher concentrations caused minimal inhibition, suggesting that the IC50 is considerably >30 μm (Fig. 7, A and B). This is in contrast to the homologous rat α3β4 nAChR where an IC50 of 0.75 μm was reported (13). Interestingly, we observed some inhibitory activity of AuIB at the hα3β2 subtype. However, inhibition was slight with ACh-evoked current amplitude inhibited by <50% in the presence of 30 μm AuIB, similar to that observed for hα3β4 (Fig. 7B).
Molecular modeling and docking simulation was also performed with α-conotoxin AuIB, indicating that many of the non-conserved residues in the complementary face of the agonist binding site are also involved in the binding of this peptide, although in a different way (Fig. 7, C and D, and Table 5). For example, residue Asp14 of AuIB forms a hydrogen bond with β4 Arg113, whereas at the α3β2 nAChR, β2 Lys79 interacts with Asp14. Residue Phe9 of AuIB interacts with several of the non-conserved residues, similar to Asn9 of RegIIA.
AuIB F9 has previously been identified as the key residue for the specific binding of AuIB to the rat α3β4 nAChR (33). AuIB Phe9 interacts with Trp57 and Lys59 residues of the WLK pocket in rat β4 loop D, (designated Trp59 and Lys61 in this paper), most likely via π-π stacking due to the deep insertion of its aromatic ring (33). An alanine substitution substantially reduced α3β4 inhibition and decreased subtype specificity. Additional AuIB analogues with other side chains at this position demonstrated that size, aromaticity, and hydrophobicity at position 9 of AuIB are important for interaction between the peptide and β4 subunit of the rat α3β4 pentamer (33).
Direct comparison of the orientation of RegIIA and AuIB at the α3(+)β4(−) binding site by MD simulation revealed that the β4 subunit interface is largely unchanged in conformation, whereas the orientation of the two bound α-conotoxins differs significantly. RegIIA Asn9 is in close proximity to the β4 Trp57 and β4 Lys59 side chains, but the AuIB backbone is shifted relative to RegIIA, locating AuIB Phe9 outside of the WLK pocket (Fig. 7D). Thereby, AuIB Phe9 forms contact with β4 Lys59 and β4 Leu119, but the crucial π-π interaction with β4 Trp57 is absent. As the tryptophan residue was indispensable for the inhibitory effect of AuIB at the rat homologue, and β4 Lys59 appeared to play an auxiliary role (33), the lack of interaction between AuIB F9 and β4 Trp57 could explain the loss of AuIB activity at the hα3β4 subtype. At hα3β2, our modeling revealed contacts between AuIB and β2 Trp57, Thr59, and other residues (Fig. 7C and Table 5). Although the crucial interaction of AuIB Phe9 with the Trp57 residue in loop D is present, residue Thr59 does not form an effective binding pocket for AuIB Phe9, unlike β4 Lys59. The hydrophilic side chain of β2 Thr59 is likely unfavorable to interact with the aromatic phenyl ring of AuIB Phe9. In summary, our modeling offers an explanation for the lower inhibitory activity of AuIB at hα3β2 and hα3β4 nAChRs. The characteristic binding pocket for AuIB F9 formed by the two rat β4 subunit loop D residues is absent in the human β2 and β4 counterparts, resulting in ineffective stabilization of AuIB.
In addition, nAChR subtype selectivity and wash-off kinetics has also been investigated for α-conotoxin BuIA (34, 35). A slow recovery from the BuIA block at α3β4 compared with significantly faster recovery at α3β2 has been observed in different species, including human (35). Mutational studies at rat β2 and β4 subunits have identified residues 59, 111, and 119 as critical for the off-rate differences. Similar to our findings with RegIIA, the off-rate of BuIA was slower in the β2[T59K] mutant compared with wild-type α3β2 (34).
Mutation of β2 T59K has also been shown to be critical for dihydro-β-erythroidine and neuronal bungarotoxin sensitivity of α3β2 (36), as well as affecting the affinity of the agonists, ACh and nicotine to the α2β2 nAChR (37). The high selectivity of α-conotoxin MII for the α3β2 nAChR subtype (>200-fold less active at other nAChR subunit combinations) has been mapped to three sequence segments using α3β2 chimeras. Within the segments, β2 residue Thr59 was identified as one factor determining the higher sensitivity of β2 to MII compared with β4 (38).
In summary, these findings confirm that subtype-selective nAChR antagonists often work through common mechanisms by interacting with the same structural components and sites on the receptor. Knowing the key residue interactions by which antagonists (such as α-conotoxins) can discern between structurally very similar nAChR subtypes provides us with the tools to design novel potent and highly subtype-selective drugs.
Here, we show that residues in loop 2 of α-conotoxins often form direct interactions with specific non-conserved residues in the agonist binding loops at the complementary interface of α3β2 and α3β4 nAChRs. In the future, generating toxin analogues with mutations in loop 2 to either improve or decrease binding to receptor key residues, such as Lys59 or Arg113 of the β4 subunit, might be a way to direct the selectivity of α-conotoxins to different subtypes of nAChRs. Given that hα3β4 subtype-selective α-conotoxins are scarce, the design of new peptides targeting this nAChR subtype is desirable.
Author Contributions
D. J. A., H. C., S. N. K., and R. Y. conceived and designed the study and wrote the manuscript. H. C. conducted the mutagenesis and electrophysiological studies and analyzed the results. S. N. K. and H. S. T. conducted electrophysiological studies and analyzed the results. R. Y. conducted and analyzed the molecular modeling studies. D. J. A. provided the financial support and resources. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This work was supported by the Australian Research Council Discovery Project Grant DP150103990 (to D. J. A.), Natural Science Foundation of China Grant 81502977 (to R. Y.), and Fundamental Research Funds for the Central Universities (to R. Y.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1.
- nAChR
- nicotinic acetylcholine receptor
- ACh
- acetylcholine
- Acm
- acetamidomethyl
- ECD
- extracellular domain
- Fmoc
- N-(9-fluorenyl)-methoxycarbonyl
- MD
- molecular dynamics
- CI
- confidence interval.
References
- 1. Wonnacott S. (1997) Presynaptic nicotinic ACh receptors. Trends Neurosci. 20, 92–98 [DOI] [PubMed] [Google Scholar]
- 2. Dani J. A., and Bertrand D. (2007) Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu. Rev. Pharmacol. Toxicol. 47, 699–729 [DOI] [PubMed] [Google Scholar]
- 3. Jensen A. A., Frølund B., Liljefors T., and Krogsgaard-Larsen P. (2005) Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J. Med. Chem. 48, 4705–4745 [DOI] [PubMed] [Google Scholar]
- 4. Changeux J. P. (2010) Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nat. Rev. Neurosci. 11, 389–401 [DOI] [PubMed] [Google Scholar]
- 5. Gotti C., and Clementi F. (2004) Neuronal nicotinic receptors: from structure to pathology. Prog. Neurobiol. 74, 363–396 [DOI] [PubMed] [Google Scholar]
- 6. Albuquerque E. X., Pereira E. F., Alkondon M., and Rogers S. W. (2009) Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol. Rev. 89, 73–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gotti C., Clementi F., Fornari A., Gaimarri A., Guiducci S., Manfredi I., Moretti M., Pedrazzi P., Pucci L., and Zoli M. (2009) Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem. Pharmacol. 78, 703–711 [DOI] [PubMed] [Google Scholar]
- 8. Lebbe E. K., Peigneur S., Wijesekara I., and Tytgat J. (2014) Conotoxins targeting nicotinic acetylcholine receptors: an overview. Mar. Drugs 12, 2970–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olivera B. M., Quik M., Vincler M., and McIntosh J. M. (2008) Subtype-selective conopeptides targeted to nicotinic receptors: concerted discovery and biomedical applications. Channels 2, 143–152 [DOI] [PubMed] [Google Scholar]
- 10. Muttenthaler M., Akondi K. B., and Alewood P. F. (2011) Structure-activity studies on α-conotoxins. Curr. Pharm. Des. 17, 4226–4241 [DOI] [PubMed] [Google Scholar]
- 11. Hurst R., Rollema H., and Bertrand D. (2013) Nicotinic acetylcholine receptors: from basic science to therapeutics. Pharmacol. Ther. 137, 22–54 [DOI] [PubMed] [Google Scholar]
- 12. Salas R., Sturm R., Boulter J., and De Biasi M. (2009) Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J. Neurosci. 29, 3014–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo S., Kulak J. M., Cartier G. E., Jacobsen R. B., Yoshikami D., Olivera B. M., and McIntosh J. M. (1998) α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 18, 8571–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luo S., Zhangsun D., Zhu X., Wu Y., Hu Y., Christensen S., Harvey P. J., Akcan M., Craik D. J., and McIntosh J. M. (2013) Characterization of a novel α-conotoxin TxID from Conus textile that potently blocks rat αa3β4 nicotinic acetylcholine receptors. J. Med. Chem. 56, 9655–9663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franco A., Kompella S. N., Akondi K. B., Melaun C., Daly N. L., Luetje C. W., Alewood P. F., Craik D. J., Adams D. J., and Marí F. (2012) RegIIA: an α4/7-conotoxin from the venom of Conus regius that potently blocks α3β4 nAChRs. Biochem. Pharmacol. 83, 419–426 [DOI] [PubMed] [Google Scholar]
- 16. Kompella S. N., Hung A., Clark R. J., Marí F., and Adams D. J. (2015) Alanine scan of α-conotoxin RegIIA reveals a selective α3β4 nicotinic acetylcholine receptor antagonist. J. Biol. Chem. 290, 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kompella S. N., Cuny H., Hung A., and Adams D. J. (2015) Molecular basis for differential sensitivity of α-conotoxin RegIIA at rat and human neuronal nicotinic acetylcholine receptors. Mol. Pharmacol. 88, 993–1001 [DOI] [PubMed] [Google Scholar]
- 18. Hogg R. C., Hopping G., Alewood P. F., Adams D. J., and Bertrand D. (2003) α-Conotoxins PnIA and [A10L]PnIA stabilize different states of the α7-L247T nicotinic acetylcholine receptor. J. Biol. Chem. 278, 26908–26914 [DOI] [PubMed] [Google Scholar]
- 19. Yu R., Craik D. J., and Kaas Q. (2011) Blockade of neuronal α7-nAChR by α-conotoxin ImI explained by computational scanning and energy calculations. PLoS Comput. Biol. 7, e1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magrane M., and UniProt Consortium (2011) UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011, 10.1093/database/bar009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ulens C., Hogg R. C., Celie P. H., Bertrand D., Tsetlin V., Smit A. B., and Sixma T. K. (2006) Structural determinants of selective α-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc. Natl. Acad. Sci. U.S.A. 103, 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., and Chen L. (2007) Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94-Å resolution. Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 23. Zouridakis M., Giastas P., Zarkadas E., Chroni-Tzartou D., Bregestovski P., and Tzartos S. J. (2014) Crystal structures of free and antagonist-bound states of human α9 nicotinic receptor extracellular domain. Nat. Struct. Mol. Biol. 21, 976–980 [DOI] [PubMed] [Google Scholar]
- 24. Shen M. Y., and Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olsson M. H., Søndergaard C. R., Rostkowski M., and Jensen J. H. (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 [DOI] [PubMed] [Google Scholar]
- 26. Case D. A., Darden T. A., Cheatham T. E., Simmerling C. L., Wang J., et al. (2008) AMBER 10, University of California, San Francisco, CA [Google Scholar]
- 27. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., and Simmerling C. (2006) Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miyamoto S., and Kollman P. A. (1992) Settle: an analytical version of the Shake and Rattle Algorithm for rigid water models. J. Comput. Chem. 13, 952–962 [Google Scholar]
- 29. Darden T., York D., and Pedersen L. (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 30. Corringer P. J., Le Novère N., and Changeux J. P. (2000) Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 40, 431–458 [DOI] [PubMed] [Google Scholar]
- 31. Zhangsun D., Zhu X., Wu Y., Hu Y., Kaas Q., Craik D. J., McIntosh J. M., and Luo S. (2015) Key residues in the nicotinic acetylcholine receptor β2 subunit contribute to α-conotoxin LvIA binding. J. Biol. Chem. 290, 9855–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo S., Zhangsun D., Schroeder C. I., Zhu X., Hu Y., Wu Y., Weltzin M. M., Eberhard S., Kaas Q., Craik D. J., McIntosh J. M., and Whiteaker P. (2014) A novel α4/7-conotoxin LvIA from Conus lividus that selectively blocks α3β2 vs. α6/α3β2β3 nicotinic acetylcholine receptors. FASEB J. 28, 1842–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grishin A. A., Cuny H., Hung A., Clark R. J., Brust A., Akondi K., Alewood P. F., Craik D. J., and Adams D. J. (2013) Identifying key amino acid residues that affect α-conotoxin AuIB inhibition of α3β4 nicotinic acetylcholine receptors. J. Biol. Chem. 288, 34428–34442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shiembob D. L., Roberts R. L., Luetje C. W., and McIntosh J. M. (2006) Determinants of α-conotoxin BuIA selectivity on the nicotinic acetylcholine receptor β subunit. Biochemistry 45, 11200–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Azam L., Dowell C., Watkins M., Stitzel J. A., Olivera B. M., and McIntosh J. M. (2005) α-Conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 280, 80–87 [DOI] [PubMed] [Google Scholar]
- 36. Harvey S. C., and Luetje C. W. (1996) Determinants of competitive antagonist sensitivity on neuronal nicotinic receptor β subunits. J. Neurosci. 16, 3798–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parker M. J., Harvey S. C., and Luetje C. W. (2001) Determinants of agonist binding affinity on neuronal nicotinic receptor β subunits. J. Pharmacol. Exp. Ther. 299, 385–391 [PubMed] [Google Scholar]
- 38. Harvey S. C., McIntosh J. M., Cartier G. E., Maddox F. N., and Luetje C. W. (1997) Determinants of specificity for α-conotoxin MII on α3β2 neuronal nicotinic receptors. Mol. Pharmacol. 51, 336–342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.