Abstract
Aspergillus fumigatus is a versatile fungus able to successfully exploit diverse environments from mammalian lungs to agricultural waste products. Among its many fitness attributes are dozens of genetic loci containing biosynthetic gene clusters (BGCs) producing bioactive small molecules (often referred to as secondary metabolites or natural products) that provide growth advantages to the fungus dependent on environment. Here we summarize the current knowledge of these BGCs—18 of which can be named to product—their expression profiles in vivo, and which BGCs may enhance virulence of this opportunistic human pathogen. Furthermore, we find extensive evidence for the presence of many of these BGCs, or similar BGCs, in distantly related genera including the emerging pathogen Pseudogymnoascus destructans, the causative agent of white-nose syndrome in bats, and suggest such BGCs may be predictive of pathogenic potential in other fungi.
This article is part of the themed issue ‘Tackling emerging fungal threats to animal health, food security and ecosystem resilience’.
Keywords: Aspergillus fumigatus, secondary metabolism, gene cluster, fungal infection
1. Introduction
Of the many informative biological stories uncovered from sequences of thousands of fungal genomes, one garnering extreme interest is the discovery of large numbers of secondary-metabolite (SM) encoding biosynthetic gene clusters (BGCs) in specific fungal taxa. Much of the interest is driven through a quest to find novel pharmaceuticals as many fungal SMs display valuable bioactivities, presumably evolved in their protective role against both biotic and abiotic stressors [1]. Furthermore, a subset of pathogenic fungi produces SMs that increase their virulence attributes on hosts. These SM-producing fungi include plant, insect and vertebrate pathogens.
An understanding that fungal SMs could adversely impact humans and other vertebrates can be traced back to the 1960s where the SM aflatoxin, produced by the seed pathogen Aspergillus flavus, resulted in deaths of thousands of poultry in England [2]. Toxic fungal SMs found in food and feed products are collectively known as mycotoxins. Owing to the harmful effects of these metabolites, worldwide efforts have resulted in the identification and characterization of the most common mycotoxin BGCs including not only aflatoxin (and the closely related sterigmatocystin BGC) but also fumonisin, trichothecene, ochratoxin, zearalenone, citrinin, ergot alkaloid, cyclopiazonic acid and patulin BGCs (reviewed in [3]). Although touted for their toxic, mutagenic, teratogenic or carcinogenic impacts on vertebrates, emerging data are now suggesting that these mycotoxins—and other BGC metabolites for that matter—serve as fitness factors for the producing fungi. For example, emerging data suggest that aflatoxin may act as an endogenous redox signal protecting the fungus from oxidative stress [4]. The connection of oxidative stress with BGC activation is, in fact, so common in fungi [5–7] that several reviews address the protective nature of fungal SMs and their possible use as oxidative treatments in human disorders [8,9].
Oxidative stress protection is but one property of a subset of SMs. Demonstrated bioactivities of other SMs range between metal acquisition, UV protection and antimicrobial activity (often via targeting of specific critical enzymes and hence potentially damaging to host tissues but useful in pharmaceutical settings). Considering that fungi can contain dozens of BGCs within their genomes, the potential of BGC metabolites to contribute to disease development is considerable. It is not yet possible to survey the total secondary metabolome of a single fungus to accurately predict how SMs could drive pathogenic ability, as characterization of the entire set of BGCs—including knowledge of structure and bioactivity of the SMs, BGC expression during pathogenesis and possible synergism of the multiple SMs—is needed for such a charge. Yet we can start to speculate on what BGCs might signal pathogenic potential from studies of the opportunistic human pathogen Aspergillus fumigatus.
Owing to its importance as a serious pathogen of immunocompromised patients, efforts have focused on characterizing the SM arsenal of this fungus with a view to determining any role of these compounds in virulence. These efforts have led to, arguably, A. fumigatus having the greatest number of characterized BGCs of any fungus studied with the possible exception of the genetic model Aspergillus nidulans [10]. Initial genome sequence predicted a total of 22 BGCs [11], but additional bioinformatic analysis now suggests over 30 BGCs are present in the genome of A. fumigatus [12]. The DHN-melanin cluster, first characterized in 1999 [13], has garnered considerable attention as a virulence factor in this organism [14].The second BGC to be characterized in A. fumigatus produced gliotoxin, which, like DHN-melanin, impacts virulence [15–18]. Following a reoccurring theme in studies of the roles of SM in fungal biology, both DHN-melanin and gliotoxin exhibit bioprotective properties well beyond a role as simply a virulence factor. While not directly examined in A. fumigatus, melanins in other fungi provide protection from UV radiation [19] and gliotoxin provides redox homeostasis properties to the fungus [20].
Since identification of the gliotoxin BGC, a successive wave of studies revealed the endocrocin [21], fumigaclavine [22], fumagillin [23], fumiquinazoline [24], fumisoquin [25], fumitremorgin [26,27], helvolic acid [28], hexadehydroastechrome [29], neosartoricin (fumicycline) [30], pseurotin [23], pyripyropene [31] and trypacidin [32] BGCs in this species. The two siderophore BGCs are not presented as such, but can be pieced together from several studies to yield a two-gene ferricrocin cluster composed of SidI and SidC [33,34] and a six-gene fusarinine C cluster (composed of SidJ, SidF, SidH, SidD, SitT and MirD [34–38]). Additional BGCs can be inferred from orthology to known clusters in other fungi including the conserved nidulanin A BGC described in A. nidulans [39], a fusarielin-like cluster described in Fusarium graminearum [40], and a fumonisin-like cluster (fumonisin is synthesized by many Fusarium spp.) probably producing sphingofungin.
Although there are several algorithms designed to identify fungal BGCs, including anti-SMASH [41], SMURF [42] and MIDDAS-M [43], these programs err on the side of over prediction, inability to discern superclusters and in missing non-canonical clusters (e.g. those lacking typical SM synthetases such as polyketide synthases (PKSs), non-ribosomal peptide synthetases and terpene cyclases). Therefore, it is not possible at this moment to give a precise number of total BGCs for any SM-rich fungus including A. fumigatus. With this in mind, here we consider only 26 A. fumigatus BGCs that either from previous characterization or from strong bioinformatic support, are likely to produce a predictable SM (electronic supplementary material, table S1), examine their expression in a murine model of infection and expand on the conservation of these BGCs in other fungi.
2. Results and discussion
(a). Twenty-six clusters
In addition to the 18 clusters that have been characterized either in A. fumigatus or other fungi, eight additional loci strongly support production of predictable SMs (all sequence data used, via MultiGeneBlast (MGB) analysis, to identify clusters in other fungi are presented as electronic supplementary material, Data S1). Focus on these 26 does not preclude the existence of other BGCs in A. fumigatus.
BGC 1. This five-gene cluster encodes a probably highly reduced polyketide as determined by the enzymatic domains present in the PKS. While not found in other sequenced Aspergillus species, the arrangement and gene number is conserved in the wheat pathogen Phaeosphaeria nodorum and the pneumocandin-producing fungus Glarea lozoyensis.
BGC 2 (Nidulanin A). This cluster produces the cyclic tetrapeptide nidulanin A, found in all Aspergillus and Penicillium species. Originally characterized in A. nidulans, currently there is no known property or function for this metabolite [39]. However, the non-ribosomal synthetase (NRPS) generating nidulanin is also involved in synthesis of fungisporin [44], which has been shown to have antibacterial properties [45].
BGC 3 (Ferricrocin). This two-gene cluster contains sidI and sidC that encode proteins required for synthesis of the siderophore ferricrocin in A. fumigatus. The pairing of sidI and sidC genes is common across Ascomycete genera [33,34]. Ferricrocin, along with fusarinine C, is critical for A. fumigatus virulence [46].
BGC 4. This six-gene cluster is likely to produce a polyketide with similarities to fusarielins, produced by Fusarium and Aspergillus spp. [47,48] and variations (4–6 genes) of this cluster are also found in Colletotrichum fioriniae, Pestalotiopsis fici and some Metarhizium spp. Fusarielins exhibit antibacterial, antifungal, anticancer, antiangiogenic and antiproliferative properties [49]. Existence of a fusarielin-like cluster in A. fumigatus was first noted in a study characterizing the F. graminearum fusarielin BGC [40].
BGC 5 (DHN-melanin). A well-characterized cluster producing the polyketide DHN-melanin giving the characteristic greyish green coloration of A. fumigatus conidia. A near identical cluster is found in closely related Aspergillus spp. including Aspergillus clavatus and Neosartorya fischeri. As mentioned above, numerous studies have described the importance of this metabolite in virulence in A. fumigatus [50]. In fact, a repeating theme in fungal biology is the requirement of spore melanins for virulence in numerous pathogenic fungi [51].
BGC 6 (Fumigaclavine). Fumigaclavines are bioactive alkaloids with numerous bioactivities ranging from antibacterial [52] to vasorelaxant [53]. Fumigaclavines are considered mycotoxins and purified compounds have a toxic impact on mammalian cells [54]. However, a role for these compounds in pathogenesis has not yet been shown in A. fumigatus [55]. The fumigaclavine-producing eleven-gene cluster—or a significant part of it—is also present in members of the dermatophytic genera Arthroderma and Trichophyton, the insect-pathogenic genus Metarhizium and a miscellaneous group of fungi including Byssochlamys spectabilis, G. lozoyensis and Claviceps purpurea.
BGC 7. This cluster encodes another uncharacterized highly reduced polyketide that is present in some aspergilli including the opportunistic pathogen Aspergillus terreus and some Penicillium spp.
BGC 8 (Fusarinine C). This second siderophore six-gene cluster is conserved in many fungi [56]. Similar clusters of four and five genes are also present in many species and may represent divergence or the absence of the missing genes.
BGC 9 (Hexadehydroastechrome). This eight-gene BGC, or variations of it, is present sporadically in Aspergillus spp. including A. terreus [57], the dermatophytic genera Arthroderma and Trichophyton, the plant pathogenic Fusarium oxysporum and Fusarium avenaceum, and partially in the blackleg pathogen Leptosphaeria maculans. Over-production of this metabolite enhances virulence in a murine model of A. fumigatus infection [29], and its activity is associated with iron homeostasis and alterations in siderophore synthesis [58].
BGC 10. This cluster contains an NRPS and NRPS-like gene suggesting that the metabolite will be a small peptide. A subsection of this cluster is present in the closely related N. fischeri but with little homology in other fungi.
BGC 11. This uncharacterized eight-gene BGC contains several homologues to the Fusarium fumonisin BGC [59] and thus is predicted to produce the similarly structured polyketide sphingofungin [60]. Fumonisin is a potent mycotoxin impacting ceramide synthase activity [61]. Sphingofungins act as antifungal compounds by targeting serine palmitoyltransferase [62]. This BGC is largely conserved in Arthroderma, Trichophyton and Metarhizium spp.
BGC 12. The presence of conserved enzymatic domains in the NRPS in this uncharacterized cluster suggests it will encode a small peptide 2–3 amino acids in size. This cluster is found in N. fischeri and a subset of the genes in a few other Aspergillus spp.
BGC 13. (Endocrocin). The polyketide endocrocin is another spore pigment with antimigratory effects on neutrophils [21]. Endocrocin, trypacidin and neosartoricin all belong to a class of related non-reduced polyketides with aromatic ring structures [63]. Variations of all these clusters are common in many fungi, with such clusters often encoding a spore pigment or toxin. The fungus Pseudogymnoascus destructans, causing the white-nose syndrome of bats, contains at least one cluster with considerable similarity to several genes in these non-reduced polyketide BGCs [63].
BGC 14 (Trypacidin). Another spore pigment, this compound is produced by a 13-gene BGC [32,64]. The endocrocin and trypacidin BGCs show redundancy as both synthesize the spore pigment endocrocin [32]. Trypacidin is known and named for its antiprotozoal (e.g. Trypanosoma cruzi) activity [65]. Trypacidin is toxic to lung cells [66].
BGC 15 (Helvolic acid). Following on trypacidin, helvolic acid also exhibits antiprotozoal activity [67] and is produced by several fungal spp. in addition to A. fumigatus, including several plant and insect pathogens and many endophytes [28,68,69]. Along with gliotoxin and fumagillin, helvolic acid exhibits cilioinhibitory activity [70] and is a potent antimicrobial [71].
BGC 16. An uncharacterized NRPS-like cluster conserved in closely related Aspergillus spp. including N. fischeri and, partially, A. clavatus. The composition of genes in this predicted BGC is less supportive of a bona fide cluster than in the cases of all other uncharacterized BGCs here presented.
BGC 17. This BGC contains the largest NRPS gene in the A. fumigatus genome with 6 adenylation and 10 condensation domains, suggestive of a 6–10 residue peptide. This BGC appears limited to A. fumigatus and N. fischeri. Although the metabolite produced by this BGC has not been characterized, disruption of the NRPS results in a hypervirulent strain as assessed in insect and murine models. The authors suggest that BGC17 product may be structural in nature [72].
BGC 18 (Fumisoquin). This newly described BGC [25] is also found in Penicillium solitum, F. graminearum and F. pseudograminearum.
BGC 19. An uncharacterized NRPS cluster immediately adjacent to the gliotoxin BGC. As with BGC 17, this cluster appears limited to A. fumigatus and N. fischeri.
BGC 20 (Gliotoxin). Variations of this 13-gene BGC are conserved in numerous fungi including may plant pathogens [15]. Gliotoxin, belonging to a class of natural products known as epipolythiopiperazines, contributes to the virulence of A. fumigatus and a similar epipolythiopiperazine, sirodesmin, is a potent phytotoxin [73].
BGC 21 (Fumiquinazoline). Various fumiquniazolines are reported to be produced by Aspergillus, Penicillium and Acremonium spp. [74] with one report of antibacterial properties of fumiquinazoline F [75]. This BGC, expressed in spores of A. fumigatus [24], is also found in the saprophyte Pseudogymnoascus pannorum.
BGC 22 (Pyripyropene A). Pyripyropenes are produced by several Aspergillus and Penicillium spp. [76]. MGB locates the BGC to these genera and the nematode endoparasite, Hirsutella minnesotensis where it is likely a virulence factor, surmising from the several patents claiming its use as a nematicide [77]. Pyripyropene is of considerable interest as a potent and selective acyl-coenyzmeA::acholesterol acyltransferase 2 (ACAT2) inhibitor [78].
BGC 23 (Neosartoricin). This polyketide cluster is conserved in many species, again including the dermatophytic genera Arthroderma and Trichophyton and insect pathogen Metarhizium. The orchid mycorrhizal species Oidiodendron maius also appears to contain this or a similar BGC.
BGC 24 (Fumitremorgin). Similar to the pyripyropene BGC, the fumitremorgin gene cluster is not only present in A. fumigatus and N. fischeri but also H. minnesotensis. Studies have identified antifungal and insecticidal properties of this and related metabolites [79].
BGC 25 (Fumagillin and pseurotin). The genes for these two SMs are intertwined in a supercluster [26]. This BGC is conserved in part in Metarhizium spp., Scedosporium apiospermum, P. solitum and Colletotrichum sublineola. Fumagillin is a potent inhibitor of methionine aminopeptidases, is considered a possible therapeutic targeting angiogenesis due to this property [80] and is currently used to treat microsporidian fungal infections [81]. Several biological activities have been attributed to pseurotin including antibacterial properties [52] and inhibition of IgE production [82].
BGC 26. This uncharacterized BGC is likely to produce a polyketide-terpene hybrid, possibly with some similarity to the Aspergillus flavus toxin aflatrem [83], and is found in N. fischeri.
How might these BGC metabolites affect virulence of A. fumigatus or other pathogenic fungi? Gene-deletion studies from six of the BGCs (3, 5, 8, 9, 17 and 20) have shown impacts on virulence in murine models of invasive aspergillosis. Another eight BGC (6, 13, 14, 15, 21, 22, 24 and 25) produce metabolites that have toxic effects on various organisms ranging from bacteria to mammals, which may indicate contributions of these metabolites to virulence, perhaps in a synergistic manner. One unexpected finding from our MGB analysis was the conservation of many of these clusters in other pathogenic fungi; in particular, the presence of many of the A. fumigatus BGCs in the insect-pathogenic genus Metarhizium, the dermatophytic genera Arthroderma and Trichophyton, but also in several plant-pathogenic species. Supporting a case for a role of these BGC metabolites in virulence, the products (pyripyropene and fumitremorgin) of two clusters conserved in the nematode parasite H. minnesotensis show insecticidal properties.
Although relatively low in BGC numbers (approx. one dozen), it is interesting to note that the emerging bat pathogen P. destructans contains a BGC with similarities to the endocrocin BGC. Endocrocin, as noted above, is an A. fumigatus spore metabolite that inhibits neutrophil migration. Because studies have documented an aberrant bat immune response to infections by P. destructans (formerly Geomyces destructans [84–86]), it is intriguing to speculate that some P. destructans SMs might be playing a role in these responses.
(b). Expression studies
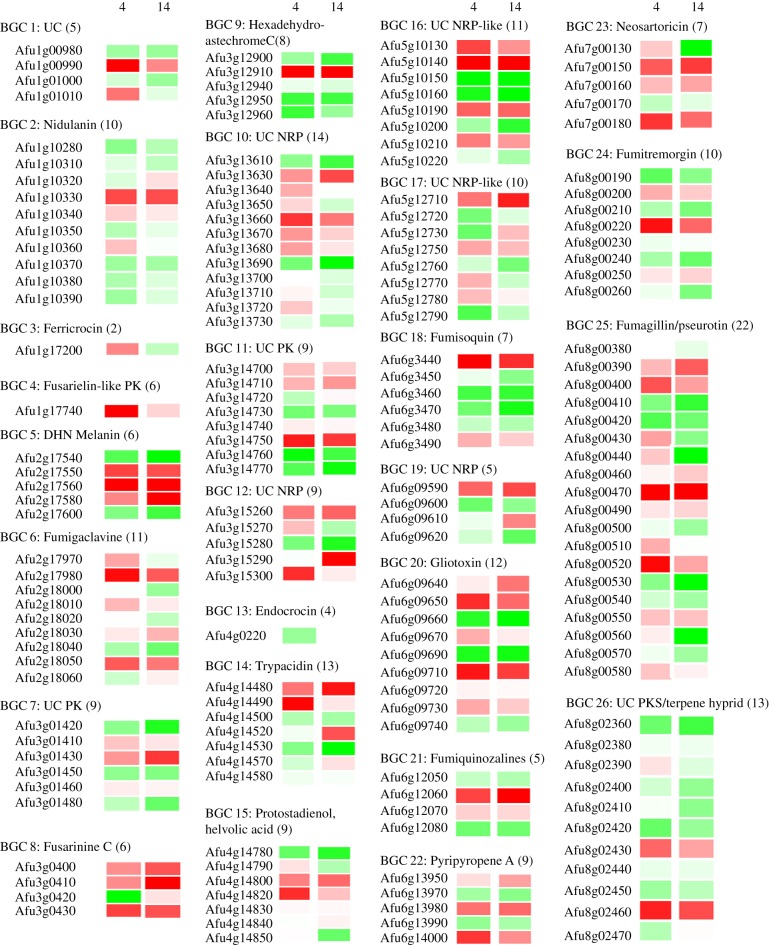
Our previous transcriptome analyses revealed that simultaneous contributions of multiple A. fumigatus SMs are likely during colonization of the mammalian host niche [87] whereby comparative analysis of A. fumigatus gene expression in laboratory culture versus the lungs of neutropenic mice during initiation of growth revealed coordinate upregulation of BGCs 5, 8, 7, 12, 20, 24 and 25 during infection of the mammalian lung. Despite the technical complexity of the experimentation, the boundaries of BGCs were clearly delineated, adding weight to the relevance of their clustered groupings to transcriptional regulation. In this study, we took an alternative view, asking whether genes of predicted BGCs were (i) expressed and (ii) prioritized over other metabolic gene functions during morphogenesis and colonization of the host environment. We used quantile normalized transcript abundance (calculated per gene as a function of all other expressed genes in the analysis) as a proxy measure of overall transcript abundance, thereby assessing gene expression levels for all BGC genes at 4 and 14 h post-infection (figure 1). Of the total cohort of n = 226 genes (electronic supplementary material, table S1), detectable expression of n = 171 (76%) was achievable (figure 1) and of these, 80 genes (47%) were expressed at significantly higher levels than the population median. Concordant with our previous analyses [87] and those assessing pathogenicity of SM-deficient null mutants [46,50,54,55,66,70,82], greater than or equal to 50% of detectable transcripts arising from BGCs 3, 5, 6, 13, 14, 15 and 25 exhibited abundances, which significantly exceeded the median value of that of the sampled gene population at one or both time-points of the analysis. A similar picture was observed for the majority of other BGCs, with the notable exceptions of BGCs 2, 9, 18, 24, 15, 11 and 26, which might be of limited relevance to murine infection or, alternatively, have relevance to time points of infection occurring later than those we studied here (as, notably, overexpression of the BGC 9 product increases virulence in the murine model of IA [29]). Concordant with roles for metal ion homeostasis in mammalian pathogenicity, genes of the siderophore BGCs 3 and 8 are highly expressed. Afu1g17200 of BCG3, known to encode an enzyme required for synthesis of conidial siderophores and germination of A. fumigatus spores in iron-depleted environments [36] was preferentially expressed at 4 h but not 14 h post-infection, as were many genes of the uncharacterized BGC 10 (figure 1). The relevance of BGCs 5 and 8 (directing biosynthesis of DHN-melanin and fusarinine C, respectively) has been conclusively demonstrated in neutropenic hosts via analysis of null mutants [36,50]. Although DHN-melanin is often regarded as a critical spore-associated metabolite we found heightened transcript abundance of several key biosynthetic genes at both the 4 h and 14 h time points, while the relevance of fusarinine C appears to escalate with time. Genes of the gliotoxin BGC 20 were predominantly upregulated, although, as we have previously discussed and noted [87], gliotoxin is not a virulence factor in neutropenic hosts. It is likely that the stresses encountered in the host environment provoke the expression of a bespoke ‘blend’ of SM production, the net effect of which is to undermine host defenses thus creating a hospitable niche. The remaining critical question to be addressed is the impact of synergistic SM activities upon host physiology and innate immunity.
Figure 1.
Expression of BGC genes during colonization of the mammalian lung. Transcript abundances of BGC genes at 4 h (n = 6 technical replicates) and 14 h (n = 4 technical replicates) post-infection of the neutropenic murine host (mean value of transcript abundance from a pooled sample of n = 7 mice per time-point). Quantile normalized transcript abundance, per gene, per time-point, calculated as a function of all expressed genes in the analysis, was used as a proxy measure of standing in the overall ranking of transcript abundance. Transcript abundance, at each time-point, relative to all expressed A. fumigatus genes in the analysis is represented as greater (red) or lower (green) than the population median. Numbers in parentheses indicate the number of genes per cluster predicted by MultiGeneBlast.
3. Conclusion
While products of BGCs are not the only factors that contribute to pathogenicity of any fungus, certain specific SMs are known virulence factors and many can modify host responses to pathogenic fungi. These metabolites have largely been understudied with regard to their impact on pathogenesis, in part, due to the immense work required to identify and characterize a complete secondary metabolome. Some BGCs encode metabolites that are fitness factors that enable a fungus to obtain nutrients and cofactors (e.g. metal acquiring metabolites) or protect from stress (e.g. oxidative, UV, other microbes) in any environment, and do not easily predict/indicate degree of virulence or pathogenicity. The complex expression of specific BGCs within a single species is also likely to be a factor impacting pathogenic abilities yet we are at a point where we cannot confidently predict which BGC couplings are synergistic but only speculate on this possibility. Nevertheless, the identification of conserved BGCs—especially those that have been demonstrated to exhibit fitness or bioactive properties—in more than one fungus lends credibility to a view that such BGCs may constitute part of the arsenal for both persistent and emergent fungal pathogens.
4. Material and methods
The MGB was used to detect homologous clusters across distantly related fungal species [88]. MGB architecture searches were carried out with the per cent identity threshold set to 25%, the synteny weight set to 0 and the maximum intergenic distance set to 110% of the span of the corresponding A. fumigatus cluster with a minimum bound of 25 kb. Other parameters were set to default. The input for each search was a multiFASTA file of the amino acid sequences of the proteins predicted to be encoded by the genes in the region of an A. fumigatus predicted cluster or hypothetical cluster variant, erring on the side of over-inclusion to detect cluster boundaries. A custom fungal database, previously described, was used for these searches [63].
(a). Murine infections
A previously published method for examination of fungal gene expression in neutropenic mice was used [87]. Briefly, intraperitoneal injections of cyclophosphamide (150 mg kg−1, Endoxana, Asta Medica) were conducted on days −3 and −1, with a single subcutaneous dose of hydrocortisone acetate (112.5 mg kg−1, Hydrocortistab, Sovereign Medical) on day −1. Aspergillus fumigatus spores from clinical isolate Af293 were prepared and intranasally administered to mice as described previously [87]. For each of the 4 and 14 h time-points seven mice were infected and then humanely sacrificed. Bronchoalveolar lavage (BAL) was performed using three to four aliquots of 37° sterile saline per mouse. BAL fluids (BALFs) were snap frozen in liquid nitrogen.
(b). Aspergillus fumigatus RNA extraction, amplification and hybridization
The BALF samples were centrifuged at 14 000 r.p.m. for 5 min and the pellet was washed with 500 µl ice-cold water. BALFs were pooled, suspended in 450 µl ME-RLC buffer (Qiagen) and ground in liquid nitrogen with a pestle and mortar. RNA was then extracted using RNeasy® Kit (Qiagen). Quality of RNA was checked by Nanodrop ND-1000 Spectrophotometer (Nanodrop, Wilmington, DE, USA). RNA with an A260/280 and an A260/230 ratio > 1.9 was used for the experiments.
Fungal RNA was doubly amplified prior [87] to array hybridization. A 70-mer oligotide glass slide DNA microarray platform was used (Pathogen Functional Genomic Resource Centre, JCVI, Rockville) and doubly amplified RNA derived from 4 and 14 h time points was hybridized, respectively, in technical duplicate or triplicate. Washed slides were scanned and data captured using GenePix 4000b (Axon Instruments, Foster City, CA, USA) semiconfocal microarray scanner and associated GenePix Pro 3.0 software. Images were saved as .TIF files and TIGR Spotfinder [89] was used to quantitate fluorescent intensities. Poor-quality spots were identified manually and flagged. After Spotfinder analysis, results were exported as TIGR software-compatible .MEV files following local background subtraction.
(c). Microarray normalization
A single-channel approach was applied using quantile normalization to extract, from dual channel co-hybridizaton data [90,91], a comparative, ranked view of Af293 transcript abundance. From .MEV files, fluorescence intensity values were captured on a gene-by-gene basis, corresponding to either (with duplicate spots) n = 6 technical replicates for the 4 h time point or (with duplicate spots) n = 4 technical replicates for the 14 h time point. Values were quantile normalized across all replicates [92] using GeneSpring v. 11.0.5 (Agilent Technologies). Following normalization, spot intensity values were averaged between technical replicates at each time point, producing a single measurement of transcript abundance per gene at each time point. Quantile-normalized transcript abundance, per gene, per time point, calculated as a function of all expressed genes in the analysis, was used as a proxy measure of standing in overall ranking of transcript abundance.
Supplementary Material
Supplementary Material
Acknowledgement
The authors thank Dr Jonathan M. Palmer for the creation of a fungal-specific MultiGeneBlast database.
Ethics
Murine infections were performed in accordance with local and international ethical governance, under UK Home Office Project Licence PPL/70/6487, in dedicated facilities at Imperial College London.
Data accessibility
Raw and quantile normalized data have been uploaded to the Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ under the title ‘Secondary metabolite arsenal of an opportunistic pathogenic fungus’. Accession number pending. Fasta format sequence data used to perform MGB analysis have been uploaded as the electronic supplementary material, Data S1.
Authors' contributions
K.T. carried out the MultiGeneBlast data analysis; T.C.C. performed the murine experimentation and devised the analytical methodology for transcriptome analyses, E.B. and N.P.K. conceived of the study, coordinated the study and helped draft the manuscript. All authors contributed to writing of the manuscript and gave final approval for publication.
Competing interests
We have no competing interests.
Funding
This work was supported by NIH grant R01 AI065728-01 to N.P.K., and in part, by grants to E.B. from the Medical Research Council (G0501164 and R118757) and British Biotechnology and Biosciences Research Council (BB/G009619/1 and T.C.C. studentship).
References
- 1.Keller NP. 2015. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 11, 671–677. ( 10.1038/nchembio.1897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nesbitt BF, O'Kelly J, Sargeant K, Sheridan ANN. 1962. Aspergillus Flavus and turkey X disease: toxic metabolites of Aspergillus flavus. Nature 195, 1062–1063. ( 10.1038/1951062a0) [DOI] [PubMed] [Google Scholar]
- 3.Throckmorton K, Isham NC, Ghannoum MA, Keller N. 2015. Mycotoxins. In Manual of clinical microbiology, 11 edn, pp. 2188–2195. American Society of Microbiology; ( 10.1128/9781555816728) [DOI] [Google Scholar]
- 4.Roze VL, Laivenieks M, Hong S-Y, Wee J, Wong S-S, Vanos B, Awad D, Ehrlich CK, Linz EJ. 2015. Aflatoxin biosynthesis is a novel source of reactive oxygen species—a potential redox signal to initiate resistance to oxidative stress? Toxins 7, 1411–1430. ( 10.3390/toxins7051411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fréalle E, Aliouat-Denis C-M, Delhaes L, Hot D, Dei-Cas E. 2013. Transcriptomic insights into the oxidative response of stress-exposed Aspergillus fumigatus. Curr. Pharm. Des. 19, 3713–3737. ( 10.2174/1381612811319200011) [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Wu F, Liu L, Liu X, Che Y, Keller NP, Guo L, Yin WB. 2015. The bZIP transcription factor PfZipA regulates secondary metabolism and oxidative stress response in the plant endophytic fungus Pestalotiopsis fici. Fungal Genet. Biol. 81, 221–228. ( 10.1016/j.fgb.2015.03.010) [DOI] [PubMed] [Google Scholar]
- 7.Schmidt-Heydt M, Stoll D, Schütz P, Geisen R. 2015. Oxidative stress induces the biosynthesis of citrinin by Penicillium verrucosum at the expense of ochratoxin. Int. J. Food Microbiol. 192, 1–6. ( 10.1016/j.ijfoodmicro.2014.09.008) [DOI] [PubMed] [Google Scholar]
- 8.White PA, et al. 2014. Antioxidant activity and mechanisms of action of natural compounds isolated from lichens: a systematic review. Molecules 19, 14 496–14 527. ( 10.3390/molecules190914496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avalos J, Limón MC. 2015. Biological roles of fungal carotenoids. Curr. Genet. 61, 309–324. ( 10.1007/s00294-014-0454-x) [DOI] [PubMed] [Google Scholar]
- 10.Yaegashi J, Oakley BR, Wang CCC. 2014. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J. Ind. Microbiol. Biotechnol. 41, 433–442. ( 10.1007/s10295-013-1386-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin RM, Fedorova ND, Jin WB, Cramer RA, Wortman JR, Kim HS, Nierman WC, Keller NP. 2007. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 3, e50 ( 10.1371/journal.ppat.0030050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. 2013. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 13, 91 ( 10.1186/1471-2180-13-91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai HF, Wheeler MH, Chang YC, Kwon-Chung KJ. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181, 6469–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayry J, et al. 2014. Surface structure characterization of Aspergillus fumigatus conidia mutated in the melanin synthesis pathway and their human cellular immune response. Infect. Immun. 82, 3141–3153. ( 10.1128/IAI.01726-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardiner DM, Howlett BJ. 2005. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol. Lett. 248, 241–248. ( 10.1016/j.femsle.2005.05.046) [DOI] [PubMed] [Google Scholar]
- 16.Bok JW, Chung D, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Kirby KA, Keller NP. 2006. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 74, 6761–6768. ( 10.1128/IAI.00780-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cramer RA, et al. 2006. Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5, 972–980. ( 10.1128/EC.00049-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spikes S, et al. 2008. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 197, 479–486. ( 10.1086/525044) [DOI] [PubMed] [Google Scholar]
- 19.Al-Laaeiby A, Kershaw MJ, Penn TJ, Thornton CR. 2016. Targeted disruption of melanin biosynthesis genes in the human pathogenic fungus Lomentospora prolificans and its consequences for pathogen survival. Int. J. Mol. Sci. 17, 444 ( 10.3390/ijms17040444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolan SK, O'Keeffe G, Jones GW, Doyle S. 2015. Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 23, 419–428. ( 10.1016/j.tim.2015.02.005) [DOI] [PubMed] [Google Scholar]
- 21.Berthier E, et al. 2013. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLoS Pathog. 9, e1003289 ( 10.1371/journal.ppat.1003289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson SL, Panaccione DG. 2012. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia 104, 804–812. ( 10.3852/11-310) [DOI] [PubMed] [Google Scholar]
- 23.Wiemann P, Guo C-J, Palmer JM, Sekonyela R, Wang CCC, Keller NP. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl Acad. Sci. USA 110, 17 065–17 070. ( 10.1073/pnas.1313258110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim FY, Ames B, Walsh C, Keller N. 2014. Coordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell. Microbiol. 16, 1267–1283. ( 10.1111/cmi.12284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baccile JA, et al. 2016. Plant-like biosynthesis of isoquinoline alkaloids in Aspergillus fumigatus. Nat. Chem. Biol. 12, 419–424. ( 10.1038/nchembio.2061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato N, Suzuki H, Okumura H, Takahashi S, Osada H. 2013. A point mutation in ftmD blocks the fumitremorgin biosynthetic pathway in Aspergillus fumigatus strain Af293. Biosci. Biotechnol. Biochem. 77, 1061–1067. ( 10.1271/bbb.130026) [DOI] [PubMed] [Google Scholar]
- 27.Maiya S, Grundmann A, Li S-M, Turner G. 2006. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem 7, 1062–1069. ( 10.1002/cbic.200600003) [DOI] [PubMed] [Google Scholar]
- 28.Lodeiro S, Xiong Q, Wilson WK, Ivanova Y, Smith ML, May GS, Matsuda SPT. 2009. Protostadienol biosynthesis and metabolism in the pathogenic fungus Aspergillus fumigatus. Org. Lett. 11, 1241–1244. ( 10.1021/ol802696a) [DOI] [PubMed] [Google Scholar]
- 29.Yin W-B, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. 2013. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J. Am. Chem. Soc. 135, 2064–2067. ( 10.1021/ja311145n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao Z, Rao G, Li C, Abil Z, Luo Y, Zhao H. 2013. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth. Biol. 2, 662–669. ( 10.1021/sb400058n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh T, Tokunaga K, Matsuda Y, Fujii I, Abe I, Ebizuka Y, Kushiro T. 2010. Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat. Chem. 2, 858–864. ( 10.1038/nchem.764) [DOI] [PubMed] [Google Scholar]
- 32.Throckmorton K, Lim FY, Kontoyiannis DP, Zheng W, Keller NP. 2016. Redundant synthesis of a conidial polyketide by two distinct secondary metabolite clusters in Aspergillus fumigatus. Environ. Microbiol. 18, 246–259. ( 10.1111/1462-2920.13007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisendle M, Oberegger H, Zadra I, Haas H. 2003. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 49, 359–375. ( 10.1046/j.1365-2958.2003.03586.x) [DOI] [PubMed] [Google Scholar]
- 34.Gründlinger M, Yasmin S, Lechner BE, Geley S, Schrettl M, Hynes M, Haas H. 2013. Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol. 88, 862–875. ( 10.1111/mmi.12225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gründlinger M, Gsaller F, Schrettl M, Lindner H, Haasa H. 2013. Aspergillus fumigatus SidJ mediates intracellular siderophore hydrolysis. Appl. Environ. Microbiol. 79, 7534–7536. ( 10.1128/AEM.01285-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schrettl M, et al. 2007. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 3, 1195–1207. ( 10.1371/journal.ppat.0030128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrettl M, et al. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70, 27–43. ( 10.1111/j.1365-2958.2008.06376.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrettl M, et al. 2010. HapX-Mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6, e1001124 ( 10.1371/journal.ppat.1001124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen MR, et al. 2013. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl Acad. Sci. USA 110, E99–E107. ( 10.1073/pnas.1205532110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sørensen JL, et al. 2012. Production of novel fusarielins by ectopic activation of the polyketide synthase 9 cluster in Fusarium graminearum. Environ. Microbiol. 14, 1159–1170. ( 10.1111/j.1462-2920.2011.02696.x) [DOI] [PubMed] [Google Scholar]
- 41.Weber T, et al. 2015. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. ( 10.1093/nar/gkv437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. ( 10.1016/j.fgb.2010.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umemura M, Koike H, Machida M. 2015. Motif-independent de novo detection of secondary metabolite gene clusters-toward identification from filamentous fungi. Front. Microbiol. 6 ( 10.3389/fmicb.2015.00371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ali H, et al. 2014. A non-canonical NRPS is involved in the synthesis of fungisporin and related hydrophobic cyclic tetrapeptides in Penicillium chrysogenum. PLoS ONE 9, e98212 ( 10.1371/journal.pone.0098212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Himaja M, Elizabeth L, Moonjit D, Asif K. 2015. Synthesis and antimicrobial evaluation of N-methylated analog of fungisporin. Univ. J. Pharm. 4, 10–14. [Google Scholar]
- 46.Schrettl M, Haas H. 2011. Iron homeostasis-Achilles’ heel of Aspergillus fumigatus? Curr. Opin. Microbiol. 14, 400–405. ( 10.1016/j.mib.2011.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sørensen JL, Akk E, Thrane U, Giese H, Sondergaard TE. 2013. Production of fusarielins by Fusarium. Int. J. Food Microbiol. 160, 206–211. ( 10.1016/j.ijfoodmicro.2012.10.016) [DOI] [PubMed] [Google Scholar]
- 48.Hung PN, Zhang D, Lee U, Jung SK, Hong DC, Byeng WS. 2007. Dehydroxychlorofusarielin B, an antibacterial polyoxygenated decalin derivative from the marine-derived fungus Aspergillus sp. J. Nat. Prod. 70, 1188–1190. ( 10.1021/np060552g) [DOI] [PubMed] [Google Scholar]
- 49.Fujimoto H, Aoyama H, Noguchi-Yachide T, Hashimoto Y, Kobayashi H. 2008. Fusarielin A as an anti-angiogenic and anti-proliferative agent: basic biological characterization. Chem. Pharm. Bull. 56, 298–304. ( 10.1248/cpb.56.298) [DOI] [PubMed] [Google Scholar]
- 50.Heinekamp T, Thywißen A, Macheleidt J, Keller S, Valiante V, Brakhage AA. 2013. Aspergillus fumigatus melanins: interference with the host endocytosis pathway and impact on virulence. Front. Microbiol. 3 ( 10.3389/fmicb.2012.00440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gómez BL, Nosanchuk JD. 2003. Melanin and fungi. Curr. Opin. Infect. Dis. 16, 91–96. [DOI] [PubMed] [Google Scholar]
- 52.Pinheiro EAA, Carvalho JM, Santos DCP Feitosa AO, Marinho PSB, Guilhon GMSP, Souza ADL, Silva FMA, Marinho AMR. 2013. Antibacterial activity of alkaloids produced by endophytic fungus Aspergillus sp. EJC08 isolated from medical plant Bauhinia guianensis. Nat. Prod. Res. 27, 1633–1638. ( 10.1080/14786419.2012.750316) [DOI] [PubMed] [Google Scholar]
- 53.Ma H-Y, Song Y-C, Mao Y-Y, Jiang JH, Tan R-X, Luo L. 2006. Endophytic fungal metabolite fumigaclavine C causes relaxation of isolated rat aortic rings. Planta Med. 72, 387–392. ( 10.1055/s-2005-916235) [DOI] [PubMed] [Google Scholar]
- 54.Li Y-X, Himaya SWA, Dewapriya P, Zhang C, Kim S-K. 2013. Fumigaclavine C from a marine-derived fungus Aspergillus fumigatus induces apoptosis in MCF-7 breast cancer cells. Mar. Drugs 11, 5063–5086. ( 10.3390/md11125063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Latif H, Gross M, Fischer D, Lierz M, Usleber E. 2015. Immunochemical analysis of fumigaclavine mycotoxins in respiratory tissues and in blood serum of birds with confirmed aspergillosis. Mycotoxin Res. 31, 177–183. ( 10.1007/s12550-015-0228-4) [DOI] [PubMed] [Google Scholar]
- 56.Lehner SM, Atanasova L, Neumann NKN, Krska R, Lemmens M, Druzhinina IS, Schuhmacher R. 2013. Isotope-assisted screening for iron-containing metabolites reveals a high degree of diversity among known and unknown siderophores produced by Trichoderma spp. Appl. Environ. Microbiol. 79, 18–31. ( 10.1128/AEM.02339-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bok JW, et al. 2015. Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics 16, 426 ( 10.1186/s12864-015-1561-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiemann P, et al. 2014. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front. Microbiol. 5, 1742 ( 10.3389/fmicb.2014.00530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stępień Ł. 2014. The use of Fusarium secondary metabolite biosynthetic genes in chemotypic and phylogenetic studies. Crit. Rev. Microbiol. 40, 176–185. ( 10.3109/1040841X.2013.770387) [DOI] [PubMed] [Google Scholar]
- 60.VanMiddlesworth F, et al. 1992. Sphingofungins A, B, C, and D; a new family of antifungal agents. I. Fermentation, isolation, and biological activity. J. Antibiot. (Tokyo) 45, 861–867. ( 10.7164/antibiotics.45.861) [DOI] [PubMed] [Google Scholar]
- 61.Nesic K, Ivanovic S, Nesic V. 2014. Fusarium toxins: secondary metabolites of Fusarium fungi. Rev. Environ. Contam. Toxicol. 228, 101–120. ( 10.1007/978-3-319-01619-1_5) [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi S. 1998. Catalytic asymmetric syntheses of antifungal sphingofungins and their biological activity as potent inhibitors of serine palmitoyltransferase (SPT). J. Am. Chem. Soc. 120, 908–919. ( 10.1021/ja9730829) [DOI] [Google Scholar]
- 63.Throckmorton K, Wiemann P, Keller NP. 2015. Evolution of chemical diversity in a group of non-reduced polyketide gene clusters: using phylogenetics to inform the search for novel fungal natural products. Toxins (Basel) 7, 3572–3607. ( 10.3390/toxins7093572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mattern DJ, et al. 2015. Identification of the antiphagocytic trypacidin gene cluster in the human-pathogenic fungus Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 99, 10 151–10 161. ( 10.1007/s00253-015-6898-1) [DOI] [PubMed] [Google Scholar]
- 65.Balan J, Ebringer L, Nemec P, Kovac S, Dobias J. 1963. Antiprotozoal antibiotics. II. Isolation and characterization of Trypacidin, a new antibiotic, active against Trypanosoma cruzi and Toxoplasma gondii. J. Antibiot. (Tokyo) 16, 157–160. [PubMed] [Google Scholar]
- 66.Gauthier T, et al. 2012. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS ONE 7, e29906 ( 10.1371/journal.pone.0029906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ganaha M, et al. 2016. In vitro antitrypanosomal activity of the secondary metabolites from the mutant strain IU-3 of the insect pathogenic fungus Ophiocordyceps coccidiicola NBRC 100683. Chem. Pharm. Bull. 64, 988–990. ( 10.1248/cpb.c16-00220) [DOI] [PubMed] [Google Scholar]
- 68.Hittalmani S, Mahesh HB, Mahadevaiah C, Prasannakumar MK. 2016. De novo genome assembly and annotation of rice sheath rot fungus Sarocladium oryzae reveals genes involved in Helvolic acid and Cerulenin biosynthesis pathways. BMC Genomics 17, 271 ( 10.1186/s12864-016-2599-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singkaravanit S, Kinoshita H, Ihara F, Nihira T. 2010. Cloning and functional analysis of the second geranylgeranyl diphosphate synthase gene influencing helvolic acid biosynthesis in Metarhizium anisopliae. Appl. Microbiol. Biotechnol. 87, 1077–1088. ( 10.1007/s00253-010-2556-9) [DOI] [PubMed] [Google Scholar]
- 70.Amitani R, Taylor G, Elezis EN, Llewellyn-Jones C, Mitchell J, Kuze F, Cole PJ, Wilson R. 1995. Purification and characterization of factors produced by Aspergillus fumigatus which affect human ciliated respiratory epithelium. Infect. Immun. 63, 3266–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ratnaweera PB, Williams DE, de Silva ED, Wijesundera RLC, Dalisay DS, Andersen RJ. 2014. Helvolic acid, an antibacterial nortriterpenoid from a fungal endophyte, Xylaria sp. of orchid Anoectochilus setaceus endemic to Sri Lanka. Mycology 5, 23–28. ( 10.1080/21501203.2014.892905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.O'Hanlon KA, Cairns T, Stack T, Schrettl M, Bignell EM, Kavanagh K, Miggin SM, O'Keeffe G, Larsen TO, Doyle S. Targeted disruption of nonribosomal peptide synthetase pes3 augments the virulence of Aspergillus fumigatus. Infection and immunity 79, 3978–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gardiner DM, Cozijnsen AJ, Wilson LM, Pedras MSC, Howlett BJ. 2004. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 53, 1307–1318. ( 10.1111/j.1365-2958.2004.04215.x) [DOI] [PubMed] [Google Scholar]
- 74.Belofsky GN, Anguera M, Jensen PR, Fenical W, Köck M. 2000. Oxepinamides A-C and fumiquinazolines H–I: bioactive metabolites from a marine isolate of a fungus of the genus Acremonium. Chemistry 6, 1355–1360. ( 10.1002/(SICI)1521-3765(20000417)6:8%3C1355::AID-CHEM1355%3E3.0.CO;2-S) [DOI] [PubMed] [Google Scholar]
- 75.Garcia Silva M, Araçari Jacometti Cardoso Furtado N, Tallarico Pupo M, José Vieira Fonseca M, Said S, Alves Da Silva Filho A, Kenupp Bastos J. 2004. Antibacterial activity from Penicillium corylophilum Dierckx. Microbiol. Res. 159, 317–322. ( 10.1016/j.micres.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 76.Hu J, Furutani A, Yamamoto K, Oyama K, Mitomi M, Anzai H. 2014. Characterization of two acetyltransferase genes in the pyripyropene biosynthetic gene cluster from Penicillium coprobium. Biotechnol. Biotechnol. Equip. 28, 818–826. ( 10.1080/13102818.2014.960140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dieleman C, Knieriem T, Krapp M, Kierkus PC, Xu W, Benton K. 2013. Composition containing a pyripyropene insecticide and a base. Patent no. PCT/EP2011/065848.
- 78.Zhan Y, Zhang X-W, Xiong Y, Li B-L, Nan F-J. 2016. Design and synthesis of simple, yet potent and selective non-ring-A pyripyropene A-based inhibitors of acyl-coenzyme A: cholesterol acyltransferase 2 (ACAT2). Org. Biomol. Chem. 14, 747–751. ( 10.1039/C5OB02019K) [DOI] [PubMed] [Google Scholar]
- 79.Li XJ, Zhang Q, Zhang AL, Gao JM. 2012. Metabolites from Aspergillus fumigatus, an endophytic fungus associated with Melia azedarach, and their antifungal, antifeedant, and toxic activities. J. Agric. Food Chem. 60, 3424–3431. ( 10.1021/jf300146n) [DOI] [PubMed] [Google Scholar]
- 80.Ehlers T, Furness S, Robinson TP, Zhong HA, Goldsmith D, Aribser J, Bowen JP. 2016. Methionine aminopeptidase type-2 inhibitors targeting angiogenesis. Curr. Top. Med. Chem. 16, 1478–1488. ( 10.2174/1568026615666150915121204) [DOI] [PubMed] [Google Scholar]
- 81.van den Heever JP, Thompson TS, Curtis JM, Ibrahim A, Pernal SF. 2014. Fumagillin: an overview of recent scientific advances and their significance for apiculture. J. Agric. Food Chem. 62, 2728–2737. ( 10.1021/jf4055374) [DOI] [PubMed] [Google Scholar]
- 82.Ishikawa M, Ninomiya T, Akabane H, Kushida N, Tsujiuchi G, Ohyama M, Gomi S, Shito K, Murata T. 2009. Pseurotin A and its analogues as inhibitors of immunoglobuline E production. Bioorg. Med. Chem. Lett. 19, 1457–1460. ( 10.1016/j.bmcl.2009.01.029) [DOI] [PubMed] [Google Scholar]
- 83.Nicholson MJ, Koulman A, Monahan BJ, Pritchard BL, Payne GA, Scott B. 2009. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75, 7469–7481. ( 10.1128/AEM.02146-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meteyer CU, Barber D, Mandl JN. 2012. Pathology in euthermic bats with white nose syndrome suggests a natural manifestation of immune reconstitution inflammatory syndrome. Virulence 3, 583–588. ( 10.4161/viru.22330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rapin N, Johns K, Martin L, Warnecke L, Turner JM, Bollinger TK, Willis CKR, Voyles J, Misra V. 2014. Activation of innate immune-response genes in little brown bats (Myotis Lucifugus) infected with the fungus Pseudogymnoascus destructans. PLoS ONE 9, e112285 ( 10.1371/journal.pone.0112285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore MS, Reichard JD, Murtha TD, Nabhan ML, Pian RE, Ferreira JS, Kunz TH. 2013. Hibernating little brown myotis (Myotis lucifugus) show variable immunological responses to white-nose syndrome. PLoS ONE 8, e58976 ( 10.1371/journal.pone.0058976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McDonagh A, et al. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 4, e1000154 ( 10.1371/journal.ppat.1000154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Medema MH, Takano E, Breitling R. 2013. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223. ( 10.1093/molbev/mst025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saeed AI, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- 90.‘t Hoen PAC, Turk R, Boer JM, Sterrenburg E, de Menezes RX, van Ommen G-JB, den Dunnen JT. 2004. Intensity-based analysis of two-colour microarrays enables efficient and flexible hybridization designs. Nucleic Acids Res. 32, e41 ( 10.1093/nar/gnh038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bossers K, Ylstra B, Brakenhoff RH, Smeets SJ, Verhaagen J, van de Wiel MA. 2010. Intensity-based analysis of dual-color gene expression data as an alternative to ratio-based analysis to enhance reproducibility. BMC Genomics 11, 112 ( 10.1186/1471-2164-11-112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bolstad BM, Irizarry R, Strand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. ( 10.1093/bioinformatics/19.2.185) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and quantile normalized data have been uploaded to the Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ under the title ‘Secondary metabolite arsenal of an opportunistic pathogenic fungus’. Accession number pending. Fasta format sequence data used to perform MGB analysis have been uploaded as the electronic supplementary material, Data S1.