Abstract
Agricultural ecosystems are composed of genetically depauperate populations of crop plants grown at a high density and over large spatial scales, with the regional composition of crop species changing little from year to year. These environments are highly conducive for the emergence and dissemination of pathogens. The uniform host populations facilitate the specialization of pathogens to particular crop cultivars and allow the build-up of large population sizes. Population genetic and genomic studies have shed light on the evolutionary mechanisms underlying speciation processes, adaptive evolution and long-distance dispersal of highly damaging pathogens in agro-ecosystems. These studies document the speed with which pathogens evolve to overcome crop resistance genes and pesticides. They also show that crop pathogens can be disseminated very quickly across and among continents through human activities. In this review, we discuss how the peculiar architecture of agro-ecosystems facilitates pathogen emergence, evolution and dispersal. We present four example pathosystems that illustrate both pathogen specialization and pathogen speciation, including different time frames for emergence and different mechanisms underlying the emergence process. Lastly, we argue for a re-design of agro-ecosystems that embraces the concept of dynamic diversity to improve their resilience to pathogens. This article is part of the themed issue ‘Tackling emerging fungal threats to animal health, food security and ecosystem resilience’.
Keywords: fungal pathogens, pathogen emergence, pathogen evolution, agricultural ecosystems, dynamic diversity, population genetics
1. Introduction
Agricultural ecosystems (agro-ecosystems) were created by humans during the invention of agriculture approximately 12 000 years ago to generate a reliable food supply that could be easily stored (e.g. cereal grains), enabling the creation of cities and the rise of civilizations. Since their origin in the Fertile Crescent, agro-ecosystems have spread globally to cover approximately 40% of the land surface, with most of this expansion occurring during the past 300 years to feed the rapidly growing human population [1]. One consequence of this expansion was an enormous reduction in plant and animal diversity globally as species-rich natural ecosystems were converted to species-poor agro-ecosystems [2,3]. To illustrate the temporal and spatial scales involved, in only 300 years agro-ecosystems expanded such that approximately 540 million ha are now planted annually to only three crop species, maize, rice and wheat (FAO Statistics), compared with the approximately 600 million ha of tropical rainforests that emerged over millions of years.
The overwhelming majority of agro-ecosystems are planted to monocultures, which for thousands of years were composed of locally adapted but genetically diverse land races [4]. During the past 100 years, and especially since the green revolution that started approximately 50 years ago [5], these land races were largely replaced by genetically uniform but broadly adapted and high-yielding dwarf cultivars, giving rise to fields typically composed of a single clone or a population of near clones that carry very little genetic diversity [4]. Another consequence of the expansion of agro-ecosystems has been a great reduction in environmental heterogeneity. Over millennia, farmers invented technologies to make their planted fields more environmentally homogeneous to favour their crops and enable mechanization, including soil tillage, controlled irrigation and applications of fertilizers and pesticides. During the past 100 years, the large-scale mechanization of planting and harvesting operations further increased the environmental homogeneity present in agro-ecosystems worldwide [2], so that a field planted to modern wheat is now remarkably similar across Asia, the Americas and Australia. By way of comparison, the tropical rainforest of South America is completely different in species composition, genetic make-up and environmental heterogeneity compared with the tropical rainforests in Africa or Asia.
The homogeneous genetic and physical environments of agro-ecosystems created a selective environment that differs greatly from that found in natural ecosystems, with the agro-ecosystem favouring the emergence of new, host-specialized, ‘domesticated’ crop pathogens that evolve more rapidly and are more virulent compared to their ‘wild’ ancestors. The increased planting density and genetic uniformity of host populations in agro-ecosystems compared with natural ecosystems makes pathogen transmission easier, enabling an increase in pathogen virulence in the agro-ecosystem [6]. These factors also increase pathogen effective population size, providing more pathogen genetic diversity by increasing the total number of mutations available in agro-ecosystems while lowering the effects of genetic drift [7]. The higher host density in agro-ecosystems allows crop pathogens to exist at a higher density, also increasing the likelihood of multi-infections in the same plant by different genotypes (strains) of the same pathogen [8–10]. Multi-infections also favour the development of higher virulence as a result of competition among strains for the same host resources [11]. The higher host and pathogen density found in agro-ecosystems also increases the likelihood of co-infection by different pathogen species, which also selects for higher virulence as a result of competition among pathogen species for the same host resources [12]. Co-infection by different pathogen species also increases the likelihood of horizontal gene transfer, as already described for several crop pathogens [13–15] that acquired genes affecting virulence, in some cases from other pathogens infecting the same host [13]. Finally, the global trade in agricultural products accelerated the movement of crop pathogens around the world and led to the rapid global spread of novel, highly damaging pathogens (e.g. Phytophthora infestans causing potato late blight [16,17]), as illustrated very recently by the introduction of the wheat blast pathogen Pyricularia graminis-tritici into Asia [18]. This global transport of pathogens created cosmopolitan, globally disseminated pathogens as well as meta-populations composed of locally adapted pathogen populations.
The first green revolution enabled the continuing growth of the human population during the past 60 years but was based on a narrowed base of crop germplasm encoding improved traits coupled with higher inputs of fertilizers and pesticides and increased mechanization [4]. The current human population is now dependent on this strategy for food production and food security. The forecasted growth of the human population will require an additional 60% increase in staple food crop production by 2050 [19]. Until now, increased demand for food was met mostly by converting natural ecosystems into agro-ecosystems, a strategy referred to as ‘extensification’ of agriculture [3]. Introduction of existing crops into natural ecosystems provided another mechanism for pathogen emergence through host jumps [20]. Continuing the extensification strategy to increase food production will require converting an additional billion hectares to agro-ecosystems by 2050 [3]. To avoid further loss of natural ecosystems and the accompanying losses in natural habitats and species associated with extensification, it will be necessary to increase food production from the existing agro-ecosystems, a strategy called ‘intensification’ [3]. Given that approximately 15% of all current crop production is lost to disease [21], diminishing these losses provides an obvious strategy to increase food production without further degrading natural ecosystems. But protecting crops from pathogens is likely to require a significant re-engineering of agro-ecosystems globally to make them more resilient to disease and less favourable to the emergence of new pathogens or rapid evolution of more damaging traits (e.g. higher virulence and fungicide resistance) in existing pathogens. Possible approaches to re-engineering of agro-ecosystems to lessen the disease burden will be discussed below.
The development of sustainable plant protection strategies requires insight into the biology and evolution of the corresponding pathogens. In particular, we would like to know how new pathogens emerge and evolve in agro-ecosystems. Central questions to be addressed include: from where do new pathogens emerge (what is their centre of origin)? How do new pathogens emerge (what has driven the process of speciation or emergence)? How is the new pathogen dispersed and what were its geographical migration routes (how was the pathogen dispersed over time and space)? Does the new pathogen exchange genetic material with other pathogen species on nearby host plants, which could become sources of new adaptive traits? And lastly, what is the reproductive mode of the new pathogen (sexual versus clonal versus mixed reproduction)?
Population genetic analyses have been used to address these questions for a number of important crop pathogens, including Zymoseptoria tritici [22,23], Rhynchosporium commune [24,25], Magnaporthe (Pyricularia) oryzae [26], Parastagonospora nodorum [27], Venturia inaequalis [28] and P. infestans [16,17]. These studies have relied on detailed spatial samplings of the pathogen and genotyping of many strains. Importantly, these studies have also included sampling of ‘wild’ pathogen populations, i.e. closely related pathogens infecting non-crop plant hosts found in the same environment where the crop grows, to make inference regarding the evolutionary history of the crop pathogen possible. Evolutionary analyses used in these studies have built on coalescence theory to fit demographic models to observed patterns of genetic diversity. By comparing distinct models, these types of analyses make it possible to estimate parameters such as the time of divergence between populations, the population size of ancestral populations and the amount of genetic exchange occurring between ancestral and extant populations [29]. From such studies, we know that some important crop pathogens have emerged from pathogen populations initially found on wild hosts, e.g. the septoria tritici leaf blotch pathogen Z. tritici [7,29], the rice blast pathogen P. oryzae [26] and the apple scab pathogen V. inaequalis [30]. The domesticated crop-infecting species acquired the traits needed to invade the new host niche (i.e. the new crop host and its corresponding agro-ecosystem) and were subsequently propagated and dispersed with the crop host. In all three examples, gene flow between the ancestral wild population and the crop-infecting population has been either absent or very limited, supporting the emergence of new host-specific pathogen species.
Several additional studies demonstrated that interspecific hybridization and horizontal gene transfer could also play important roles in the emergence of new pathogens, e.g. the causal agent of Dutch elm disease Ophiostoma novo-ulmi [31], the poplar rust fungus Melampsora x columbiana [32] and the tan spot pathogen Pyrenophora tritici-repentis [13]. Long-distance dispersal of pathogens, facilitated by agricultural trade and transport of infected plant material as well as the uniform distribution of agro-ecosystems across large spatial scales, supports the spread of new pathogens originating from distinct geographical locations [33,34]. Thereby, current agricultural practices continuously sustain the formation and spread of new pathogens. Below, we provide four examples of pathogen emergence and speciation in some detail to highlight the extraordinary potential of plant pathogens to emerge, evolve and spread rapidly in agro-ecosystems.
2. Example 1: the recent emergence and spread of wheat blast: a new threat to global wheat production
The wheat blast disease caused by P. graminis-tritici was first reported in Brazil in 1985 [35] and has since spread across the wheat-growing region of Brazil and into the neighbouring countries of Bolivia, Paraguay and Argentina, causing crop losses of up to 40–100% [36–39]. Wheat blast is now considered a major quarantine disease and a threat to wheat crops in the USA [40,41] and other wheat-growing regions. It was recently reported in Bangladesh, causing crop losses of up to 100%, and now threatens wheat crops in India [18]. It was originally thought that the wheat blast disease emerged through a host jump from Pyricularia strains infecting rice, but by 1993 it was already clear that wheat blast was caused by two populations of Pyricularia, one that can infect both rice and wheat under greenhouse conditions and one that infects only wheat [42]. Recent population genetic analyses based on 11 microsatellite loci of 184 Pyricularia strains sampled from wheat and rice fields across Brazil found high population differentiation between wheat- and rice-infecting populations, with very little gene flow between the two host populations and with all 69 wheat-infecting isolates avirulent on rice [43]. These analyses also detected extensive gene flow among wheat fields across Brazil and indicated a mixed reproductive system including sexual recombination as well as asexual reproduction. By way of comparison, field populations of the rice blast pathogen P. oryzae are typically composed of a small number of clonal lineages and exhibit no evidence for sexual recombination [44]. Several races of P. graminis-tritici specialized to infect different Brazilian wheat cultivars were discovered among the wheat blast strains, suggesting the evolution of additional host specialization since the original appearance of the wheat-infecting pathogen population 30 years ago [43]. It was proposed that the host-specialized clones originated through sexual recombination, were then selected to become specialized to infect particular wheat cultivars, and were then spread across Brazil on infected seed [43]. Resistance to fungicides also emerged rapidly and is now widespread across Brazil [36].
The findings of little gene flow between rice- and wheat-infecting populations and avirulence of P. graminis-tritici strains on rice suggest that the wheat-infecting population was derived from an as-yet-unknown grass-infecting population of Pyricularia. To identify the original host of the wheat blast pathogen, Pyricularia strains were sampled from wheat and rice fields across Brazil as well as from weeds and pasture grasses growing in sympatry near those fields, yielding more than 500 strains for further analyses. More than 3300 bp of DNA sequence from 10 housekeeping genes were obtained from 128 of these strains, representing sympatric populations sampled across seven states in Brazil. Phylogenetic analyses based on these sequences identified a unique clade infecting wheat that was named P. graminis-tritici [45]. Comparative population genomics analyses that included more than 20 Pyricularia strains representing the known geographical and host diversity across Brazil enabled differentiation of many distinct lineages within the Pyricularia species complex and showed that the closest relative of the wheat pathogen was found on the widely grown pasture grass Urochloa (D. Croll, P. Ceresini, J. Maciel, B. A. McDonald 2016, unpublished data; https://github.com/crolllab/wheat-blast). Strains originating from the pasture grass Urochloa, grown on more than 90 million ha in Brazil, were identical to strains sampled from wheat fields for SSRs, DNA sequences of housekeeping genes and pathotypes. By way of comparison, the genome sequences of rice-infecting strains from around the world were distantly related and exhibited very little diversity (https://github.com/crolllab/wheat-blast). These findings suggest that the wheat blast pathogen emerged through a host jump from the Pyricularia population infecting Urochloa approximately 30 years ago. Urochloa was introduced into Brazil from Africa around 60 years ago [46]. Hence, it is possible that the closest ancestor of the Brazilian wheat blast pathogen originated in Africa.
3. Example 2: hybridization of crop plants driving hybrid speciation of fungal pathogens: a new disease of the recently created cereal crop triticale
Rice and wheat have been cultivated for thousands of years, but new crop species continue to be developed by plant breeders who select on wild plant populations or make crosses among existing crop plants. The creation of new crop species has now been shown to drive the rapid speciation of new crop pathogens. For example, the cereal crop triticale is a human-generated hybrid of wheat and rye that has been cultivated in Europe only since the 1960s [47]. A major new disease of triticale is powdery mildew caused by the ascomycete fungus Blumeria graminis. Blumeria graminis consists of several formae specialis that are specialized to infect different cereal hosts. Triticale is infected by the forma specialis mildew pathogen B.g. triticale. The disease was first reported in 2001, suggesting a very recent emergence of this pathogen.
Menardo and co-workers investigated the evolutionary origin of B.g. triticale using a comparative population genomics approach. They sequenced multiple B. graminis isolates sampled from wheat, rye and triticale and conducted a detailed analysis of the distribution of single nucleotide polymorphisms (SNPs) [48]. The three formae specialis mildew pathogens can be distinguished using fixed SNPs, which clearly separate the three pathogens. However, B.g. triticale shows an interesting genomic mosaic of segments with fixed SNPs originating from either B.g. tritici or B.g. secalis in addition to SNPs specific only to B.g. triticale. This genomic pattern is consistent with a recent hybrid origin of B.g. triticale, which probably originated from a cross between B.g. tritici and B.g. secalis. Hence, the wheat–rye hybrid triticale selected for a new pathogen that emerged through a hybridization event between the wheat- and rye-infecting formae specialis.
Identification of the new hybrid pathogen raised three important questions: where did the hybrid emerge? Did hybridization of B.g. tritici and B.g. secalis occur once or is it an ongoing process with continuous gene flow between the two original pathogens? Which are the virulence determinants that allowed B.g. triticale to emerge as a new triticale pathogen? The population genomic data allowed the authors to address the first two questions: based on phylogeographic and admixture analyses of isolates originating from different continents, they showed that the triticale-infecting pathogen emerged very recently in Central Europe. The hybrid lineage originated from a small number of hybridization events and establishment of the hybrid population involved some backcrossing with the wheat-infecting formae specialis B.g. tritici.
Menardo and co-workers applied a comparative transcriptome approach to identify virulence determinants in B.g. triticale. B.g. triticale can infect wheat but not rye, and transcriptomic data were therefore generated only for the wheat and triticale host–pathogen interactions [48]. However, the infection experiments of the hybrid pathogen on wheat and triticale did not reveal genes differentially expressed in the two hosts, suggesting that virulence may depend on the effects of multiple genes [48]. It is also possible that particular positive or negative interactions among genes originating from the two parental species B.g. tritici and B.g. secalis contribute to the novel virulence phenotype of B.g. triticale [48,49]. B.g. tritici populations are known to vary in the composition and diversity of effector candidate genes [50]. It is possible that the number of successful hybrids has been restricted to a limited number of crosses among parents that carried exactly the right combination of virulence traits to enable successful infection of the new host. Once the virulent hybrid emerged, further propagation and dispersal were facilitated by the extensive availability of susceptible hosts in agricultural ecosystems.
4. Example 3: clonal speciation and rapid adaptive evolution in the absence of sex
For the examples outlined above, sexual recombination was instrumental in the generation of new genotypes that were favoured by natural selection and propagated on a new host [48]. By recombination, parental genomes are re-assorted to generate new combinations of alleles in the progeny population. Recombination furthermore plays a fundamental role in adaptive evolution in sexually reproducing organisms by enhancing the efficiency of both purifying and positive selection [51]. This occurs as recombination breaks down the linkage between loci carrying favourable and non-favourable mutations thereby allowing the fixation of advantageous alleles and the removal of deleterious alleles. Organisms that propagate clonally are characterized by a high extent of linkage disequilibrium because all alleles in the genome are essentially linked. Theory predicts that low levels of genetic diversity will also characterize populations of clonal species [51]. However, as summarized below, studies of plant pathogen populations show that some species can circumvent the disadvantageous consequences of clonal propagation. Clonal pathogens have been shown capable of generating high levels of genotypic diversity by different mechanisms, including extensive chromosome rearrangements mediated by transposable elements and aneuploidy formation (see review in [34]).
Many fungi, including a large number of crop pathogens, are known to propagate clonally or only rarely undergo sexual recombination [52]. Even in the absence of sexual recombination, fungal crop pathogens can exhibit sufficient genetic diversity to allow them to rapidly overcome new host resistances or evolve resistance against new fungicides [52,53]. This common observation is at odds with evolutionary theory that predicts low levels of genetic diversity and slow rates of evolution in clonal species [50,54]. Genome-based studies have shed light on mechanisms of evolution and rapid adaptation in clonal pathogen species, highlighting exceptionally high rates of genome plasticity, particular patterns of genome compartmentalization, horizontal gene transfer and parasexual recombination as mechanisms facilitating rapid evolution of clonal crop pathogens. In general, transposable elements appear to play an essential role in genome evolution in these species.
The vascular wilt fungus Verticilium dahliae is known to reproduce mainly by clonal propagation. Verticilium dahliae invades the water-conducting xylem vessels of susceptible plants and produces conidia that are transported in the xylem to distant plant parts. Control of V. dahliae is difficult owing to the limited availability of genetic resistances and the inability of fungicides to reach the pathogen inside xylem vessels. An intriguing question has been how a clonal pathogen like V. dahliae is able to diversify its repertoire of effector genes to infect multiple plant species.
Population genomic sequencing of 11 isolates originating from North America and Europe revealed the relatively low number of 236 785 genome-wide SNPs [55]. For comparison, population genomic sequencing of 13 isolates of Z. tritici sampled from one region in Australia showed more than 800 000 SNPs [56]. Although the number of polymorphic nucleotides was found to be relatively low, the 11 isolates of V. dahliae were found to exhibit a dramatic variation in their genome composition and genome content. The V. dahliae genome is comprised of lineage-specific blocks of DNA that are exclusive to one or a few isolates. These lineage-specific fragments are characterized by a high content of transposable elements and a low density of protein-coding genes. However, the few genes present in the lineage-specific regions are enriched in plant-induced genes, including genes encoding effectors and determinants of host specificity [56,57]. Faino and co-workers investigated the origin of the lineage-specific fragments using whole-genome assemblies obtained by single molecule real-time sequencing [58]. They showed that lineage-specific fragments are generated mainly by segmental genomic duplications followed by extensive sequence divergence. Active transposable elements located in the lineage-specific fragments probably contribute to the rapid evolution of these regions and the genes located within them. As these regions are enriched in effectors, the activity of the transposable elements can be linked directly to the diversification of effectors and thereby to the evolution of new virulence determinants. Transposable elements have also been associated with effector gene evolution in other clonal plant pathogens, including Fusarium oxysporum [59], M. (P.) oryzae [60] and the oomycete potato late blight pathogen P. infestans [61].
The detailed genome studies of V. dahliae and other clonal crop pathogens illustrate the potential of these species to circumvent the negative consequences of asexual reproduction. Transposable elements, high rates of genomic plasticity and reduced selective constraints on sequence evolution allow these species to evolve as rapidly as their sexual counterparts infecting crop hosts in agro-ecosystems (see also the review in [62]). It remains to be seen if the same processes, to the same extent, play a role in adaptation of asexual pathogens in natural ecosystems.
5. Example 4: rapid global dissemination of new clones carrying novel traits that increase adaptation to agro-ecosystems
A major challenge in the control of plant pathogens is the ability of many species to disseminate over long distances. Many plant pathogens produce air-borne spores that can be carried hundreds of kilometres by wind (reviewed in [33]). Even soil-borne pathogens can be carried over long distances by water movement or wind-blown soil. Human-mediated spread through trade and travel also plays a critical role in the global dissemination of many plant pathogens, as illustrated most recently by the introduction of wheat blast into Asia from South America.
The wheat yellow rust fungus Puccinia striiformis exemplifies how human activities contributed to the rapid spread of a plant pathogen among continents [63]. Puccinia striiformis originated in the Himalayas and was introduced from there into Europe and Asia. It spread to North and South America at the beginning of the twentieth century and has more recently been disseminated from wheat fields in Europe and North America to wheat fields in Africa and Australia [63]. Almost all populations of P. striiformis are mainly clonal; however, the presence of sexually reproducing populations in the Himalayas indicates that the original pathogen life cycle involved a sexual stage. The sexual stage of many rust fungi, including P. striiformis, occurs on an alternative host, in the case of P. striiformis on barberry (Berberis vulgaris) [64].
The virulence of P. striiformis isolates is highly correlated with temperature [65]. This reflects the fact that germination of the asexual urediospores is temperature limited and disease development in wheat fields is correlated with spore germination. The severe stripe rust epidemic in the USA in 2000 was caused by the emergence of new clone-producing spores that could germinate at increased temperatures. This temperature adaptation allowed the pathogen to invade hitherto disease-free wheat fields in the southern USA and cause devastating yield losses [65]. The clone that had adapted to warmer temperatures was found in Australia only two years later, documenting the high risk associated with inter-continental spread of new pathogen strains through human activities [66]. Emergence of new P. striiformis clones with increased aggressiveness occurs periodically when existing clones acquire new mutations enabling them to overcome new host resistances [67].
To infer the global migration routes of P. striiformis, Ali and co-workers characterized a global collection of P. striiformis isolates [63]. Besides reconstructing the global migration routes from the Himalayas to Europe and Asia and later to the ‘New World’, the authors also investigated the relationships among global stripe rust populations. Interestingly, the global population genetic structure of P. striiformis is characterized by strong differentiation among geographically separated populations, even though clones can be disseminated over long distances. New clones with novel virulence traits can emerge and spread without completely replacing the existing P. striiformis populations. It appears probable that the clonal nature of the wheat-infecting rust populations is retained owing to the absence of an alternative host for sexual recombination, preventing gene flow among introduced genotypes and existing populations of the pathogen, thereby maintaining the genetic differentiation between strains [63]. Nevertheless, reconstructing the migration routes of the rust pathogen documents that aggressive clones emerge from local populations and travel across and among continents not only by wind but also by human activities.
6. How to re-engineer agro-ecosystems to make them more resilient to infectious diseases?
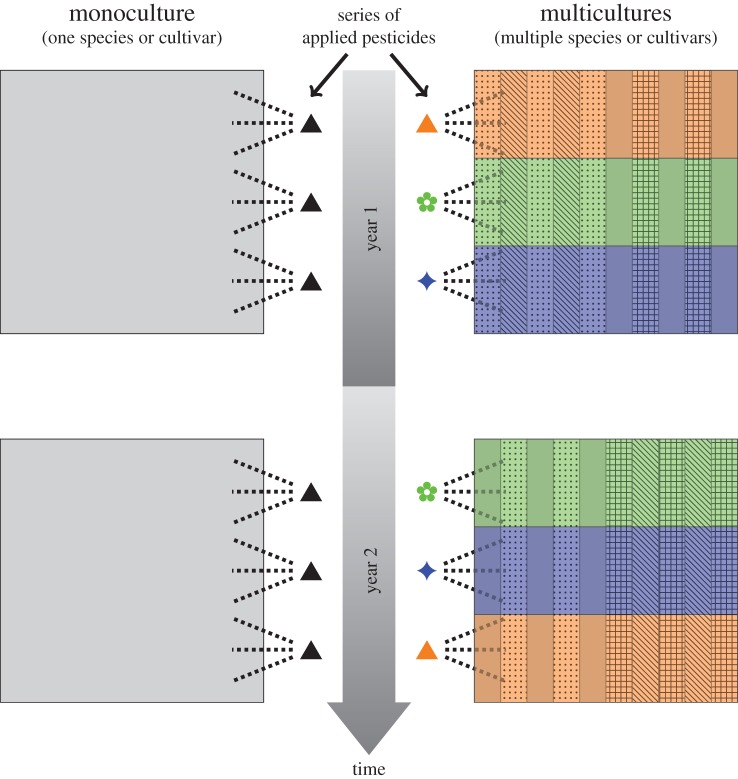
The fundamental trade-off in all agro-ecosystems is that the environmental and genetic uniformity that enables mechanization and increases the efficiency of food production also facilitates pathogen evolution and increases the risk of significant losses owing to large-scale disease epidemics. The long-term solution will be to change the overall structure of agro-ecosystems to make them more heterogeneous, both environmentally and genetically, over both time and space, in order to impose trade-offs on the corresponding pathogen populations [68]. An over-riding principle is the need to increase agro-ecosystem diversity in a dynamic way that fluctuates regularly (e.g. annually or every few years) in order to impose disruptive selection on populations of crop pathogens and force them to make trade-offs among traits (figure 1). This ‘dynamic diversity’ can be implemented in many ways [67–70], including improved deployment strategies for resistance genes and resistant cultivars, more frequent turnover of crop varieties that carry different resistance genes, improved deployment strategies for fungicides, more frequent crop rotations that include more species, smaller fields planted to individual crops and growing greater numbers of crop species per unit area under cultivation. The overall objective is to break up the adaptive landscape into smaller units that change on a regular basis to present the corresponding populations of crop pathogens with evolutionary dilemmas that lead to disruptive selection (figure 1). For example, an effector gene mutation that enables a pathogen strain to overcome a recently deployed major resistance (R) gene present in a new cultivar may decrease fitness on a different cultivar lacking that R-gene. The decreased fitness may reflect a trade-off that lowers overall pathogen reproduction, transmission or virulence on the cultivar lacking the R-gene [69–72]. Similarly, a mutation in a gene that decreases sensitivity to one fungicide may increase sensitivity to a different fungicide [73,74]. These examples illustrate opportunities for imposing pathogen trade-offs by increasing diversity in the agro-ecosystem landscape. One strategy for applying these concepts while maintaining the advantages of mechanization is given below.
Figure 1.
Schematic illustration of agro-ecosystem field compositions based on monoculture (left) and multicultures (right). In the monoculture agro-ecosystem, one crop species is planted into a large field and treated several times during the growing season with the same pesticide. The same crop is planted and the same pesticide is applied in the following year. The resulting homogeneous environment (one selective environment per field shown in the figure) rapidly selects for specialized pathogen strains or populations that are highly adapted to this monoculture agro-ecosystem. In the multiculture agro-ecosystem, multiple crop species or cultivars of the same crop species (indicated by different patterns) are grown simultaneously in wide rows (to facilitate mechanization) in the same field. The total area planted with one crop species or crop cultivar is considerably smaller compared with the monoculture field. A series of pesticides with different modes of action is applied in wide rows (to facilitate mechanization) in an alternating pattern (orange, green and blue) over both time and space. In the following year, the same crop species or cultivars can be planted in the same field, but the respective positions of the rows can be altered and the pesticide treatments can be applied in a new alternating pattern. The net effect of the multiculture approach compared to monoculture is to confront the pathogen population with a dynamically diverse selective landscape (12 different selective environments per field shown in the figure) encompassing smaller spatial and temporal scales that forces trade-offs among traits, slowing or preventing the emergence of highly virulent and broadly adapted pathogen populations.
Combining different management practices (integrated disease management [75]) is also likely to slow pathogen evolution towards increased virulence, host specialization and fungicide resistance. All management practices that reduce pathogen effective population size, such as improved sanitation through seed treatments and removal or treatment of infected plant stubble, will decrease the amount of genetic diversity and reduce the evolutionary potential in the corresponding pathogen population. Practices that restrict gene or genotype flow over long distances, such as improved quarantines or mandatory sterilization of straw, soil or grain that is shipped among countries, will slow movement of highly adapted pathogen strains among similar agro-ecosystems on different continents (e.g. movement of the F. oxysporum wilt pathogen causing the Panama wilt disease, which destroys banana plantations globally [76]) or damaging traits such as fungicide resistance or increased virulence [67,76]. Identification and removal of alternative hosts, growing in or near agro-ecosystems, that facilitate pathogen sexual recombination or that act as over-seasoning hosts can further slow the rate of evolution in pathogen populations. Combining disease management practices also reduces the risk of a catastrophic yield loss under conditions when one of the control practices fails. For example, when fungicide resistance emerges, the pathogen strains that are resistant to the fungicide may be held in check by a host resistance gene or a biological control agent. It will become increasingly important to consider the long-term consequences of short-term crop management decisions [77]. For example, large-scale adoption of no-till or low-till agriculture is important to preserve soil, but the resulting increase in crop stubble left on the soil surface allows many stubble-borne, necrotrophic pathogens such as Z. tritici to greatly increase their effective population size by improving survival through saprophytic growth between growing seasons [67].
The introduction of dynamic diversity into agro-ecosystems should encompass both spatial and temporal components of diversity that do not conflict with the advantages of mechanization and the further intensification of agronomic practices in existing fields [78]. Obvious manipulations of spatial components of diversity include smaller average field sizes and a larger number of crop species grown per unit area of agriculture. Less obvious strategies include planting strips of the same plant species that are genetically different (e.g. ‘strip mixtures’ composed of different wheat cultivars), or strips of different crop species that easily coexist (e.g. alternating strips of wheat and barley that have similar maturity and height). Planting multi-species row mixtures (often called ‘intercropping’) has been shown to have many desirable properties and is commonly done on a small scale that is not mechanized worldwide [79], mainly in subsistence agriculture or home gardens. Intercropping typically requires significant labour inputs and does not scale up easily, but planting the diversity in strips that match the width of existing machinery could enable efficient mechanization of planting, tillage and harvesting operations. Additional levels of diversity within a field can be implemented by overlaying additional selective agents. For example, two or more fungicides with different modes of action could be applied in alternating strips at a 90° angle to the direction in which the alternating rows of two or more genetically diverse hosts were planted (figure 1). This simple re-organization of existing planting and pesticide applications would generate a patchwork of four or more independent host/chemical environments coexisting in the same field, yet remain fully mechanized and could easily be scaled up or down in size. Adding more host cultivars or species and more fungicides would quickly increase the degree of diversity that confronts the pathogen population at the field scale where most pathogen evolution occurs (figure 1). Other approaches to increasing genetic diversity through plant breeding strategies [69] or through physical mixing of different cultivars or isogenic lines carrying different R-genes have already been thoroughly described and shown to lower disease [80] as well as to slow pathogen evolution [81].
Obvious temporal components that increase diversity in agro-ecosystems include annual rotations among different crop species, e.g. maize followed by oilseed rape followed by wheat. Many such rotations were developed to reflect local soils and climatic conditions and were used for centuries, but have fallen into disuse during the past few decades, mainly for economic reasons. Less obvious strategies for increasing diversity over time include alternating applications of fungicides or biocontrol agents, whereby fungicides or biocontrol agents with different modes of action are applied at different points in time during crop development. Over longer time periods, temporal diversity could include planting different crop cultivars carrying different R-genes during the crop rotation. For example, a different wheat cultivar could be planted every third year in the maize–oilseed rape–wheat rotation mentioned earlier. This wheat cultivar could be chosen to have a different combination of R-genes compared with earlier wheat cultivars, presenting the corresponding populations of wheat pathogens with a different evolutionary puzzle to solve every third year. More complex and longer-term temporal diversity strategies become available when considering agro-forestry, where long-lived perennial trees are grown in combination with annual crops [79]. An example strategy based on row mixtures that combines spatial and temporal components to achieve dynamic diversity is illustrated in figure 1.
These examples illustrate that many options are available for increasing dynamic diversity in agro-ecosystems. We anticipate that further intensification of food production in agro-ecosystems using traditional approaches (e.g. increased inputs of fertilizers and pesticides, technologies associated with precision agriculture, increased use of transgenic cultivars leading to further decreases in genetic diversity) will not be sustainable because of the resulting increase in losses owing to adaptation in the corresponding pest and pathogen populations. A truly sustainable intensification of food production will require re-engineering existing agro-ecosystems to introduce dynamic diversity in order to suppress the rapid emergence of new and highly damaging plant pathogens.
Authors' contributions
B.A.M. and E.H.S. conceived and drafted the manuscript.
Competing interests
The authors have no competing interests.
Funding
B.A.M. acknowledges research funding from the Swiss National Science Foundation (31003A_134755 and 31003A_155955). Research in the group of E.H.S. is funded by the Max Planck Society and by a personal grant from the State of Schleswig-Holstein, Germany.
References
- 1.Foley JA, et al. 2011. Solutions for a cultivated planet. Nature 478, 337–342. ( 10.1038/nature10452) [DOI] [PubMed] [Google Scholar]
- 2.Benton TG, Vickery JA, Wilson JD. 2003. Farmland biodiversity: is habitat heterogeneity the key? Trends Ecol. Evol. 18, 182–188. ( 10.1016/S0169-5347(03)00011-9) [DOI] [Google Scholar]
- 3.Tilman D, et al. 2001. Forecasting agriculturally driven global environmental change. Science 292, 281–284. ( 10.1126/science.1057544) [DOI] [PubMed] [Google Scholar]
- 4.van de Wouw M, Kik C, van Hintum T, van Treuren R, Visser B. 2010. Genetic erosion in crops: concept, research results and challenges. Plant Genet. Resour. 8, 1–15. ( 10.1017/S1479262109990062) [DOI] [Google Scholar]
- 5.Pingali PL. 2012. Green revolution: impacts, limits, and the path ahead. Proc. Natl Acad. Sci. USA 109, 12 302–12 308. ( 10.1073/pnas.0912953109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Read AF. 2016. The evolution of virulence. Trends Microbiol. 2, 73–76. ( 10.1016/0966-842X(94)90537-1) [DOI] [PubMed] [Google Scholar]
- 7.Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, McDonald BA, Wang J, Schierup MH. 2011. The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 21, 2157–2166. ( 10.1101/gr.118851.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linde CC, Zhan J, McDonald BA. 2002. Population structure of Mycosphaerella graminicola: from lesions to continents. Phytopathology 92, 946–955. ( 10.1094/PHYTO.2002.92.9.946) [DOI] [PubMed] [Google Scholar]
- 9.Keller SM, McDermott JM, Pettway RE, Wolfe MS, McDonald BA. 1997. Gene flow and sexual reproduction in the wheat glume blotch pathogen Phaeosphaeria nodorum (Anamorph Stagonospora nodorum). Phytopathology 87, 353–358. ( 10.1094/PHYTO.1997.87.3.353) [DOI] [PubMed] [Google Scholar]
- 10.McDonald BA, Zhan J, Burdon JJ. 1999. Genetic structure of Rhynchosporium secalis in Australia. Phytopathology 89, 639–645. ( 10.1094/PHYTO.1999.89.8.639) [DOI] [PubMed] [Google Scholar]
- 11.van Baalen M, Sabelis MW. 1995. The dynamics of multiple infection and the evolution of virulence. Am. Nat. 146, 881–910. ( 10.1086/285830) [DOI] [Google Scholar]
- 12.Alizon S, de Roode JC, Michalakis Y. 2013. Multiple infections and the evolution of virulence. Ecol. Lett. 16, 556–567. ( 10.1111/ele.12076) [DOI] [PubMed] [Google Scholar]
- 13.Friesen TL, et al. 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. ( 10.1038/ng1839) [DOI] [PubMed] [Google Scholar]
- 14.Gardiner DM, et al. 2012. Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8, e1002952 ( 10.1371/journal.ppat.1002952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonald MC, Oliver RP, Friesen TL, Brunner PC, McDonald BA. 2013. Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 199, 241–251. ( 10.1111/nph.12257) [DOI] [PubMed] [Google Scholar]
- 16.Goodwin SB, Cohen BA, Deahl KL, Fry WE. 1994. Migration from northern Mexico as the probable cause of recent genetic changes in populations of Phytophthora infestans in the United States and Canada. Phytopathology 84, 553–558. ( 10.1094/Phyto-84-553) [DOI] [PubMed] [Google Scholar]
- 17.Goss EM, Cardenas ME, Myers K, Forbes GA, Fry WE, Restrepo S, Grünwald NJ. 2011. The plant pathogen Phytophthora andina emerged via hybridization of an unknown Phytophthora species and the Irish potato famine pathogen, P. infestans. PLoS ONE 6, e24543 ( 10.1371/journal.pone.0024543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Callaway E. 2016. Devastating wheat fungus appears in Asia for first time. Nature 532, 421–422. ( 10.1038/532421a) [DOI] [PubMed] [Google Scholar]
- 19.Fischer RA, Byerlee D, Edmeades GO.2014. Crop yields and global food security: will yield increase continue to feed the world? Canberra, Australia: Australian Centre for International Agricultural Research. (Available to download from http://aciar.gov.au/publication/mn158. )
- 20.Linde CC, Zala M, McDonald BA. 2009. Molecular evidence for recent founder populations and human-mediated migration in the barley scald pathogen Rhynchosporium secalis. Mol. Phylogenet. Evol. 51, 454–464. ( 10.1016/j.ympev.2009.03.002) [DOI] [PubMed] [Google Scholar]
- 21.Oerke E-C. 2006. Crop losses to pests. J. Agric. Sci. 144, 31–43. ( 10.1017/S0021859605005708) [DOI] [Google Scholar]
- 22.Zhan J, Pettway RE, McDonald BA. 2003. The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet. Biol. 38, 286–297. ( 10.1016/S1087-1845(02)00538-8) [DOI] [PubMed] [Google Scholar]
- 23.Banke S, McDonald BA. 2005. Migration patterns among global populations of the pathogenic fungus Mycosphaerella graminicola. Mol. Ecol. 14, 1881–1896. ( 10.1111/j.1365-294X.2005.02536.x) [DOI] [PubMed] [Google Scholar]
- 24.Zaffarano PL, McDonald BA, Linde CC. 2008. Rapid speciation following recent host shifts in the plant pathogenic fungus Rhyncosporim. Evolution 62, 1418–1436. ( 10.1111/j.1558-5646.2008.00390.x) [DOI] [PubMed] [Google Scholar]
- 25.Zaffarano PL, McDonald BA, Linde CC. 2009. Phylogeographical analyses reveal global migration patterns of the barley scald pathogen Rhynchosporium secalis. Mol. Ecol. 18, 279–293. ( 10.1111/j.1365-294X.2008.04013.x) [DOI] [PubMed] [Google Scholar]
- 26.Couch BC, Fudal I, Lebrun M-H, Tharreau D, Valent B, van Kim P, Nottéghem J-L, Kohn LM. 2005. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170, 613–630. ( 10.1534/genetics.105.041780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stukenbrock EH, Banke S, McDonald BA. 2006. Global migration patterns in the fungal wheat pathogen Phaeosphaeria nodorum. Mol. Ecol. 15, 2895–2904. ( 10.1111/j.1365-294X.2006.02986.x) [DOI] [PubMed] [Google Scholar]
- 28.Leroy T, Lemaire C, Dunemann F, Le Cam B. 2013. The genetic structure of a Venturia inaequalis population in a heterogeneous host population composed of different Malus species. BMC Evol. Biol. 13, 1–10. ( 10.1186/1471-2148-13-64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stukenbrock EH, Banke S, Javan-Nikkhah M, McDonald BA. 2007. Origin and domestication of the fungal wheat pathogen Mycosphaerella graminicola via sympatric speciation. Mol. Biol. Evol. 24, 398–411. ( 10.1093/molbev/msl169) [DOI] [PubMed] [Google Scholar]
- 30.Lemaire C, De Gracia M, Leroy T, Michalecka M, Lindhard-Pedersen H, Guerin F, Gladieux P, Le Cam B. 2016. Emergence of new virulent populations of apple scab from nonagricultural disease reservoirs. New Phytol. 209, 1220–1229. ( 10.1111/nph.13658) [DOI] [PubMed] [Google Scholar]
- 31.Brasier CM, Kirk SA. 2010. Rapid emergence of hybrids between the two subspecies of Ophiostoma novo-ulmi with a high level of pathogenic fitness. Plant Pathol. 59, 186–199. ( 10.1111/j.1365-3059.2009.02157.x) [DOI] [Google Scholar]
- 32.Newcombe G, Stirling B, McDonald S, Bradshaw HD. 2000. Melampsora icolumbiana, a natural hybrid of M. medusae and M. occidentalis. Mycol. Res. 104, 261–274. ( 10.1017/S0953756299001665) [DOI] [Google Scholar]
- 33.Brown JKM, Hovmøller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541. ( 10.1126/science.1072678) [DOI] [PubMed] [Google Scholar]
- 34.Stukenbrock EH. 2016. The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology 106, 104–112. ( 10.1094/PHYTO-08-15-0184-RVW) [DOI] [PubMed] [Google Scholar]
- 35.Igarashi S, Utiamada CM, Igarashi LC, Kazuma AH, Lopes RS. 1986. Pyricularia em trigo. 1. Ocorrência de Pyricularia sp. no estado do Paraná. Fitopatol Bras 11, 351–352. [Google Scholar]
- 36.Castroagudín VL, Ceresini PC, de Oliveira SC, Reges JTA, Maciel JLN, Bonato AL V, Dorigan AF, McDonald BA. 2015. Resistance to QoI fungicides is widespread in Brazilian populations of the wheat blast pathogen Magnaporthe oryzae. Phytopathology 105, 284–294. ( 10.1094/PHYTO-06-14-0184-R) [DOI] [PubMed] [Google Scholar]
- 37.Maciel JLN. 2011. Magnaporthe oryzae, the blast pathogen: current status and options for its control. Plant Sci. Rev. 264, 233–240. [Google Scholar]
- 38.Silva CP, Nomura E, Freitas EG, Brugnaro C, Urashima AS. 2009. Eficiência de tratamentos alternativos no controle de Pyricularia grisea em sementes de trigo. Trop. Plant Pathol. 34, 127–131. ( 10.1590/S1982-56762009000200009) [DOI] [Google Scholar]
- 39.Duveiller E, Hodson D, Tiedmann A. 2010. Wheat blast caused by Magnaporthe grisea: a reality and new challenge for wheat research. In 8th Int. Wheat Conf., pp. 247–248. St. Petersburg, Russia: N.I. Vavilov Research Institute of Plant Industry.
- 40.Duveiller E, Singh RP, Nicol JM. 2007. The challenges of maintaining wheat productivity: pests, diseases, and potential epidemics. Euphytica 157, 417–430. ( 10.1007/s10681-007-9380-z) [DOI] [Google Scholar]
- 41.Kohli MM, Mehta YR, Guzman E, Viedma L, Cubilla LE. 2011. Pyricularia blast–a threat to wheat cultivation. Czech. J. Genet. Plant Breed 47, S130–S134. [Google Scholar]
- 42.Urashima AS, Igarashi S, Kato H. 1993. Host range, mating type, and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis. 77, 1211–1216. ( 10.1094/PD-77-1211) [DOI] [Google Scholar]
- 43.Maciel JLN, Ceresini PC, Castroagudin VL, Zala M, Kema GHJ, McDonald BA. 2014. Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology 104, 95–107. ( 10.1094/PHYTO-11-12-0294-R) [DOI] [PubMed] [Google Scholar]
- 44.Saleh D, et al. 2012. Sex at the origin: an Asian population of the rice blast fungus Magnaporthe oryzae reproduces sexually. Mol. Ecol. 21, 1330–1344. ( 10.1111/j.1365-294X.2012.05469.x) [DOI] [PubMed] [Google Scholar]
- 45.Castroagudin VL, et al. 2016. Wheat blast disease caused by Pyricularia graminis-tritici sp. nov. bioRxiv. ( ) [DOI]
- 46.Naumova NT, Hayward DM, Wagenvoort M. 1999. Apomixis and sexuality in diploid and tetraploid accessions of Brachiaria decumbens. Sex Plant Reprod. 12, 43–52. ( 10.1007/s004970050170) [DOI] [Google Scholar]
- 47.Mergoum M, Singh PK, Peña RJ, Lozano-del Río AJ, Cooper KV, Salmon DF, Macpherson HG. 2009. Triticale: a ‘new’ crop with old challenges. In Cereals (ed. Carena JM.), pp. 267–287. New York, NY: Springer US. [Google Scholar]
- 48.Menardo F, et al. 2016. Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nat. Genet. 48, 201–205. ( 10.1038/ng.3485) [DOI] [PubMed] [Google Scholar]
- 49.Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM. 1996. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272, 741 ( 10.1126/science.272.5262.741) [DOI] [PubMed] [Google Scholar]
- 50.Wicker T, et al. 2013. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096. ( 10.1038/ng.2704) [DOI] [PubMed] [Google Scholar]
- 51.Marais G, Charlesworth B. 2003. Genome evolution: recombination speeds up adaptive evolution. Curr. Biol. 13, R68–R70. ( 10.1016/S0960-9822(02)01432-X) [DOI] [PubMed] [Google Scholar]
- 52.Taylor JW, Hann-Soden C, Branco S, Sylvain I, Ellison CE. 2015. Clonal reproduction in fungi. Proc. Natl Acad. Sci. USA 112, 8901–8908. ( 10.1073/pnas.1503159112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Moore M, Milus EA, Long DL, Line RF, Marshall D, Jackson L. 2002. Wheat stripe rust epidemics and races of Puccinia striiformis f. sp. tritici in the United States in 2000. Plant Dis. 86, 39–46. ( 10.1094/PDIS.2002.86.1.39) [DOI] [PubMed] [Google Scholar]
- 54.Hahn M. 2014. The rising threat of fungicide resistance in plant pathogenic fungi: Botrytis as a case study. J. Chem. Biol. 7, 133–141. ( 10.1007/s12154-014-0113-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Jonge R, Bolton MD, Kombrink A, van den Berg GCM, Yadeta KA, Thomma BPHJ. 2013. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res. 23, 1271–1282. ( 10.1101/gr.152660.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDonald MC, McGinness L, Hane JK, Williams AH, Milgate A, Solomon PS. 2016. Utilizing gene tree variation to identify candidate effector genes in Zymoseptoria tritici. Genes Genom. Genet. 6, 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crow JF, Kimura M. 1965. Evolution in sexual and asexual populations. Am. Nat. 99, 439–450. ( 10.1086/282389) [DOI] [Google Scholar]
- 58.Faino L, Seidl MF, Shi-Kunne X, Pauper M, van den Berg GCM, Wittenberg AHJ, Thomma BPHJ.2016. How transposons drive evolution of virulence in a fungal pathogen. bioRxiv. ( ) [DOI] [PMC free article] [PubMed]
- 59.Ma L-J, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. ( 10.1038/nature08850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chuma I, et al. 2011. Multiple translocation of the AVR-pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7, e1002147 ( 10.1371/journal.ppat.1002147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raffaele S, et al. 2010. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330, 1540–1543. ( 10.1126/science.1193070) [DOI] [PubMed] [Google Scholar]
- 62.Seidl MF, Thomma BPHJ. 2014. Sex or no sex: evolutionary adaptation occurs regardless. Bioessays 36, 335–345. ( 10.1002/bies.201300155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ali S, Gladieux P, Leconte M, Gautier A, Justesen AF, Hovmøller MS, Enjalbert J, de Vallavieille-Pope C. 2014. Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici. PLoS Pathog. 10, e1003903 ( 10.1371/journal.ppat.1003903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin Y, Szarbo LJ, Carson M. 2010. Century-old mystery of Puccinia striiformis life history solved with the identification of Berberis as an alternate host. Phytopathology 100, 432–435. ( 10.1094/PHYTO-100-5-0432) [DOI] [PubMed] [Google Scholar]
- 65.Milus EA, Kristensen K, Hovmøller MS. 2009. Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp. tritici causing stripe rust of wheat. Phytopathology 99, 89–94. ( 10.1094/PHYTO-99-1-0089) [DOI] [PubMed] [Google Scholar]
- 66.Hovmøller MS, Yahyaoui AH, MIilus EA, Justesen AF. 2008. Rapid global spread of two aggressive strains of a wheat rust fungus. Mol. Ecol. 17, 3818–3826. ( 10.1111/j.1365-294X.2008.03886.x) [DOI] [PubMed] [Google Scholar]
- 67.Hovmøller MS, Sørensen CK, Walter S, Justesen AF. 2011. Diversity of Puccinia striiformis on cereals and grasses. Annu. Rev. Phytopathol. 49, 197–217. ( 10.1146/annurev-phyto-072910-095230) [DOI] [PubMed] [Google Scholar]
- 68.Zhan J, Thrall PH, Papaïx J, Xie L, Burdon JJ. 2015. Playing on a pathogen's weakness: using evolution to guide sustainable plant disease control strategies. Annu. Rev. Phytopathol. 53, 19–43. ( 10.1146/annurev-phyto-080614-120040) [DOI] [PubMed] [Google Scholar]
- 69.McDonald BA. 2014. Using dynamic diversity to achieve durable disease resistance in agricultural ecosystems. Trop. Plant Pathol. 39, 191–196. ( 10.1590/S1982-56762014000300001) [DOI] [Google Scholar]
- 70.Thrall PH, Burdon JJ. 2003. Evolution of virulence in a plant host–pathogen metapopulation. Science 299, 1735–1737. ( 10.1126/science.1080070) [DOI] [PubMed] [Google Scholar]
- 71.Alizon S, Hurford A, Mideo N, Van Baalen M. 2009. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 22, 245–259. ( 10.1111/j.1420-9101.2008.01658.x) [DOI] [PubMed] [Google Scholar]
- 72.Sacristan S, Garcia-Arenal F. 2008. The evolution of virulence and pathogenicity in plant pathogen populations. Mol. Plant Pathol. 9, 369–384. ( 10.1111/j.1364-3703.2007.00460.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morais D, Gélisse S, Laval V, Sache I, Suffert F. 2016. Inferring the origin of primary inoculum of Zymoseptoria tritici from differential adaptation of resident and immigrant populations to wheat cultivars. Eur. J. Plant Pathol. 145, 393–404. ( 10.1007/s10658-015-0853-y) [DOI] [Google Scholar]
- 74.Leroux P, Albertini C, Gautier A, Gredt M, Walker A. 2007. Mutations in the CYP51 gene correlated with changes in sensitivity to sterol 14α-demethylation inhibitors in field isolates of Mycosphaerella graminicola. Pest Manag. Sci. 63, 688–698. ( 10.1002/ps.1390) [DOI] [PubMed] [Google Scholar]
- 75.McDonald BA, Mundt CC. 2016. How knowledge of pathogen population biology informs management of Septoria tritici blotch. Phytopathology 106, 948–955. [DOI] [PubMed] [Google Scholar]
- 76.Ordonez N, Seidl MF, Waalwijk C, Drenth A, Kilian A, Thomma BPHJ, Ploetz RC, Kema GHJ. 2015. Worse comes to worst: bananas and Panama disease—when plant and pathogen clones meet. PLoS Pathog. 11, e1005197 ( 10.1371/journal.ppat.1005197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhan J, Thrall PH, Burdon JJ. 2014. Achieving sustainable plant disease management through evolutionary principles. Trends Plant Sci. 19, 570–575. ( 10.1016/j.tplants.2014.04.010) [DOI] [PubMed] [Google Scholar]
- 78.Lin BB. 2011. Resilience in agriculture through crop diversification: adaptive management for environmental change. Bioscience 61, 183–193. ( 10.1525/bio.2011.61.3.4) [DOI] [Google Scholar]
- 79.Malézieux E, Crozat Y, Dupraz C, Laurans M, Makowski D, Ozier-Lafontaine H, Rapidel B, De Tourdonnet S, Valantin-Morison M. 2009. Mixing plant species in cropping systems: concepts, tools and models: a review. Agron. Sustain. Dev. 29, 43–62. ( 10.1051/agro:2007057) [DOI] [Google Scholar]
- 80.Mundt CC. 2002. Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Phytopathol. 40, 381–410. ( 10.1146/annurev.phyto.40.011402.113723) [DOI] [PubMed] [Google Scholar]
- 81.Zhan J, McDonald BA. 2013. Field-based experimental evolution of three cereal pathogens using a mark–release–recapture strategy. Plant Pathol. 62, 106–114. ( 10.1111/ppa.12130) [DOI] [Google Scholar]