Abstract
Fungal pathogens severely impact global food and fibre crop security. Fungal species that cause plant diseases have mostly been recognized based on their morphology. In general, morphological descriptions remain disconnected from crucially important knowledge such as mating types, host specificity, life cycle stages and population structures. The majority of current fungal species descriptions lack even the most basic genetic data that could address at least some of these issues. Such information is essential for accurate fungal identifications, to link critical metadata and to understand the real and potential impact of fungal pathogens on production and natural ecosystems. Because international trade in plant products and introduction of pathogens to new areas is likely to continue, the manner in which fungal pathogens are identified should urgently be reconsidered. The technologies that would provide appropriate information for biosecurity and quarantine already exist, yet the scientific community and the regulatory authorities are slow to embrace them. International agreements are urgently needed to enforce new guidelines for describing plant pathogenic fungi (including key DNA information), to ensure availability of relevant data and to modernize the phytosanitary systems that must deal with the risks relating to trade-associated plant pathogens.
This article is part of the themed issue ‘Tackling emerging fungal threats to animal health, food security and ecosystem resilience’.
Keywords: cryptic diversity, DNA barcoding, fungal taxonomy
1. Introduction
Global travel and trade in food and fibre products have become a way of life and underpin the global economy. Current estimates of a world population of 9.1 billion people projected by 2050, changing diets and consumption patterns, and the increasing inability of some regions of the world to produce sufficient food for local consumption (www.fao.org) suggest that (i) intercontinental travel and trade of agricultural and forestry produce will increase and (ii) production of produce will need to become more efficient to meet ever increasing needs. Thus pre- and post-harvest losses, whatever the cause, will be increasingly intolerable.
Importing countries are vulnerable to accidental introductions of new and potentially devastating plant pathogenic fungi [1–3]. Quarantine systems, including trade restrictions from areas where pathogens occur, required treatment of goods and inspections for infected material are all intended to reduce this risk. Given the increase in travel, as well as the volume of trade around the world, including in live plants and fresh produce, the capacity to apply these systems is wholly inadequate, even in the most resourced countries such as the USA [4]. Furthermore, the rate at which goods are being moved around the world is also increasing. For example, bananas grown in South America are served in European households within days of harvest and this is true for many other plant products globally. This implies that the window of opportunity to intercept, identify and act on a potential new invasion is minute at best.
Currently, applied quarantine systems are complicated by the fact that most well-known fungal plant pathogenic species are primarily known from a specific suite of disease symptoms and general morphology based on only a part of their life cycle [5]. Many pathogens remain undetected as latent infections in apparently healthy tissue and they are unlikely to be detected in routine inspections [6,7]. Furthermore, many of the fungi that cause major problems remain unknown or unnamed until well after they begin to cause major losses and will neither be sought nor detected in quarantine inspections. Once they are studied more intensively, especially where modern molecular tools are applied, many of the fungi that are detected are often found to represent species complexes that would earlier have passed unnoticed [8]. Pathogen detection that relies on visual plant symptoms and pathogen morphology is quite evidently unable to effectively cope with the threats posed by fungi found in traded plants or plant products.
Quarantine systems have traditionally relied on fungal names. These names are increasingly being shown as simplistic and ineffectual as representatives of the relevant information associated with a particular pathogen. Beyond the species level, knowledge relating to mating types, and even clones of particular pathogens, is crucially important when seeking to understand or manage fungal invasions [9,10]. The same is true for the variable presence or absence of small dispensable chromosomes carrying genes involved in pathogenicity, which can influence the ability of a species to infect a specific host, e.g. in Alternaria alternata [11] and Fusarium oxysporum [12]. To further complicate matters, quarantine lists with names as actionable organisms are often found only in inaccessible national databases or government publications. These are frequently not linked to relevant data; neither are they consistent with modern taxonomic treatments of the species in question.
Although it is unknown how many species of fungi occur on the Earth, and estimates range anywhere from 1.5 to several million [13–15], it is reasonable to conclude that the majority of species have not yet been seen or recorded [16,17]. Communication relating to these species by means of fungal names that remain largely linked to the phenotype, and detached from the genotypic, ecological and other data [16,18], is clearly insufficiently informative to deal with risks associated with increasing volumes and rates of trade in food and fibre. This approach also poses a serious threat to the global security of plant production and the environment alike. In this review, we consider several of these issues and approaches that could help to traverse seemingly unnecessary barriers to efficient identification procedures and management of fungal threats linked to global trade in agricultural and forestry products.
2. One fungus, but which name?
Scientific names remain the foundation of how we communicate regarding species of phytopathogenic fungi; also with regards to quarantine. Names are ideally linked to knowledge of the biology, distribution, ecology, host range, control and risks associated with fungal pathogens. The concept of pleomorphism relates to the fact that many ascomycetous fungi are known by either their sexual, asexual or synasexual morphs, to which different names have been attributed based on their morphology; commonly referred to as dual nomenclature [11,19,20].
In practice, this dual nomenclature has meant that a single fungus could be listed on the quarantine list of a country under any one of three valid names (e.g. apple scab caused by Venturia inaequalis, asexual morph Fusicladium pomi or synasexual morph Spilocaea pomi [21]). This is in a conservative scenario. Where known synonyms are considered, the list of names can be disturbingly long; all of which confuse the literature and they commonly persist in national quarantine lists. Other than having different names to contend with, quarantine officers are also faced with the difficult reality that many reported asexual–sexual relationships have never actually been experimentally confirmed and can also be incorrect.
Following the ‘One Fungus: One Name’ symposium, and the publication of the ‘Amsterdam Declaration on Fungal Nomenclature’, several radical changes were proposed to the code of nomenclature that governs the naming of fungi [19]. This subsequently led to the amendment of the International Code of Nomenclature for algae, fungi and plants (ICN) (Article 59) to abolish the use of dual nomenclature, as well as other sensible changes including registration of nomenclatural details of fungal novelties in databases such as MycoBank [22], the acceptance of electronic publication, and English (as alternative to Latin) descriptions of new names [19,20].
Moving to the application of single names for plant pathogens was strongly supported by the plant pathology community [5], which needed to have meaningful names for species associated with important plant diseases. The concomitant changes to the ICN code, together with the increased understanding of systematic relationships among fungi based on more representative DNA sequence-based phylogenies, have resulted in a large number of taxonomic revisions in recent years. While these name changes might have caused confusion for plant health and quarantine practitioners in the short term, the more accurate application of generic names based on DNA-based data will ensure longer-term stability in the use of names. Appropriate names of fungal pathogens are also important for fundamental plant pathology research, such as various ‘omics’ approaches aimed at understanding the mechanisms of plant–pathogen interactions through comparisons among related species. Past taxonomic treatments lead to confusion in this regard; for example, genome comparisons of ‘Mycosphaerella’ that were later shown to represent members of different genera, e.g. Zymoseptoria tritici [23,24], Pseudocercospora fijiensis [25,26] and Dothistroma septosporum [27,28].
Unfortunately, many genera and species remain to be revised or they are devoid of critical data that would allow for accurate identification and phylogenetic placement. Between 2000 and 2013, 1833 fungal genera were described for which only 155 (8.4%) have type specimens linked to reliably annotated ITS nrDNA sequence data in public databases [29]. This implies that the number of newly described fungi lacking DNA data continues to increase rather than decrease. In an attempt to alleviate this problem, ‘The Genera of Fungi’ project was launched, with the aim of sequencing, restudying and/or recollecting the type species of genera of fungi, focusing on a subset of names that are currently accepted [29,30]. Furthermore, to help plant pathologists to know which generic names they should apply to pleomorphic genera, committees under the auspices of the International Commission for the Taxonomy of Fungi have been tasked with preparing lists of accepted names recommended for use (e.g. [31,32]). These names will be evaluated by the Nomenclature Committee for Fungi, and formally accepted or not at the Nomenclature Session of the 2017 International Botanical Congress to be held in Shenzhen, Southern China.
Another major constraint to appropriate naming is the general lack of appropriate, well-characterized reference specimens and/or cultures of quarantine and related species in publicly accessible collections (see review by [33]). Such biological resource centres are also under constant threat from decreasing budgets and increasing costs. These invaluable reference specimens and cultures are critically important for the establishment of reliable identification systems. Their loss would represent a huge impediment for future generations of mycologists, plant pathologists and other end users.
The above-mentioned efforts to clarify the names of fungal pathogens and link their phenotypes to genotypic information are critical to ensure a useful framework for efficient identification and communication of fungi. This is also essential to ensure the continued discovery and characterization of the millions of fungi that are estimated to remain unnamed. It is, therefore, important that plant pathologists take note of these efforts and support them with urgency.
3. Cryptic species, mating types and clones
A key question in considering global biosecurity is when to stop considering a pathogen as an ‘actionable organism’, e.g. when is it accepted as established in a country? It is important here to recognize that a species is not ‘one dimensional’ as is suggested by a name on a list. By contrast, it represents a complex or pool of different ‘sexes’ or mating types, virulence factors and genes. All these influence its response to hosts, environments and a wide range of other factors. Furthermore, many pathogens represent cryptic species that pose a particular problem with regard to understanding invasion and potential quarantine procedures (e.g. [8,34,35]).
Fungi can reproduce either asexually or sexually. In the latter case, they typically have two or more sexual mating types that are needed for sexual recombination to occur [36,37]. Understanding these cycles is critical for disease management. This is because it significantly affects the ability of fungal pathogens to overcome resistance mechanisms of their hosts; with sexually reproducing strains having an ecological advantage to infect and invade [9]. Mating types should, therefore, have significant relevance for the status of quarantine organisms. For instance, D. septosporum, the causal agent of the devastating pine disease Red Band Needle Blight, has been introduced into many countries. By generating the mating-type primers for this pathogen, Groenewald et al. [38] were able to show that although the species was introduced into South Africa, Australia and New Zealand, both mating types were present in South Africa (i.e. sexual recombination possible), but that only a single mating type can be found in Australia and New Zealand (i.e. sexual recombination not possible). It is clearly not only the species, but also the mating types that are of quarantine concern.
Specific and even clonal lineages in a pathogen population have relevance to quarantine. This is well illustrated in bananas, which represent one of the important global staple food crops, having evolved in the Indo-Malayan archipelago. Panama disease, which is caused by F. oxysporum f.sp. cubense (Foc), appears also to have originated in Southeast Asia [39]. Based on molecular studies, it appears that Foc is a haploid asexual pathogen with a clonal population structure, and that temporal and spatial dispersal of devastating disease linked to Tropical Race 4 is actually due to a single clone [39]. If additional clones were thus to move from Southeast Asia, the disease would become even more difficult to manage because a broader range of cultivars are likely to be affected. A further complicating factor is that species in the F. oxysporum complex also undergo horizontal gene and chromosome transfer as a means for lineages to broaden their host range, and this can influence their pathogenicity [40].
In some genera of phytopathogenic fungi, names mask variation in host specificity and pathogenicity that is present below the species level. Although such variation in plant pathogenic fungi is often found to represent several cryptic species, the opposite situation also occurs. One case in point is the A. alternata species complex, to which the quarantine species A. mali, causal agent of Alternaria blotch of apple, belongs. Woudenberg et al. [11] employed whole-genome and multi-gene analysis to reduce 35 Alternaria morpho-species to synonymy under the older name, A. alternata. The authors concluded that it is the presence or absence of the gene cluster that codes for a specific toxin that is of quarantine concern, and not necessarily a specific synonym of A. alternata.
McTaggart et al. [10] recently called for ‘gene-based biosecurity’. They point out that our knowledge of genes that underlie complex traits such as pathogenicity is growing exponentially. Prediction of lifestyle (e.g. biotrophic versus saprotrophic), the presence of pathogenicity factors and other elements in the genome are known to be linked to pathogenicity. It will still be some time before such information is known for a sufficient number of fungi to rely solely on genome scans. But it is relevant to consider the fact that such an approach would allow much more predictive and preventative action than any name-based biosecurity system. Until such information is available for all actionable quarantine organisms, a more detailed identification system is urgently needed and in many cases it is already feasible.
4. Latent or endophytic fungal infections
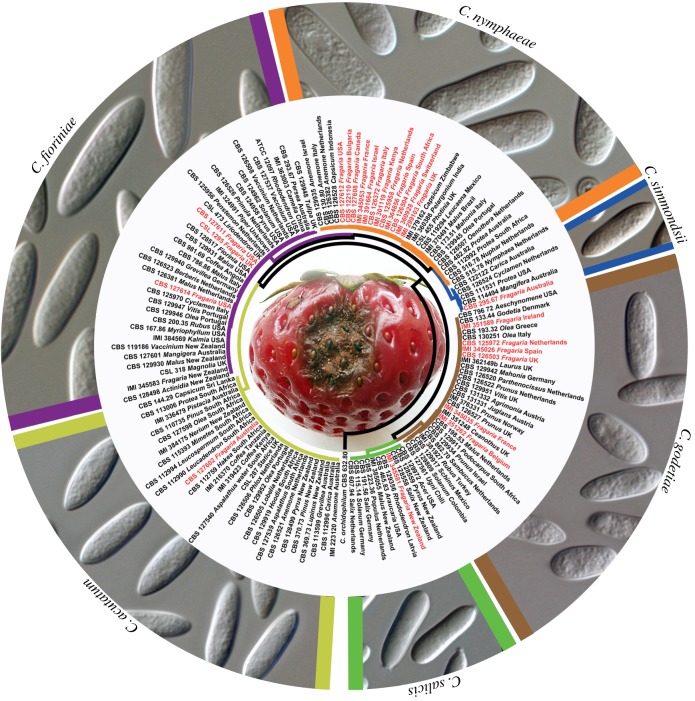
Apart from systematic problems to identify fungal threats in traded plants and plant products, biosecurity is also currently unable to deal with the cryptic nature of fungal infections. There are a great many plant pathogenic fungi that cause latent infections. These represent a particularly difficult challenge for international trade and associated quarantine measures. Latent infections involve a parasitic relationship between a pathogen and a host that might remain asymptomatic for some period of time, even years, but that eventually induces disease symptoms [41]. In this situation, a pathogen remains latent until environmental or nutritional conditions or the stage of maturity of the host or pathogen allow it to produce symptoms of disease [42]. A few pertinent case studies are provided in figures 1–3.
Figure 1.
Parsimony phylogeny depicting the host range and geographical distribution of strawberry-associated species belonging to the Colletotrichum acutatum species complex. Strains from Fragaria are indicated in red text and the branches and micromorphology photos of the different fungal species are colour coded. The alignment is based on a subset of the six-locus alignment of Damm et al. [43], see TreeBASE (study number 12762). Anthracnose disease of strawberry is a particularly serious problem for commercial fruit production [44], which resulted in C. acutatum being listed as a regulated plant quarantine pest by the European and Mediterranean Plant Protection Organization (EPPO) and the EU Council Directive 2000/29 Annexes I and II, from which it was removed in 2009. Sources of inoculum include infected plants, weeds and other hosts [45,46], while the pathogen is also well known to survive via latent infections on strawberries [47]. To further complicate matters, Damm et al. [43] recently separated the morpho-species C. acutatum into 31 taxa, of which 21 were shown to represent novel species. Under these circumstances, it is difficult to imagine how quarantine can be applied through attempts at visual inspection for symptoms or morphological identification of species.
Figure 2.
Species of Botryosphaeriaceae associated with cankers on (a–d) Acacia, Protea, Eucalyptus and Pinus. Diseases caused by Botryosphaeriaceae mostly follow the onset of stress [7]. Many species of Botryosphaeriaceae are known to exist via localized, latent infections in their hosts, which appears to be a common characteristic of this group [7,48]. Once introduced into a new area as latent infection or endophyte on one host, they can easily move to other hosts where these otherwise ‘innocent’ endophytes become serious pathogens. Some species of Botryosphaeriaceae can remain latent for many years as localized infections deep inside woody or other tissues. Diplodia sapinea, for example, is a common global pathogen of Pinus spp., but is also well known to exist as latent infections in wood of stems, branches, twigs, seed cones and (to a limited extent) seed [49–52]. It has evidently been introduced with its host multiple times around the world, which illustrates the extent to which quarantine systems have failed to halt the movement of such latent infections [49,52].
Figure 3.
Foliar diseases caused by (a,b) Pseudocercospora eumusae and P. fijiensis on Musa, (c) Dothistroma septosporum on Pinus and (d) Pseudocercospora angolensis on Citrus. The Mycosphaerellaceae comprises one of the largest families in the Phylum Ascomycota, in which some species have evolved as latent pathogens, saprophytes or symbionts. For example, D. septosporum, the causative agent of Red Band Needle Blight disease, is an important pathogen of Pinus spp. [27], which has also been isolated from asymptomatic pine needles. Species of Pseudocercospora are commonly associated with leaf spots, with some taxa such as P. angolensis on Citrus [53], and P. fijiensis, P. musicola and P. eumusae on Musa [26] being of major quarantine concern. Other than these examples, a great number of species from diverse genera in the Mycosphaerellaceae are commonly isolated as latent pathogens, occurring on a wide range of asymptomatic host plants [25].
The key issue illustrated by the three case studies (and there are many others that could be used) is that many genera include important plant pathogens that have a latent phase in their life cycles. This easily leads to unwanted introductions, further complicated by the fact that these pathogens frequently also have wide host ranges and thus spread throughout local plant communities. The only way to overcome this problem is through the application of molecular based detection. DNA barcoding technologies and data sharing abilities for such an approach already exist [54,55], but are not used widely yet for quarantine purposes. For this goal to be realized, much work is required to firstly provide a solid taxonomic framework (as discussed above). And there will be a need for human capacity development within quarantine structures to utilize this information.
5. Conclusion
Global trade in plant products faces major challenges related to fungal pathogens that threaten food and fibre security, as well as ecosystem health. Unfortunately, these challenges are exacerbated by inefficiencies in the systematic and physical identification of fungi, which is due to the reliance on outdated taxonomic information and systems, as well as our inability to recognize the cryptic fungal infections. Given the enormity of the risks, it is unfortunate that there is a general lack of global urgency to incorporate already existing tools to deal with them. These tools would make it possible to implement a barcoding-based information and identification system to screen plants and plant products that are traded internationally.
One of the major issues that hamper progress towards an effective DNA-based barcoding system for biosecurity is the present ICN, which governs the naming of fungi, and essentially allows plant pathogenic fungal species to be described without DNA data. This leaves researchers and practitioners trying to play ‘catch-up’ at huge additional cost, having to recollect isolates to provide molecular data for previously described plant pathogens. A potential remedy would be for the International Commission for the Taxonomy of Fungi to implement a set of guidelines that authors, editors and reviewers could follow to ensure that, wherever possible, relevant genotypic data are provided to supplement novel species descriptions of suspected or known plant pathogens. The current absence of such guidelines hampers both progress and the application of broadly accepted best practices in fungal identification and description. This is not only to the detriment of mycology, but also of global food and fibre production and ecosystem health.
A major constraint to effective plant quarantine is the poor linkage between resources that carry layers of information regarding plant pathogens. Unfortunately, there is a general lack of support to maintain and link databases such as Q-bank (http://www.q-bank.eu/), MycoBank (http://www.mycobank.org/), Index Fungorum (http://www.indexfungorum.org), UNITE (https://unite.ut.ee/), GenBank (http://www.ncbi.nlm.nih.gov/) and the ARS-USDA fungus–host distribution database (http://nt.ars-grin.gov/fungaldatabases/fungushost/fungushost.cfm), to name but a few. Each of these databases includes unique information about species, their identification, strains, hosts and much more. Linking them, and supporting their expansion, appears to be ‘low-hanging fruit’ from a global quarantine and plant health management perspective. Doing so would immediately unlock large volumes of data for important pathogens globally. Only specialists who understand the intricacies of navigating this maze of data resources can currently access much of this information.
As handy as DNA barcodes can be as tools for species recognition, the real value of these data collections will emerge once the fungal genomes have been analysed and linked to function, e.g. using secondary metabolites to infer ecology, the identification of pathogenicity factors, transposable elements, as well as life cycle and population structure [10,56]. There is a growing realization that not only future biological studies but also future quarantine and management systems will be reliant on this information. If we are serious about reducing the impact of fungal pathogens on trade in food and fibre, a fundamental change in how we operate will be required. Names, morphology and visual inspection for fungal pathogens are simply not sufficient to deal with the problem.
Competing interests
We declare we have no competing interests.
Funding
We received no funding for this study.
References
- 1.Desprez-Loustau M-L, Robin C, Buée M, Courtecuisse R, Garbaye J, Suffert F, Sache I, Rizzo DM. 2007. The fungal dimension of biological invasions. Trends Ecol. Evol. 22, 472–480. ( 10.1016/j.tree.2007.04.005) [DOI] [PubMed] [Google Scholar]
- 2.Hantula J, Müller MM, Uusivuori J. 2014. International plant trade associated risks: laissez-faire or novel solutions. Environ. Sci. Policy 37, 158–160. ( 10.1016/j.envsci.2013.09.011) [DOI] [Google Scholar]
- 3.Wingfield MJ, Brockerhoff EG, Wingfield BD, Slippers B. 2015. Planted forest health: the need for a global strategy. Science 349, 832–836. ( 10.1126/science.aac6674) [DOI] [PubMed] [Google Scholar]
- 4.Liebhold AM, Brockerhoff EG, Garrett LJ, Parke JL, Britton KO. 2012. Live plant imports: the major pathway for forest insect and pathogen invasions of the US. Front. Ecol. Environ. 10, 135–143. ( 10.1890/110198) [DOI] [Google Scholar]
- 5.Wingfield MJ, de Beer ZW, Slippers B, Wingfield BD, Groenewald JZ, Lombard L, Crous PW. 2012. One fungus, one name promotes progressive plant pathology. Mol. Plant Pathol. 13, 604–613. ( 10.1111/j.1364-3703.2011.00768.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer C-L, Skinner W. 2002. Mycosphaerella graminicola: latent infection, crop devastation and genomics. Mol. Plant Pathol. 3, 63–70. ( 10.1046/j.1464-6722.2002.00100.x) [DOI] [PubMed] [Google Scholar]
- 7.Slippers B, Wingfield MJ. 2007. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol. Rev. 21, 90–106. ( 10.1016/j.fbr.2007.06.002) [DOI] [Google Scholar]
- 8.Crous PW, Groenewald JZ. 2005. Hosts, species and genotypes: opinions versus data. Australas. Plant Pathol. 34, 463–470. ( 10.1071/AP05082) [DOI] [Google Scholar]
- 9.McDonald BA, Linde C. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Ann. Rev. Phytopathol. 40, 349–379. ( 10.1146/annurev.phyto.40.120501.101443) [DOI] [PubMed] [Google Scholar]
- 10.McTaggart AR, van der Nest MA, Steenkamp ET, Roux J, Slippers B, Shuey LS, Wingfield MJ, Drenth A. 2016. Fungal genomics challenges the dogma of name-based biosecurity. PLoS Pathog. 12, e1005475 ( 10.1371/journal.ppat.1005475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woudenberg JHC, Seidl MF, Groenewald JZ, de Vries M, Stielow JB, Thomma BPHJ, Crous PW. 2015. Alternaria section Alternaria: species, formae speciales or pathotypes? Stud. Mycol. 82, 1–21. ( 10.1016/j.simyco.2015.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonkers W, et al. 2014. EBR1 genomic expansion and its role in virulence of Fusarium species. Environ. Microbiol. 16, 1982–2003. ( 10.1111/1462-2920.12331) [DOI] [PubMed] [Google Scholar]
- 13.Hawksworth DL, Rossman AY. 1997. Where are all the undescribed fungi? Phytopathology 87, 888–891. ( 10.1094/PHYTO.1997.87.9.888) [DOI] [PubMed] [Google Scholar]
- 14.Blackwell M. 2011. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98, 426–438. ( 10.3732/ajb.1000298) [DOI] [PubMed] [Google Scholar]
- 15.Crous PW, Hawksworth DL, Wingfield MJ. 2015. Identifying and naming plant-pathogenic fungi: past, present, and future. Annu. Rev. Phytopathol. 53, 247–267. ( 10.1146/annurev-phyto-080614-120245) [DOI] [PubMed] [Google Scholar]
- 16.Hibbett D. 2016. The invisible dimension of fungal diversity. Science 351, 1150–1151. ( 10.1126/science.aae0380) [DOI] [PubMed] [Google Scholar]
- 17.Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proc. Natl Acad. Sci. USA 113, 5970–5975. ( 10.1073/pnas.1521291113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibbett DS, Taylor JW. 2013. Fungal systematics: is a new age of enlightenment at hand? Nat. Rev. Microbiol. 11, 129–133. ( 10.1038/nrmicro2963) [DOI] [PubMed] [Google Scholar]
- 19.Hawksworth DL. 2011. A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys 1, 7–20. ( 10.3897/mycokeys.1.2062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor JW. 2011. One fungus=one name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2, 113–120. ( 10.5598/imafungus.2011.02.02.01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schubert K, Ritschel A, Braun U. 2003. A monograph of Fusicladium s. lat. (hyphomycetes). Schlechtendalia 9, 1–132. [Google Scholar]
- 22.Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22. [Google Scholar]
- 23.Quaedvlieg W, Kema GHJ, Groenewald JZ, Verkley GJ, Seifbarghi S, Razavi M, Mirzadi Gohari A, Mehrabi R, Crous PW. 2011. Zymoseptoria gen. nov.: a new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia 26, 57–69. ( 10.3767/003158511X571841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stukenbrock EH, Quaedvlieg W, Javan-Nikhah M, Zala M, Crous PW, McDonald BA. 2012. Zymoseptoria ardabiliae and Z. pseudotritici, two progenitor species of the septoria tritici leaf blotch fungus Z. tritici (synonym: Mycosphaerella graminicola). Mycologia 104, 1397–1407. ( 10.3852/11-374) [DOI] [PubMed] [Google Scholar]
- 25.Crous PW, Braun U, Hunter GC, Wingfield MJ, Verkley GJM, Shin H-D, Nakashima C, Groenewald JZ. 2013. Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 75, 37–114. ( 10.3114/sim0005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang TC, Salvucci A, Crous PW, Stergiopoulos I. 2016. Comparative genomics of the Sigatoka disease complex on banana suggests a link between parallel evolutionary changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and increased virulence on the banana host. PLoS Genet. 12: e1005904 ( 10.1371/journal.pgen.1005904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnes I, Van der Nest A, Mullett MS, Crous PW, Drenkhan R, Musolin DL, Wingfield MJ. 2016. Neotypification of Dothistroma septosporum and epitypification of D. pini, causal agents of Dothistroma needle blight of pine. For. Pathol. ( 10.1111/efp.12304) [DOI] [Google Scholar]
- 28.de Wit PJ, et al. 2012. The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8, e1003088 ( 10.1371/journal.pgen.1003088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crous PW, et al. 2014. The genera of fungi: fixing the application of type species of generic names. IMA Fungus 5, 141–160. ( 10.5598/imafungus.2014.05.01.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirk PM, et al. 2013. A without-prejudice list of generic names of fungi for protection under the International Code of Nomenclature for algae, fungi, and plants. IMA Fungus 4, 381–443. ( 10.5598/imafungus.2013.04.02.17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossman AY, et al. 2015. Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 6, 507–523. ( 10.5598/imafungus.2015.06.02.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Réblová M, et al. 2016. Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7, 131–153. ( 10.5598/imafungus.2016.07.01.08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stackebrandt E, Smith D, Casaregola S, Varese GC, Verkleij G, Lima N, Bridge P. 2014. Deposit of microbial strains in public service collections as part of the publication process to underpin good practice in science. SpringerPlus 3, 208 ( 10.1186/2193-1801-3-208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez G, Slippers B, Wingfield MJ, Wingfield BD, Carnegie AJ, Burgess TI. 2012. Cryptic species, native populations and biological invasions by a eucalypt forest pathogen. Mol. Ecol. 21, 4452–4471. ( 10.1111/j.1365-294X.2012.05714.x) [DOI] [PubMed] [Google Scholar]
- 35.Sakalidis ML, Slippers B, Wingfield BD, Hardy GEStJ, Burgess TI. 2013. The challenge of understanding the origin, pathways and extent of fungal invasions: global populations of the Neofusicoccum parvum-N. ribis species complex. Divers. Distrib. 19, 873–883. ( 10.1111/ddi.12030) [DOI] [Google Scholar]
- 36.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. 2011. Sex in fungi. Annu. Rev. Genet. 45, 405–430. ( 10.1146/annurev-genet-110410-132536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor JW, Hann-Soden C, Branco S, Sylvain I, Ellison CE. 2015. Clonal reproduction in fungi. Proc. Natl Acad. Sci. USA 112, 8901–8908. ( 10.1073/pnas.1503159112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groenewald M, et al. 2007. Characterization and distribution of mating type genes in the Dothistroma needle blight pathogens. Phytopathology 97, 825–834. ( 10.1094/PHYTO-97-7-0825) [DOI] [PubMed] [Google Scholar]
- 39.Ordonez N, Seidl MF, Waalwijk C, Drenth A, Kilian A, Thomma BP, Ploetz RC, Kema GH. 2015. Worse comes to worst: bananas and panama disease—when plant and pathogen clones meet. PLoS Pathog. 11, e1005197 ( 10.1371/journal.ppat.1005197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehrabi R, et al. 2011. Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiol. Rev. 35, 542–554. ( 10.1111/j.1574-6976.2010.00263.x) [DOI] [PubMed] [Google Scholar]
- 41.Verhoeff K. 1974. Latent infections by fungi. Annu. Rev. Phytopathol. 12, 99–110. ( 10.1146/annurev.py.12.090174.000531) [DOI] [Google Scholar]
- 42.Agrios GN. 2005. Plant pathology, 5th edn New York, NY: Academic Press. [Google Scholar]
- 43.Damm U, Cannon PF, Woudenberg JHC, Crous PW. 2012. The Colletotrichum acutatum species complex. Stud. Mycol. 73, 37–113. ( 10.3114/sim0010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freeman S, Katan T. 1997. Identification of Colletotrichum species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathology 87, 516–521. ( 10.1094/PHYTO.1997.87.5.516) [DOI] [PubMed] [Google Scholar]
- 45.McInnes TB, Black LL, Gatti JM. 1992. Disease-free plants for management of strawberry anthracnose crown rot. Plant Pathol. 76, 260–264. ( 10.1094/pd-76-0260) [DOI] [Google Scholar]
- 46.Parikka P, Pääskynkivi E, Lemmetty A. 2006. Survival of Colletotrichum acutatum in dead plant material and soil in Finland. Acta Hortic. 708, 131–134. ( 10.17660/ActaHortic.2006.708.20) [DOI] [Google Scholar]
- 47.Parikka P, Lemmetty A. 2004. Tracing latent infection of Colletotrichum acutatum on strawberry by PCR. Eur. J. Plant Pathol. 110, 393–398. ( 10.1023/B:EJPP.0000021073.67137.d2) [DOI] [Google Scholar]
- 48.Slippers B, Crous PW, Denman S, Coutinho TA, Wingfield BD, Wingfield MJ. 2004. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96, 83–101. ( 10.2307/3761991) [DOI] [PubMed] [Google Scholar]
- 49.Burgess T, Wingfield BD, Wingfield MJ. 2001. Comparison of genotypic diversity in native and introduced populations of Sphaeropsis sapinea isolated from Pinus radiata. Mycol. Res. 105, 1331–1339. ( 10.1017/S0953756201005056) [DOI] [Google Scholar]
- 50.Smith H, Wingfield MJ, Crous PW, Coutinho TA. 1996. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. S. Afr. J. Bot. 62, 86–88. ( 10.1016/S0254-6299(15)30596-2) [DOI] [Google Scholar]
- 51.Smith H, Wingfield MJ, de Wet J, Coutinho TA. 2000. Genotypic diversity of Sphaeropsis sapinea from South Africa and Northern Sumatra. Plant Dis. 84, 139–142. ( 10.1094/PDIS.2000.84.2.139) [DOI] [PubMed] [Google Scholar]
- 52.Bihon W, Slippers B, Burgess T, Wingfield MJ, Wingfield BD. 2011. Sources of Diplodia pinea endophytic infections in Pinus patula and P. radiata seedlings in South Africa. For. Pathol. 41, 370–375. ( 10.1111/j.1439-0329.2010.00691.x) [DOI] [Google Scholar]
- 53.Quaedvlieg W, Groenewald JZ, de Jesús Yáñez-Morales M, Crous PW. 2012. DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia 29, 101–115. ( 10.3767/003158512X661282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schoch CL, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl Acad. Sci. USA 109, 6241–6246. ( 10.1073/pnas.1117018109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao R, Zhang G. 2013. Potential of DNA barcoding for detecting quarantine fungi. Phytopathology 103, 1103–1107. ( 10.1094/PHYTO-12-12-0321-R) [DOI] [PubMed] [Google Scholar]
- 56.Orton ES, Deller S, Brown JKM. 2011. Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 12, 413–424. ( 10.1111/j.1364-3703.2010.00688.x) [DOI] [PMC free article] [PubMed] [Google Scholar]